Key Points
This study describes a method for the comparison of gene expression data of any type of cancer cells with their corresponding normal cells.
Our analyses reveal novel disease entities, identify common deregulated transcriptional networks, and predict survival.
Abstract
Gene expression profiling has been used extensively to characterize cancer, identify novel subtypes, and improve patient stratification. However, it has largely failed to identify transcriptional programs that differ between cancer and corresponding normal cells and has not been efficient in identifying expression changes fundamental to disease etiology. Here we present a method that facilitates the comparison of any cancer sample to its nearest normal cellular counterpart, using acute myeloid leukemia (AML) as a model. We first generated a gene expression-based landscape of the normal hematopoietic hierarchy, using expression profiles from normal stem/progenitor cells, and next mapped the AML patient samples to this landscape. This allowed us to identify the closest normal counterpart of individual AML samples and determine gene expression changes between cancer and normal. We find the cancer vs normal method (CvN method) to be superior to conventional methods in stratifying AML patients with aberrant karyotype and in identifying common aberrant transcriptional programs with potential importance for AML etiology. Moreover, the CvN method uncovered a novel poor-outcome subtype of normal-karyotype AML, which allowed for the generation of a highly prognostic survival signature. Collectively, our CvN method holds great potential as a tool for the analysis of gene expression profiles of cancer patients.
Introduction
Global gene expression profiling (GEP) has been used for more than a decade to uncover the underlying transcriptional programs of normal and malignant cells. In cancer, GEP has been used successfully to identify cancer subtypes, to stratify patients into responders vs nonresponders, and to predict survival but has, to a large extent, failed to uncover genes that are causally involved in cancer initiation and maintenance.1-8 These genes are obviously of great interest because they constitute potential targets for therapeutic intervention.
The reasons for the failure of GEP to uncover targets of therapeutic relevance are potentially many and may differ between different cancer types. However, as most studies compare cancer with cancer, a lot of the detected transcriptional changes between different cancer samples may arise from differences in cell type and developmental stage and, consequently, will not identify those gene expression programs that underlie the malignant phenotype. Moreover, in the few instances in which comparisons of cancer cells with normal cells have been attempted, they have generally used heterogeneous cell mixtures of the organ in question (such as whole bone marrow [BM] or CD34+ cells in acute myeloid leukemia [AML]), which is not ideal when the aim is to precisely map changes in gene expression between cancer cells and their nearest normal counterpart. Given the caveats with the standard procedures of analyzing large-scale GEP data sets from cancer patients, we hypothesized that a method that facilitates a comparison of gene expression profiles between cancer samples and their nearest normal counterpart holds the potential to significantly improve disease stratification, the identification of novel disease subtypes, prognostication, and the identification of gene expression changes underlying the malignant phenotype.
Here we present such a method and apply it retrospectively to 5 publically available data sets from AML patients.9-15 We show that comparison of cancer vs normal (CvN) is equivalent to cancer vs cancer (CvC) in terms of GEP-based prediction of AML subtypes harboring different aberrant karyotypic lesions (AK-AML). Moreover, applying this method to data sets of AML patients with normal karyotype AML (NK-AML) lead to identification of a novel AML subtype associated with poor outcome. Finally, the method provides a list of deregulated genes and common transcriptional programs that differ from those of normal progenitors. These may potentially underlie the malignant phenotype in different AML subtypes.
Materials and methods
GEP of normal hematopoietic stem and progenitor cells
BM samples were aspirated from the posterior iliac crest of healthy subjects, according to the standard protocol of the Department of Hematology, Rigshospitalet University of Copenhagen, and in accordance with the Declaration of Helsinki. Normal hematopoietic stem and progenitor cells (HSCs/HPCs), as well as mature myeloid cells representing successive developmental stages of the hematopoietic differentiation, were purified from BM samples of healthy subjects by flow cytometry-based cell sorting and subjected to GEP (see the supplemental Methods, available on the Blood Web site). All microarray files of our newly generated data set are available at the Gene Expression Omnibus database (accession number GSE42519).
Bioinformatics analyses
Gene expression-based map of the normal hematopoietic hierarchy.
The normalized16 and batch-corrected17 data set of normal blood and BM populations was filtered to exclude probe sets with low variance and low expression (see Table 1 for sample list and origin18-22 and the supplemental Methods for data normalization and batch correction). Probe sets with a standard deviation 3 times lower than the average standard deviation of the entire data set and with an average log2 expression below 6 were excluded from the analysis, yielding a total of 2119 probe sets (1367 unique gene symbols). These were used in a principal component analysis (PCA)23 to generate a map of the normal hematopoietic hierarchy by projection of the data onto the first 2 principal components (ie, the 2 directions, which explain the most variance in the 2119 dimensional space of standardized gene expression values).
Mapping of AML samples onto the gene expression-based map of the normal hematopoietic hierarchy.
Each AML sample was normalized together with the data set of normal cells, using the same procedure described earlier. The normal populations closest to each individual AML sample were identified by a 2-step approach. First, we computed the Euclidian distances between the AML sample and all normal populations, using the expression profiles projected to the first 6 principal components, which explain more than 90% of the variation for more than 95% of the samples. Second, for each individual AML sample, we selected the 50% most varying probe sets within the 15 closest normal samples and subsequently mapped the AML sample in this reduced local gene expression space. Using PCA projections in these steps reduces noise and computation time while preserving relevant information. For each AML sample mapped in the gene expression landscape of normal hematopoiesis, we next calculated a weighted average gene expression profile of the 5 nearest normal samples. The weights were based on the Euclidian distance between normal and AML samples in the reduced second PCA and were set to decrease exponentially with distance and, subsequently, renormalize to sum to 1. This approach gives more weight to populations closest to the AML sample. Finally, gene expression changes between individual AML samples and their corresponding individual average-weighted normal counterpart were computed.
For the standard CvC method, we generated an average GEP profile based on all the samples in a given AML data set. We used this to compute the log2 fold-changes for individual AML samples.
Gene set overlap analysis.
We created gene sets using the 1% most up- and downregulated genes for each AML sample compared with its computed normal counterpart. Using a hypergeometric test, we calculated the significance of the overlap between gene sets of up- and downregulated genes against various AML-subtype signatures, as well as curated gene signatures (C2), gene ontology signatures (C5), and oncogenic signatures (C6) from the MSigDB molecular signature database (www.broadinstitute.org/gsea/msigdb/).
