Key Points
Immune stimulation of cyclin D1 transgenic mice bearing Bim-deficient B cells induces an MCL phenotype.
The induced lymphoma of EμCycD1CD19CREBimfl/fl mice highlights the collaborative roles of Bim deletion and cyclin D1 expression in MCL.
Abstract
Mantle cell lymphoma (MCL) is a highly aggressive B-cell lymphoma resistant to conventional chemotherapy. Although defined by the characteristic t(11;14) translocation, MCL has not been recapitulated in transgenic mouse models of cyclin D1 overexpression alone. Indeed, several genetic aberrations have been identified in MCL that may contribute to its pathogenesis and chemoresistance. Of particular interest is the frequent biallelic deletion of the proapoptotic BCL-2 family protein BIM. BIM exerts its pro-death function via its α-helical BH3 death domain that has the dual capacity to inhibit antiapoptotic proteins such as BCL-2 and MCL-1 and directly trigger proapoptotic proteins such as the mitochondrial executioner protein BAX. To evaluate a functional role for Bim deletion in the pathogenesis of MCL, we generated cyclin D1–transgenic mice harboring Bim-deficient B cells. In response to immunization, EμCycD1CD19CREBimfl/fl mice manifested selective expansion of their splenic mantle zone compartment. Three distinct immune stimulation regimens induced lymphomas with histopathologic and molecular features of human MCL in a subset of mice. Thus, deletion of Bim in B cells, in the context of cyclin D1 overexpression, disrupts a critical control point in lymphoid maturation and predisposes to the development of MCL. This genetic proof of concept for MCL pathogenesis suggests an opportunity to reactivate the death pathway by pharmacologic mimicry of proapoptotic BIM.
Introduction
Mantle cell lymphoma (MCL) accounts for 5% to 10% of all non-Hodgkin lymphomas and has an aggressive clinical course.1 Median survival is approximately 3 years, with few long-term survivors.1 Virtually all cases of MCL contain a t(11;14)(q13;q32) translocation, which places the cyclin D1 gene under the transcriptional control of the immunoglobulin heavy chain promoter.1 This translocation results in the overexpression of cyclin D1, which phosphorylates the retinoblastoma protein leading to unrestrained proliferation.2 Interestingly, transgenic mouse models of cyclin D1 overexpression alone do not recapitulate an MCL phenotype.3,4 Indeed, comparative genomic hybridization and chromosomal banding studies have identified many additional genetic aberrations in MCL.5,6 In particular, the proapoptotic BCL-2 family member BIM showed biallelic deletion in 5 of 12 MCL cell lines and loss of expression in 7 of 22 patient samples by tissue microarray analysis.5 These data suggest that BIM may function as a critical tumor suppressor, whose loss in the context of cyclin D1 overexpression may contribute to the pathogenesis of MCL.
Apoptotic deregulation is a prominent feature of MCL. For example, in addition to loss of the proapoptotic BH3-only protein BIM, overexpression of antiapoptotic BCL-2 family members is also common.7-10 High-level expression of BCL-2, BCL-XL, and MCL-1 has been documented in a variety of MCL cell lines.7-10 Moreover, in primary specimens, BCL-2 overexpression is seen in almost 97% of cases,11 with 15% showing specific genomic gains or amplifications at the BCL-2 locus.12 Overexpression of BCL-XL has also been reported in MCL cells and linked to constitutive activation of the nuclear factor–kB pathway.13 High expression levels of MCL-1 correlate with high-grade morphology and a more aggressive disease course.14 Several studies have implicated interactions between cyclin D1 and BAX or BCL-2 in MCL pathophysiology.12,15 These findings highlight that BCL-2 family deregulation may serve as a critical control point for MCL pathogenesis and a potential opportunity for therapeutic intervention using pharmacologic modulators of the pathway.16,17 Nevertheless, genetic proof of a direct, collaborative role between cyclin D1 expression and BCL-2 family deregulation in the genesis of MCL has not been established.
The development of an MCL mouse model that reflects the human pathophysiology of the disease has been a longstanding challenge. Overexpression of cyclin D1 alone not only fails to produce lymphoproliferative disease, but also results in little to no alteration in B- or T-cell development.3,4 In combination with transgenic overexpression of N-myc or C-myc, cyclin D1 accelerates the formation of B-cell lymphomas, but these tumors are more similar to those produced in mice engineered to overexpress Myc alone.3,4 Moreover, overexpression or rearrangement of Myc is infrequent in MCL, occurring only in the uncommon blastoid variant.18 Overexpression of a mutant cyclin D1 that is constitutively nuclear is sufficient to produce B-cell lymphomas by 13 to 14 months of age. However, these tumors also manifest blastoid morphology with high levels of Myc expression.19 In addition, cyclin D1 mutations that result in constitutive nuclear expression are rare in human MCL.20 A third model of blastoid MCL was created by overexpression of Myc and interleukin-14α.21 Although these mice recapitulate many features of MCL, including cyclin D1 and CD5 positivity,21 their underlying genetics, transgenic overexpression of Myc and interleukin-14α, and the absence of a syntenic equivalent translocation for cyclin D1, bear little resemblance to human MCL. A fourth model of blastoid/pleomorphic MCL was derived from a mouse cell line created by irradiating interleukin 3–dependent BaF3 pro-B lymphocytes with inducible expression of cyclin D1.12 When transplanted into RAG2−/−γc−/− mice, cyclin D1–positive tumors emerged and contained similar genomic alterations as human MCL, as defined by array comparative genomic hybridization. However, the cells were both negative for CD5 and exhibited large, pleomorphic cell morphology. Finally, lymphoma with features of nonblastoid MCL resulted from treating 12- to 14-month-old cyclin D1 transgenic mice with pristane, an autoimmune disease–inducing agent.22
Here, we report the generation and characterization of a genetically engineered mouse model of MCL. The development of disease was triggered by immune stimulation in the context of both cyclin D1 overexpression and deletion of proapoptotic Bim in B cells, recapitulating 2 common cytogenetic abnormalities seen in human MCL. Thus, we find that apoptosis deregulation is a predisposing factor for mantle cell lymphomagenesis, with proapoptotic BIM functioning as a tumor suppressor.
Methods
Mice
The mouse cyclin D1 complementary DNA (a gift from P. Scizinski, Dana-Farber Cancer Institute) was cloned into the pBluescript II vector, which contains a 1-kb fragment of the mouse variable chain enhancer and promoter pEμ and a 1.6-kb fragment of the human β-globin gene to provide introns and the polyadenylation signal sequence (a gift from F. Alt, Children’s Hospital Boston). Insert DNA, released by BssHII digestion, was purified and introduced by pronuclear injection into oocytes from B6SJLF1 mice (Taconic). An expressing transgenic line was interbred with CD19CRE (Jackson Laboratory, strain B6.129P2(C)-CD19tm1(cre)Cgn/J) and with Bimfl/fl mice.23
Treatment regimens for age-matched mice included (1) intraperitoneal (IP) injection (0.3 mL) of a 3.3% sheep red blood cell (SRBC) suspension in phosphate-buffered saline (PBS) (Cocalico Biologicals) followed by repeat dosing after a 2-month interval, (2) IP injection (0.5 mL) of pristane (400 mg) monthly for 3 months, (3) single-dose IP SRBC followed by pristane (400 mg IP) 1 week later, (4) single-dose IP SRBC followed by 400 cGy irradiation 1 week later, and (5) weekly IP SRBC for 4 months. All animal experiments were approved by and conform to the standards of the Dana-Farber Cancer Institute Animal Care and Use Committee.