Further information on experimental procedures and bioinformatics analyses are provided in the supplemental Methods. Here we describe methods for supervised and unsupervised classification of AML, the identification of deregulated transcriptional programs in AML compared with normal, and the generation and performance assessment of a novel survival signature for NK-AML.
Results
The gene expression-based landscape of the normal hematopoietic hierarchy
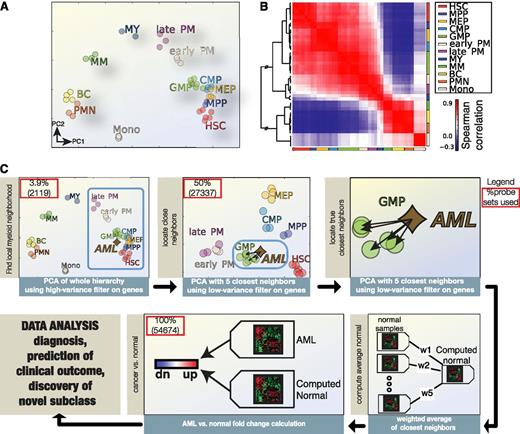
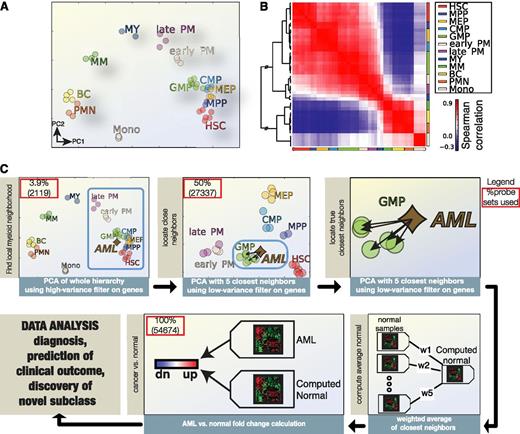
Using our own and publically available microarray data sets (Table 1) of highly purified HSCs/myeloid HPCs and their mature progeny (see supplemental Figure 1A-B for cell sorting strategy), we generated a gene expression-based landscape representing the developmental hierarchy of the hematopoietic system using the first 6 components of a PCA (Figure 1A-B; gene lists provided in supplemental Table 1A). Significantly, this landscape not only faithfully reconstructed the normal hierarchical order of myeloid differentiation but also demonstrated a tight clustering of replicates from the same normal populations, thereby highlighting both the high quality of data processing and the data itself.
CvN method: identification of the nearest normal population for individual AK-AML samples, using a gene expression-based landscape of the normal hematopoietic hierarchy. (A) PCA of gene expression profiles from the following normal purified BM populations: HSCs, multipotent progenitors (MPPs), common myeloid progenitors (CMPs), granulocyte-monocyte progenitors (GMPs), megakaryocyte-erythrocyte progenitors (MEPs), early PM, late PM, myelocytes (MY), metamyelocytes (MM), band cells (BC), polymorphonuclear neutrophilic granulocytes (PMN_BM), and monocytes (Mono). The PCA was performed on 2119 probe sets (1367 genes) selected by variance filtering. (B) Spearman correlation matrix of gene expression of the samples from A. (C) Workflow of the CvN method for the identification of the nearest normal counterpart for individual AK-AML samples (CvN method): AML samples are normalized individually together with the data set of the normal hematopoietic hierarchy shown in A and B, and the normal populations closest to AML samples are identified by a 2-step approach. First, the Euclidian distances between each individual AML sample and all the normal blood and BM populations are calculated using gene expression profiles projected onto the first 6 principal components. Next, the 50% most varying probe sets within the 15 closest normal populations are selected for each individual AML samples and used in a second PCA to map the AML sample to its 5 nearest normal populations. Subsequently, a weighted average gene expression profile based on the Euclidian distance between the 5 normal populations and AML sample is calculated. Finally, gene expression profiles of AML samples and their corresponding individual average-weighted normal population are compared with defined differentially expressed genes in individual AML samples for enrichment analysis, prognostification, and further analyses.
CvN method: identification of the nearest normal population for individual AK-AML samples, using a gene expression-based landscape of the normal hematopoietic hierarchy. (A) PCA of gene expression profiles from the following normal purified BM populations: HSCs, multipotent progenitors (MPPs), common myeloid progenitors (CMPs), granulocyte-monocyte progenitors (GMPs), megakaryocyte-erythrocyte progenitors (MEPs), early PM, late PM, myelocytes (MY), metamyelocytes (MM), band cells (BC), polymorphonuclear neutrophilic granulocytes (PMN_BM), and monocytes (Mono). The PCA was performed on 2119 probe sets (1367 genes) selected by variance filtering. (B) Spearman correlation matrix of gene expression of the samples from A. (C) Workflow of the CvN method for the identification of the nearest normal counterpart for individual AK-AML samples (CvN method): AML samples are normalized individually together with the data set of the normal hematopoietic hierarchy shown in A and B, and the normal populations closest to AML samples are identified by a 2-step approach. First, the Euclidian distances between each individual AML sample and all the normal blood and BM populations are calculated using gene expression profiles projected onto the first 6 principal components. Next, the 50% most varying probe sets within the 15 closest normal populations are selected for each individual AML samples and used in a second PCA to map the AML sample to its 5 nearest normal populations. Subsequently, a weighted average gene expression profile based on the Euclidian distance between the 5 normal populations and AML sample is calculated. Finally, gene expression profiles of AML samples and their corresponding individual average-weighted normal population are compared with defined differentially expressed genes in individual AML samples for enrichment analysis, prognostification, and further analyses.
A method for assessment of gene expression changes between AML and its nearest normal counterpart (CvN method)
We next applied a 2-step approach to identify the closest normal population for individual AML patient samples using publically available AK-AML gene expression data sets (Table 1; supplemental Table 2). In the first step, we mapped individual AML samples onto the PCA space of normal hematopoietic differentiation, using genes selected by a high-stringency variance filter (Figure 1C). Next we reduced the filter stringency to increase mapping precision and identify the 5 closest normal BM populations. Finally, the GEPs of these 5 BM populations were merged into a “virtual” distance-weighted GEP representing the closest normal counterpart of the tested AML sample and subsequently used to calculate gene expression changes between normal and the individual AML sample (Figure 1C).
Mapping of individual AML patient samples to the gene expression landscape of the normal hematopoietic hierarchy demonstrated varying normal counterparts for different AML subtypes. Whereas samples of AML patients with a complex karyotype mapped to different normal populations, ranging from HSCs to monocytes, the more defined t(15;17) AMLs predominantly mapped closely to the related granulocyte-monocyte progenitors and early promyelocytes (early PMs), representing the developmental stage of this particular AML subtype (Figure 2A-B). Other AK-AML subtypes demonstrated mapping patterns between these extremes (supplemental Figure 2).
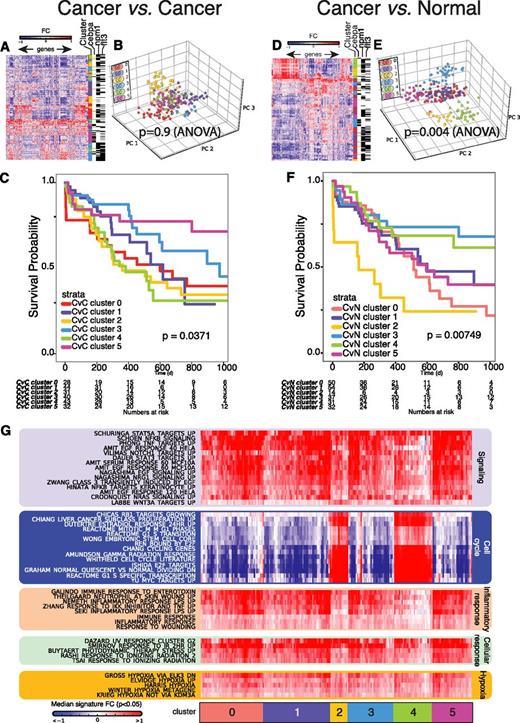
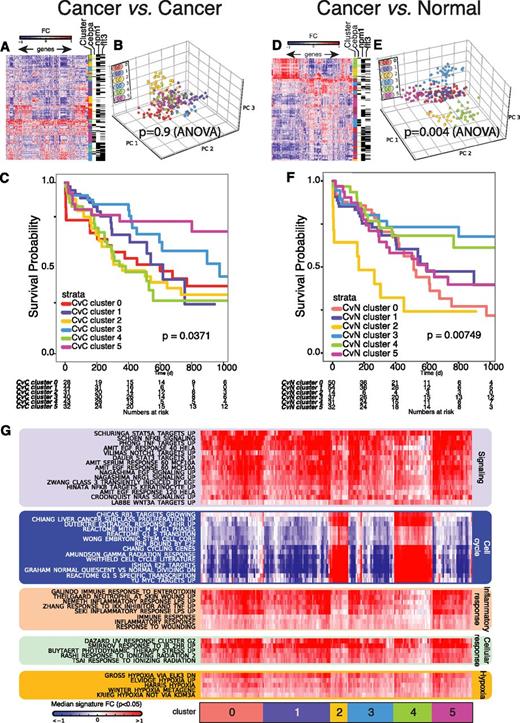
Cluster analysis of AK-AML samples using the CvN method and the CvC method. (A-B) Example of PCA plots of individual gene expression profiles of complex karyotype AML (A) and t(15;17) AML patient samples (B) projected to the gene expression-based map of the normal hematopoietic hierarchy, using the CvN method (see “Bioinformatics analyses” and Figure 1). Only the 2 first (ie, PC1 and PC2) PCs are given in the PCA plot. A line indicates the nearest normal counterpart for each of the AK-AML samples. (C-D) Unsupervised clustering of AK-AML. PCA of AK-AML based on genes identified by the CvC- (C) and CvN method (D). Genes were selected by variance (1545 and 1449 probe sets in (C) and (D); respectively). ANOVA analysis of the segregation of the clusters using the first 5 PCs reports P values of .49 for the CvC method (inter-group variance: 6.08, intra-group variance: 495.85) and .004 for the CvN method (inter-group variance: 115.58, intra-group variance: 1685.95) for the CvC and CvN methods, respectively. (E) ROC curves (classification performances) for 2 published and 1 novel AML t(11q23) gene signatures. Areas under ROC curve (AUCs) are reported in the graph. (F). Heat map representing the degree of enrichment (-log10[P value]) in the 1% upregulated genes in AML patients9 of known and novel AK-AML gene signatures.
Cluster analysis of AK-AML samples using the CvN method and the CvC method. (A-B) Example of PCA plots of individual gene expression profiles of complex karyotype AML (A) and t(15;17) AML patient samples (B) projected to the gene expression-based map of the normal hematopoietic hierarchy, using the CvN method (see “Bioinformatics analyses” and Figure 1). Only the 2 first (ie, PC1 and PC2) PCs are given in the PCA plot. A line indicates the nearest normal counterpart for each of the AK-AML samples. (C-D) Unsupervised clustering of AK-AML. PCA of AK-AML based on genes identified by the CvC- (C) and CvN method (D). Genes were selected by variance (1545 and 1449 probe sets in (C) and (D); respectively). ANOVA analysis of the segregation of the clusters using the first 5 PCs reports P values of .49 for the CvC method (inter-group variance: 6.08, intra-group variance: 495.85) and .004 for the CvN method (inter-group variance: 115.58, intra-group variance: 1685.95) for the CvC and CvN methods, respectively. (E) ROC curves (classification performances) for 2 published and 1 novel AML t(11q23) gene signatures. Areas under ROC curve (AUCs) are reported in the graph. (F). Heat map representing the degree of enrichment (-log10[P value]) in the 1% upregulated genes in AML patients9 of known and novel AK-AML gene signatures.
The CvN method is comparable to the classical CvC method in stratifying AK-AML patients
We next performed a side-by-side comparison of the CvN and CvC methods, using unsupervised standard analysis of a GEP data set derived from 4 distinct AML subtypes [inv16/t(16;16), t(11q23), t(15;17), t(8;21)]. Both methods generated distinct clusters, each representing a genetically defined AK-AML subclass (P < 1e−5; Kruskal–Wallis test), as visualized by either unsupervised PCA (Figure 2C-D) or hierarchical clustering (supplemental Figure 3A-B). However, testing the intercluster and intracluster variance of the first 5 principal components in an analysis of variance (ANOVA) test (Figure 2C-D) demonstrated that genes identified by the CvN method, but not the CvC method, form significant clusters in an unsupervised analysis (P = .004 and P = .49, respectively).