Histology and immunohistochemistry
Mouse tissues were fixed in 10% formalin for more than 24 hours, passed through graded alcohols, and embedded in paraffin. Slides containing 5-μm-thick sections were used for hematoxylin and eosin staining and immunohistochemistry (Specialized Histopathology Service, Brigham and Women’s Hospital). The slides were soaked in xylene, passed through graded alcohols, and immersed in distilled water. Immunohistochemistry was performed using the following conditions: Slides were treated with either 1 mM EDTA (pH 8.0) (for CD138 and Ki-67) or 10 mM citrate (pH 6.0) (for cyclin D1, B220, BIM, and phosphorylated retinoblastoma protein) (Invitrogen) in a steam pressure cooker (Decloaking Chamber, BioCare Medical) according to the manufacturer’s instructions, and then washed in distilled water. All subsequent steps were performed at room temperature in a hydrated chamber. Slides were treated with Peroxidase Block (DakoCytomation) for 5 minutes to quench endogenous peroxidase activity and then primary monoclonal B220 antibody (1:200) (DakoCytomation), CD138 antibody (1:1000) (BD Pharma), BIM antibody (1:50) (Cell Signaling), phosphorylated retinoblastoma protein antibody (1:100) (Cell Signaling), Ki-67 antibody (1:250) (Vector Laboratory), or cyclin D1 antibody (1:40) (Neomarkers) applied in DakoCytomation diluent for 1 hour. Slides were washed in 50 mM Tris-Cl (pH 7.4). To develop the slides, an anti-rabbit (for B220, BIM, phosphorylated retinoblastoma protein, Ki-67, and cyclin D1 antibodies) or an anti-mouse (for CD138 antibody) horseradish peroxidase–conjugated secondary reagent (Envision Detection Kit, DakoCytomation) was applied for 30 minutes, followed by washing, immunoperoxidase staining with the Diaminobenzidine Chromogen Kit (DakoCytomation), and hematoxylin counterstaining. Slides were analyzed using an Olympus BX43 microscope equipped with a 5-mP Spot idea (28.2) digital photo camera or with an Aperio Scanscope XT automated scanner and images downloaded to Adobe Photoshop software. The objective lens magnifications and numerical apertures used include: ×2/0.06, ×4/0.10, ×10/0.25, ×20/0.40, ×40/0.65, and ×100/1.25 oil.
Flow cytometry
Dissected spleens were washed in 1X PBS with 0.2% fetal bovine serum, minced, and filtered through a 45-μm cell strainer. Cells were washed several times and red blood cells lysed with Red Blood Cell Lysing Buffer (Sigma, R7757). The isolated splenocytes were incubated in 100 μL 1X PBS/0.2% fetal bovine serum with the following antibodies at 1:100 for 20 minutes at room temperature and then analyzed by flow cytometry (FACSCalibur, Becton Dickinson): anti-CD19-FITC (BD Pharmingen, 553785), anti-B220-FITC (BD Pharmingen, 553092), anti-CD23-PE (eBioscience, 12-0232-82), anti-CD5-APC (BD Pharmingen, 561895), and anti-IgM-PE (BD Pharmingen, 553409). Fluorescence-activated cell sorter data were analyzed using FlowJo software (Tree Star Inc.).
Molecular analysis
RNA and DNA were isolated with Trizol from whole spleen frozen at −80°C using a Qiagen AllPrep kit (80204) according to the manufacturer’s instructions. Fifty nanograms of genomic DNA was amplified with Phusion polymerase (Finnzymes) and primers corresponding to the FR3 of most VHJ558 family members (5′-GGAATTCGCCTGACATCTGAGGACTCTGC-3′ and 5′-GACTAGTCCTCTCCAGTTTCGGCTGAATCC-3′).24 RNA was processed and hybridized to an Illumina mouse WG-6 v2.0 chip at the Yale Center for Genome Analysis. The microarray data analyses were carried out using packages in R (Development Core Team, 2010). Raw expression data were normalized using the quantile method provided by the lumi package in R/Bioconductor.25 Differentially expressed genes were determined by Linear Models for Microarray Data26 with a false discovery rate cutoff of 0.1 and an absolute log2 fold-change >0.263. Gene Set tests were performed using quantitative gene set enrichment analysis (QuSAGE) version 1.3.127 on 3 gene set collections: (1) REACTOME gene sets from the MSigDB database v4.0 (http://www.broadinstitute.org/gsea/msigdb/download_file.jsp?filePath=/resources/msigdb/4.0/c2.cp.reactome.v4.0.symbols.gmt), (2) C6 oncogenic gene signatures from the MSigDB database v4.0 (http://www.broadinstitute.org/gsea/msigdb/download_file.jsp?filePath=/resources/msigdb/4.0/c6.all.v4.0.symbols.gmt), and (3) a gene signature defined for human MCL by Rosenwald et al.28 The microarray data have been deposited in the National Center for Biotechnology Information’s Gene Expression Omnibus29 and are accessible through Gene Expression Omnibus series accession number GSE52214 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE52214).