We next used a standard supervised classification analysis and found that genes selected by the CvN method performed as well as those selected by the CvC method for the training set9 (error rates, CvC: 2.05%; CvN: 1.55%; supplemental Figure 4), as well as in an independent AK-AML test set18 (error rates, CvC: 8.62%; CvN: 6.46%). To test whether the CvN method correctly identifies transcriptional changes specific for distinct AK-AML subtypes, we generated signatures of the most discriminatory genes for each AML subtype (supplemental Table 3; supplemental Methods).24 We next generated patient-specific signatures for each individual AML patient based on the 1% most up- and downregulated genes between AML and normal. Finally, using a hypergeometric test, we calculated the significance of the overlap between the subtype-specific CvN-defined, as well as previously reported CvC-defined, AK-AML signatures and the patient-specific signatures for upregulated genes (Figure 2E). Both types of subtype-specific AK-AML signatures displayed a strong overlap with the patient-specific signatures, but receiver-operator characteristic curves showed that the CvN-defined signatures outperformed their CvC-defined counterparts (Figure 2F; supplemental Figure 5A-D; supplemental Table 4). Significantly, this was also demonstrated in an independent data set11 (supplemental Figure 5E-I).
In conclusion, our CvN method allows for the efficient classification of AK-AML subtypes with defined cytogenetic aberrations and outperforms the CvC method in unsupervised and supervised classification analysis.
The CvN method identifies genes and common transcriptional programs potentially linked to malignant transformation and maintenance of AK-AML
Although the CvN method allows for classification of AML subtypes, its main strength lies in its potential to identify changes in gene expression between cancer and normal. We therefore identified genes that exhibit deregulated expression (|log2 FC| > 2; P < 10−5; see supplemental Table 1D-G for gene lists) between AML and normal for patients belonging to different AK-AML subtypes. This yielded complex patterns of gene expression changes between different AK-AML subtypes and their respective normal counterparts, which is also evident from the corresponding hierarchical clustering (supplemental Figure 6A-C). Of particular interest is the 1018 probe sets that are commonly deregulated (|log2 FC| > 1; P < 10−5; supplemental Table 1C) in all the 4 AK-AML subtypes. Among the genes up-regulated compared with their nearest normal counterpart, we find the RAS homology gene RHOB as well as the epigenetic regulators JMJD3 and BRD4 (supplemental Figure 7). Notably, the latter was recently identified as a therapeutic target in AML.25
The CvN method also identified high expression of EVI1 to be specifically associated with t(11q23) AML (supplemental Table 3). This concurs with the previously reported cooccurrence of t(11q23) lesions with high EVI1 expression, and it has been suggested that the latter correlates with the maturation stage of the leukemic blasts.26 To test this, we separated the t(11q23) cohort into EVI1high and EVI1low patients (supplemental Figure 8A). We next assessed the average expression values of genes belonging to a novel stem cell signature (supplemental Methods) and find the EVI1high group to exhibit a higher score, suggesting that this subgroup of t(11q23) is more immature (supplemental Figure 8B). As high EVI1 expression correlates with adverse outcome,26 we predict the EVI1high subgroup to have a poor overall survival (OS).
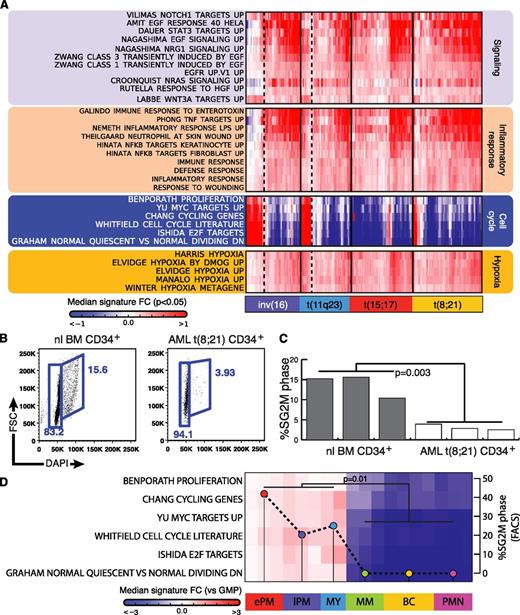
To further explore the transcriptional programs underlying the leukemic phenotypes, we used a hypergeometric test to compare the significance of the overlap between the patient-specific signatures, described earlier, and known gene expression signatures representing curated gene sets (C2), gene ontology gene sets (C5), and oncogenic signatures (C6) from the MSigDB database. Using this gene set overlap analysis, we identified the 200 best correlated MSigDB signatures for each AK-AML subclass (P < 1e−5; supplemental Table 5) and selected for signatures that, based on literature review of experimental design, represented bona fide correlates of normal cellular activities and responses (ie, cell cycle, signaling, and inflammatory and hypoxia response). We report this selection of signatures and their median log2 fold-change when compared with normal for patients derived from each AK-AML subtype (Figure 3A).
Identification of deregulated gene expression programs in AK-AML. (A) Median gene expression fold change of selected MsigDB gene signatures that overlap significantly (P < 1e-5, median, subclass-wise) with patient-specific AK-AML signatures. (B-C) Cell cycle analysis of CD34+ cells from healthy subjects (n = 3) and t(8;21) AML patients (n = 3). (D) Median gene expression fold-change (vs normal GMPs) in cell cycle-related gene signatures for purified normal BM populations together with the experimentally determined cell cycle status (cell cycle profiles were presented in Mora-Jensen et al48 ). The correlation coefficient between “percentage of cells in SG2M” and the average median fold change for the 6 cell cycle signatures was r2=0.8. The following populations are depicted: early promyelocytes (ePM), late promyelocytes (lPM), MY, MM, band cells (BC) and polymorphonuclear neutrophilic granulocytes (PMN).
Identification of deregulated gene expression programs in AK-AML. (A) Median gene expression fold change of selected MsigDB gene signatures that overlap significantly (P < 1e-5, median, subclass-wise) with patient-specific AK-AML signatures. (B-C) Cell cycle analysis of CD34+ cells from healthy subjects (n = 3) and t(8;21) AML patients (n = 3). (D) Median gene expression fold-change (vs normal GMPs) in cell cycle-related gene signatures for purified normal BM populations together with the experimentally determined cell cycle status (cell cycle profiles were presented in Mora-Jensen et al48 ). The correlation coefficient between “percentage of cells in SG2M” and the average median fold change for the 6 cell cycle signatures was r2=0.8. The following populations are depicted: early promyelocytes (ePM), late promyelocytes (lPM), MY, MM, band cells (BC) and polymorphonuclear neutrophilic granulocytes (PMN).
Strikingly, our analysis identified a predominance of transcriptional programs in all AK-AML subtypes, reflecting a low cell cycle activity combined with elevated activities of inflammatory response, hypoxia, and signaling. High cell cycle activity was most abundant among AK-AML patients with inv16/t(16;16) and t(11q23) and was low among t(15;17) and t(8;21) patients. Significantly, these findings demonstrate that our CvN method-based gene set overlap analysis allows for the identification of common sets of transcriptional programs shared by AK-AML patients of different genetic subclasses. Because of the lack of publicly available survival data for the AK-AML cohort, we were unfortunately not able to assess the relevance of differences in common transcriptional programs with respect to clinical outcome. It is, however, likely that individual patients of genetically defined AK-AML subclasses whose common transcriptional programs differ substantially (high vs low cell cycle activity, etc) also may exhibit differential survival.
Finally, to validate the functional relevance of one of the transcriptional programs identified by the CvN method, we performed cell cycle analysis on CD34+ cells from t(8;21) AK-AML patients predicted to exhibit low cell cycle activity and CD34+ populations of healthy subjects (Figure 3B-C). Indeed, this analysis demonstrated a low proliferation rate of leukemic compared with normal CD34+ cells, which is consistent with the predicted low “transcriptional” cell cycle activity of t(8;21) AK-AML. Importantly, the GEP-based prediction of proliferation could also be extended to normal myeloid progenitors (Figure 3D).
Overall, our CvN-based analysis suggests that genetically and clinically diverse AK-AML subclasses share a common set of transcriptional programs that potentially represent abnormal activity of core cellular functions associated with transformation and maintenance of the leukemic phenotype.
Comparison of NK-AML patient samples to their nearest normal counterpart identifies novel subtypes
Having demonstrated the ability of the CvN method to correctly classify subtypes of AK-AML patients and identify common transcriptional programs, we next tested its potential on a data set of NK-AML patients, including survival rates.2 NK-AML is associated with mutations in key hematopoietic and epigenetic regulators (NPM1, CEBPA, FLT3, RUNX1, TET2, DNMT3A, and others27,28 ), but only NPM1 and CEBPA mutant AML constitutes distinct subtypes approved by the World Health Organization.29 We reasoned that, similar to AK-AML, NK-AML would harbor distinct subtypes that could be identified through the CvN method.
As a first approach to estimate the number of potential subtypes in NK-AML patients, we used a similar strategy as that outlined earlier to perform hierarchical clustering on an data set of 218 NK-AML patients,2 including information on survival and mutational status of CEBPA, FLT3, and NPM1. Visual inspection of this initial clustering analysis suggested the presence of 6 subtypes in the NK-AML data set (supplemental Figure 9). To further refine the analysis, we next performed K-means clustering to assign the patients to 6 clusters using variance-selected genes (Figure 4A-B,D-E; supplemental Figure 10A-G). We note that NPM1 and FLT3 mutations did not segregate to any distinct cluster with either the CvN nor the CvC method, which likely reflects the high frequency of patients with combined NPM1 and FLT3 mutations in our NK-AML cohort. In contrast, patients with CEBPA mutations formed a distinct cluster with both methods (CvC cluster_5, Figure 4A; CvN cluster_3, Figure 4D). Although the data set does not contain information on the presence of bi- vs monoallelic CEBPA mutations, the published frequency of biallelic CEBPA AML (≈70%) suggest that these cluster contains the biallelic CEBPA AMLs (76% in CvC cluster_5; 71% in CvN cluster_3).11
The CvN method improves classification of NK-AML patients. Side-by-side comparison of clustering performance of the CvC (A-C) and CvN (D-F) methods on a NK-AML data set (GSE15434). Heat maps (hierarchical clustering) of genes identified by the CvC method (A) and CvN methods (D), using a NK-AML patient data set. Differentially expressed genes identified by each method were selected by variance (1614 and 1383 probe sets in A and D, respectively) and rescaled gene wise. An initial hierarchical clustering was used to identify the optimal number of patient clusters (n = 6; supplemental Figure 9). This was followed by K-means clustering (K = 6), which distributed the samples into 6 patient clusters (color labeled). (B,E) 3-dimensional-PCA plots of the 6 K-means-derived patient clusters identified by the CvC (B) and CvN (E). (C,F) Kaplan-Meier plots depicting the OS curves for of the 6 NK-AML clusters assessed by (C) the CvC method and (F) the CvN method (P = .04 and P = .007, respectively, χ-square). (G) Median gene expression fold change of selected MsigDB gene signatures that overlap significantly (P < 1e−5, median, subclass-wise) with patient-specific NK-AML signatures.
The CvN method improves classification of NK-AML patients. Side-by-side comparison of clustering performance of the CvC (A-C) and CvN (D-F) methods on a NK-AML data set (GSE15434). Heat maps (hierarchical clustering) of genes identified by the CvC method (A) and CvN methods (D), using a NK-AML patient data set. Differentially expressed genes identified by each method were selected by variance (1614 and 1383 probe sets in A and D, respectively) and rescaled gene wise. An initial hierarchical clustering was used to identify the optimal number of patient clusters (n = 6; supplemental Figure 9). This was followed by K-means clustering (K = 6), which distributed the samples into 6 patient clusters (color labeled). (B,E) 3-dimensional-PCA plots of the 6 K-means-derived patient clusters identified by the CvC (B) and CvN (E). (C,F) Kaplan-Meier plots depicting the OS curves for of the 6 NK-AML clusters assessed by (C) the CvC method and (F) the CvN method (P = .04 and P = .007, respectively, χ-square). (G) Median gene expression fold change of selected MsigDB gene signatures that overlap significantly (P < 1e−5, median, subclass-wise) with patient-specific NK-AML signatures.
To assess the relative performance of the 2 methods, we performed a silhouette analysis,30 which demonstrated that both the CvN and CvC clusters were robust (with the former being slightly better [P < 1e−5, t test]; supplemental Figure 10H-I). However, when the intercluster and intracluster variance of the first 3 principal components were tested by ANOVA, only the CvN method yielded significant clusters (P = .9 vs P = .004).
To determine to what extent the CvC and CvN methods used different or overlapping genes to separate their respective clusters, we next merged the lists of genes selected by the 2 methods. Of the approximately 450 genes that were used for clustering by each method, 54% were shared. Importantly, when we analyzed the contribution of CvC-specific, shared, and CvN-specific genes to cluster formation, we found that the CvN-specific genes were better than the CvC-specific genes in separating the clusters (supplemental Figure 10B-G; CvN: P = .001; CvC: P = .02; ANOVA test on first 3 principal components, using the method-specific probe sets). Hence, the residual predictive power of the CvC genes in our analysis of NK-AML is primarily driven by a subgroup of genes, which is also selected by the CvN method, thus highlighting its excellent performance.
Overall, our cluster analysis demonstrates that the CvN method is capable of identifying potential subtypes of NK-AML patients with distinct patterns of aberrantly expressed genes.
The CvN-predicted NK-AML clusters display differential OS
We next assessed the potential of the CvN and CvC clusters to predict OS in NK-AML (Figure 4C,F). Interestingly, the clusters generated by the CvN method displayed distinct distributions of OS rates among NK-AML patients, suggesting that this method is capable of extracting prognostic relevant disease entities that are not defined by specific genetic lesions but, rather, by distinct gene expression programs representing surrogates of their leukemic phenotype. Of the 6 CvN clusters, we found cluster_2 to be associated with significantly worse outcome compared with the remaining 5 clusters (Figure 4F; supplemental Table 6). Moreover, multivariate Cox regression analysis identified cluster_2 as the strongest independent prognostic factor for OS in NK-AML patients, performing better than known risk factors such as FLT3 mutations and age (Table 2). These findings were corroborated by a random forest analysis31 that highlighted cluster_2 as the most important variable for the prediction of OS (supplemental Figure 11). Collectively, this demonstrates the ability of the CvN method to identify novel prognostic relevant subtypes of NK-AML patients.
To explore potentially disrupted core cellular functions associated with the leukemic phenotype and OS of the 6 NK-AML clusters, we again generated patient-specific signatures (as described earlier) and scored the significance of the overlap for each individual NK-AML signature against the gene expression signatures from the MSigDB database. We identified the 200 most significant and positively correlated MSigDB signatures for each NK-AML cluster (P < 1e−5; supplemental Table 5) and selected for signatures based on literature review, as described earlier. This analysis allowed us to identify NK-AML patients with high (cluster_2 cluster_4) vs low (cluster_0, cluster_1, cluster_3, cluster_5) cell cycle activity compared with their normal counterparts. Similar to AK-AML, the majority of NK-AML patients shared a common transcriptional program reflecting elevated activity of inflammatory response, hypoxia, and signaling activities independent of their mutation and cluster status (Figure 4G).
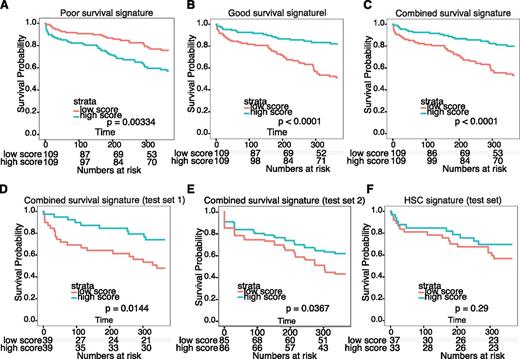
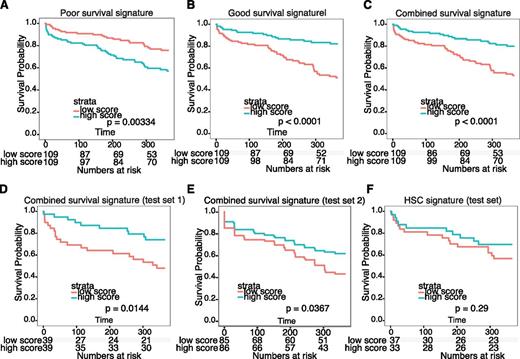
Surprisingly, cluster_2 and cluster_4, which differed widely with respect to clinical outcome, shared high cell cycle activity and did not differ markedly with respect to activity of other common transcriptional programs. Consistently, we noted that cluster_2, with a poor outcome, expressed a very similar set of deregulated genes compared with cluster_4 with a favorable outcome (|log2 FC| > 2; P < 10−5 vs normal cells; Figure 4D; supplemental Tables 1H and 4). We therefore hypothesized that the few differentially expressed genes between these clusters would be highly enriched in genes that account for chemotherapy resistance, and thus could predict OS for the entire NK-AML patient data set. To test this, we identified differentially expressed genes between cluster_2 and cluster_4 (|log2 FC| > 2; P < .05) and determined the significance by which they could predict differences in OS in the upper and lower fold-change quartiles, using the entire NK-AML patient data set (supplemental Table 1I). We next used these genes to build a poor-outcome signature, a good-outcome signature, and a combined survival signature, which were all able to efficiently allocate the entire NK-AML data set, as well as 2 independent data sets,13,15 into patients with good and poor outcome (Figure 5A-E). Importantly, the genes in these signatures are predicted to be enriched for genes directly involved in disease etiology, including resistance to chemotherapy (supplemental Table 7). On a final note, we found that a previously reported hematopoietic stem cell signature32 was unable to predict survival in the NK-AML patient data set15 (Figure 5F).
Survival signature predicts survival of patients with NK-AML. (A-C) Survival analysis based on 3 survival signatures derived from genes differentially expressed in patient cluster_2 and cluster_4. The effect of the expression of individual probe sets on survival was tested by dividing the entire data set into low- and high-scoring samples (median). Probe sets associated with poor and good OS (P < .05, moderated t test) and the ability to separate the data set (P < .05, log-rank test) were used to generate a poor outcome signature (A), good outcome signature (B), and combined survival signature (C). (D-E) Testing of the combined survival signature on 2 independent NK-AML patient data sets.13,15 (F) Testing of a previously published HSC signature32 revealed its inability to predict survival in the NK-AML patient data set in D.
Survival signature predicts survival of patients with NK-AML. (A-C) Survival analysis based on 3 survival signatures derived from genes differentially expressed in patient cluster_2 and cluster_4. The effect of the expression of individual probe sets on survival was tested by dividing the entire data set into low- and high-scoring samples (median). Probe sets associated with poor and good OS (P < .05, moderated t test) and the ability to separate the data set (P < .05, log-rank test) were used to generate a poor outcome signature (A), good outcome signature (B), and combined survival signature (C). (D-E) Testing of the combined survival signature on 2 independent NK-AML patient data sets.13,15 (F) Testing of a previously published HSC signature32 revealed its inability to predict survival in the NK-AML patient data set in D.
Collectively, our analyses demonstrate that the CvN method is able to stratify NK-AML patients into known subtypes (cluster_3 with CEBPA mutations), stratify patients into new subtypes exhibiting differential OS, and extract a set common transcriptional programs that likely represent disrupted core cellular functions underlying the leukemic phenotype (see supplemental Table 5 for additional data). In addition, our analysis might imply that increased chemotherapy resistance in NK-AML clusters with poor vs favorable outcome is primarily driven by a limited number of highly prognostically relevant genes rather than higher or lower activity of some of the common transcriptional programs.
Discussion
GEP has the potential to yield fundamental insights into the transcriptional programs of cancer cells and has thus been used for more than a decade to probe tumor phenotypes. However, with few exceptions, these analyses have all compared cancer to cancer, with the obvious risk that differences in cell type and developmental stage may render the identification of truly malignant gene expression programs impossible. Here we present a simple method, referred to as the CvN method, that allows us to identify the nearest normal counterpart for individual AML patient samples and calculate gene expression differences between AML and normal. Our method performed extremely well in classification of AK-AML and identified gene expression programs associated with distinct AK-AML subtypes. Moreover, we were able to clearly separate an NK-AML patient data set into several clusters according to transcriptional differences between individual AML patient samples and their closest normal counterpart. These clusters were associated with distinct OS and were predictive in multivariate analysis, highlighting their biological relevance.
Recent epidemiologic and clinical studies have demonstrated a higher incidence and aggressiveness of cancer in patients with diabetes and in patients with inflammatory and autoimmune diseases.33-35 Consistently, treatment with metformin reduces cancer in patients with diabetes, and inflammatory ligands are elevated and promote maintenance and proliferation of malignant cells in different cancer entities.33,36-39 In addition, NF-κB, the key transcriptional regulator of inflammatory response, was demonstrated to be constitutively activated in various types of cancers, including AML, and to play an important role in malignant transformation in mouse models.39-43 Finally, the ability of various solid cancer cells to adapt to hypoxia and switch metabolism from oxidative phosphorylation toward glycolysis has emerged as a novel hallmark of cancer that defines more aggressive cancer phenotypes.44
Consistent with these reports, a previous study45 identified a cancer signature of malignant transformation that is not only shared by various types of cancers but also overlaps significantly with gene expression signatures of chronic inflammatory conditions (colitis ulcerosa, rheumatoid arthritis, systemic lupus erythematosus, Crohn’s disease) and metabolic diseases (diabetes, obesity, hypercholesterolemia, atherosclerosis, cardiomyopathy). On the basis of their findings, the authors argued that physiological and/genetic disruption of core biological pathways maintaining normal cell functions generates a gene expression program that is common to a diverse set of human diseases.45
In line with these findings, our analysis demonstrated a significant overlap of the “common cancer signature” and AK-AML and NK-AML signatures generated by the CvN method (supplemental Table 5; HIRSCH_CELLULAR_TRANSFORMATION_SIGNATURE_UP). Strikingly, our analysis also unraveled common transcriptional programs among all AML patients that are associated with elevated signaling activity, inflammatory response, and hypoxia. Indeed, these programs might reflect a disruption of normal core cellular functions that are shared by most AML patients despite their otherwise profound clinical and genetic heterogeneity.
Significantly, our gene set overlap analysis allowed us to discriminate AML patients with a high vs low cell cycle activity compared with their normal counterpart. Whereas the majority of AK- and NK-AML patients demonstrated a low cell cycle activity combined with elevated activities of inflammatory response, hypoxia, and signaling, a minor number of patients demonstrated a program of high cell cycle activity. The latter included a significant number of AK-AML patients with inv16/t(16;16) and t(11q23) as well as all cluster_2 and cluster_4 NK-AML patients. Surprisingly, cluster_2 and cluster_4 demonstrated poor and favorable outcomes, respectively, despite comparable high cell cycle activity combined with a similar activity of inflammatory response, hypoxia, and signaling. Consistently, they shared a high number of aberrantly expressed genes compared with normal but also demonstrated a limited number of differentially expressed genes that formed the basis for a powerful NK-AML survival signature. These findings suggest that resistance to chemotherapy in NK-AML patients with poor vs favorable outcome is primarily driven by a minor number of prognostic relevant genes and not by the differential activity of common transcriptional programs. Importantly, as these common transcriptional programs likely represent disrupted core cellular functions, some of them may be relevant for future targeting.
The CvN method may, in principle, be improved by several means. As an example, the data used in the present work originate from AML data sets derived from bulk tumor material. Hence, an obvious improvement of the precision of the CvN method would be to perform the analysis on purified AML subpopulations and compare those with their respective normal counterparts. Furthermore, our method is dependent on the availability of GEPs from normal cells to construct a gene expression landscape of the hematopoietic hierarchy onto which we can map AML samples. Obviously, the precision of the mapping and the subsequent deduction of gene expression changes between cancer cells, and their corresponding normal counterpart is dependent on the number and quality of normal reference populations. Given the high density of functionally defined intermediate HPCs on the path from HSCs to mature blood cells within the hematopoietic hierarchy, its associated malignancies are ideally suited for the CvN method. However, by combining multiparameter cell sorting with highly innovative methods for the analysis of flow cytometry data, such as the recently published Cyto-Spanning Tree Progression of Density Normalized Events (CytoSPADE) method, it should be possible to isolate novel intermediate HPCs for subsequent GEP, thereby further refining the resolution of the gene expression landscape of the normal hematopoietic hierarchy.46,47 This will in turn improve the extent to which transcriptional changes between normal and cancer cells can be detected. Importantly, as no functionally characterization is required, implementation of SPADE or similar protocols may be used to isolate a suitable number of stem/progenitor cells from other organs, thereby making the CvN method amenable for analysis of solid tumors. Thus, our approach has the potential to be widely applicable to a substantial number of cancer types and promises to expand the clinical use of GEP.
The data reported in this article have been deposited in the Gene Expression Omnibus database (accession number GSE42519).
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by grants from the Danish Council for Strategic Research (09-065157, 10-092798), the Danish Cancer Society (R2-Rp1425), the NovoNordisk Foundation (11628, R168-A14079), and the Lundbeck Foundation (R34-A3620) and through a center grant from the Novo Nordisk Foundation Section for Stem Cell Biology in Human Disease. This work is based on the joint research activities under the framework of the European Program for Cooperation in Science and Technology (Action BM0801, WG1). L.B. was supported in part by the Deutsche Forschungsgemeinschaft (Heisenberg-Stipendium BU 1339/3-1). K.T.-M. is supported by a clinical research fellowship from the Novo Nordisk Foundation (R191-A15986). O.W. acknowledges funding from the Novo Nordisk Foundation (05-04-2005).
Authorship
Contribution: N.R., N.B., A. Krogh, O.W., K.T.-M., and B.T.P. conceived and designed the study; N.B. and H.M.J. provided BM samples; A. Kohlmann, C.T., L.B., J.J., and H.M.-J. collected and assembled the data; N.R. produced the figures; N.R., F.O.B., K.T.-M., and B.T.P. analyzed and interpreted the data; and N.R., B.T.P., K.T.-M., O.W., and F.O.B. wrote the manuscript.
Conflict-of-interest disclosure: A. Kohlman is employed by the Munich Leukemia Laboratory. The remaining authors declare no competing financial interests.
Correspondence: Bo Porse, Finsen Laboratory/Rigshospitalet, University of Copenhagen, Ole Maaløesvej 5, 2200 Copenhagen N, Denmark; e-mail: bo.porse@finsenlab.dk.
References
Author notes
K.T.-M. and B.T.P. contributed equally to this study.

![Figure 2. Cluster analysis of AK-AML samples using the CvN method and the CvC method. (A-B) Example of PCA plots of individual gene expression profiles of complex karyotype AML (A) and t(15;17) AML patient samples (B) projected to the gene expression-based map of the normal hematopoietic hierarchy, using the CvN method (see “Bioinformatics analyses” and Figure 1). Only the 2 first (ie, PC1 and PC2) PCs are given in the PCA plot. A line indicates the nearest normal counterpart for each of the AK-AML samples. (C-D) Unsupervised clustering of AK-AML. PCA of AK-AML based on genes identified by the CvC- (C) and CvN method (D). Genes were selected by variance (1545 and 1449 probe sets in (C) and (D); respectively). ANOVA analysis of the segregation of the clusters using the first 5 PCs reports P values of .49 for the CvC method (inter-group variance: 6.08, intra-group variance: 495.85) and .004 for the CvN method (inter-group variance: 115.58, intra-group variance: 1685.95) for the CvC and CvN methods, respectively. (E) ROC curves (classification performances) for 2 published and 1 novel AML t(11q23) gene signatures. Areas under ROC curve (AUCs) are reported in the graph. (F). Heat map representing the degree of enrichment (-log10[P value]) in the 1% upregulated genes in AML patients9 of known and novel AK-AML gene signatures.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/123/6/10.1182_blood-2013-02-485771/4/m_894f2.jpeg?Expires=1768001012&Signature=Xi3FUEbQ9HIZ9MnmAzdBdoQoPnai978Jyw98jih0JDABLoFUw7j6oZiJ-ExHaL4fr9lLbHO1DsFYqvBKRmNM0zN09EkVISrQHugKdfK5j2QdUf3OlSCfKQhZNbHOZu8cYJr~VgVP-BJmMJKLVJECjxoBUw~iLjszSQbomSq8il~1xhbcp~eHDjAwqYTI8lr2Ip2N7ANTmlcywFVpjKI~6KsmR4NuInOmrN6hHLY6tZrYWcBTLQAuiXA-sIr8r02dfTZRHsq1WwnkImiLgWR5gdbKr~nIXgR9Ssz0IHGksCdstAOUHmkHYEhrkjPL0OAyYxSjgARh~a6hAG3ORK2Q5w__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)


![Figure 2. Cluster analysis of AK-AML samples using the CvN method and the CvC method. (A-B) Example of PCA plots of individual gene expression profiles of complex karyotype AML (A) and t(15;17) AML patient samples (B) projected to the gene expression-based map of the normal hematopoietic hierarchy, using the CvN method (see “Bioinformatics analyses” and Figure 1). Only the 2 first (ie, PC1 and PC2) PCs are given in the PCA plot. A line indicates the nearest normal counterpart for each of the AK-AML samples. (C-D) Unsupervised clustering of AK-AML. PCA of AK-AML based on genes identified by the CvC- (C) and CvN method (D). Genes were selected by variance (1545 and 1449 probe sets in (C) and (D); respectively). ANOVA analysis of the segregation of the clusters using the first 5 PCs reports P values of .49 for the CvC method (inter-group variance: 6.08, intra-group variance: 495.85) and .004 for the CvN method (inter-group variance: 115.58, intra-group variance: 1685.95) for the CvC and CvN methods, respectively. (E) ROC curves (classification performances) for 2 published and 1 novel AML t(11q23) gene signatures. Areas under ROC curve (AUCs) are reported in the graph. (F). Heat map representing the degree of enrichment (-log10[P value]) in the 1% upregulated genes in AML patients9 of known and novel AK-AML gene signatures.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/123/6/10.1182_blood-2013-02-485771/4/m_894f2.jpeg?Expires=1768425155&Signature=V6F~2gkZu3HtKUJFjPzFpmEXpiwbwqSDJZQGN-kkSzpZjtYcVavNBgqaLCLcBCitxs137quZVou7naXPvslKJuW~PJMr-Ow2n1KnOCCsAcYxepEYa6IRneV3abcIgH4RXnlAUh0Kz25n6rHIUvau8nHT7qGIIPtDfaeLyLdOUyELqXPDW9duFs-3nx6D~R-fK5z3902FrfMH4vnsK6K61~JQw~r0d23cUcJm91~x9p-Gsjq4KpuVk0Vf6UwFaEd2PjiN8wnkpULkyROlIlw0il7FqWRcTZWB~EUUyqBDMNdtuBqJxwyqu7CelGjPT~ww9zQBFXd~Uj2R5Y~vJwbwCw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)