Results
Generation of EμCycD1CD19CREBimfl/fl mice
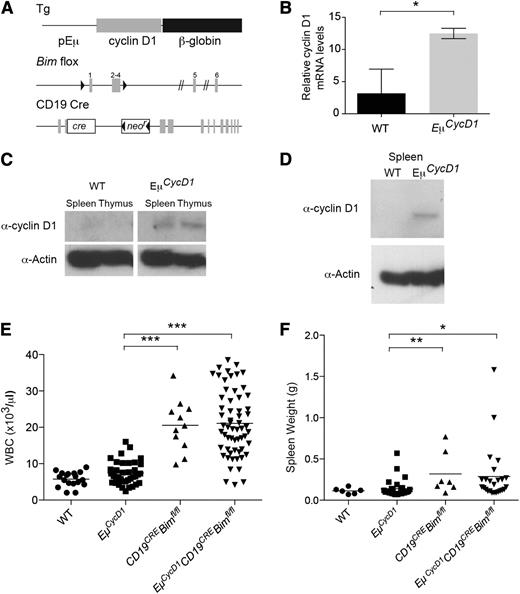
To emulate the characteristic t(11;14) translocation that causes overexpression of cyclin D1 driven by the immunoglobulin heavy chain locus in human MCL, the murine cyclin D1 open reading frame was placed under control of the immunoglobulin VH promoter and Eμ enhancer elements (Figure 1A). Four founder mice were obtained, as confirmed by polymerase chain reaction (PCR) screening and Southern analysis, and then compared for expression levels, white blood cell (WBC) count, and spleen weight (supplemental Figure 1). Line 1 exhibited increased expression of cyclin D1 by quantitative reverse transcriptase-PCR (Figure 1B), which correlated with modest protein level elevation observed by western blot analysis of whole spleen and thymus extracts (Figure 1C). In addition, cyclin D1 protein was detected in purified EμCycD1 splenic B cells stimulated for 4 days in culture with lipopolysaccharide, whereas the corresponding cells from WT mice showed no cyclin D1 expression (Figure 1D). Histologic analysis of the spleens from EμCycD1 mice did not reveal alteration of splenic architecture or a significant increase in cyclin D1–positive B cells, as evaluated by immunohistochemistry of the spleen (supplemental Figure 1).
Development and characterization of a cyclin D1 transgenic mouse line bearing Bim−/− B cells. (A) Schematic of the cyclin D1 transgene, floxed Bim locus, and CD19CRE locus. (B) Reverse transcriptase-PCR of spleen RNA for the transgenic cyclin D1 locus in wild-type (WT) and EμCycD1 mice. (C) Western blot of cyclin D1 in spleens and thymi of WT and EμCycD1 mice. (D) Western blot of cyclin D1 in B220-sorted splenocytes stimulated with lipopolysaccharide. (E) CD19CREBimfl/fl and EμCycD1CD19CREBimfl/fl mice manifest similarly elevated WBC counts at 20 to 25 weeks of age compared with EμCycD1 mice. (F) CD19CREBimfl/fl and EμCycD1CD19CREBimfl/fl mice exhibit a similar degree of splenomegaly compared with EμCycD1 mice. *P < .05; **P < .01; ***P < .001; 2-tailed Student t test. mRNA, messenger RNA.
Development and characterization of a cyclin D1 transgenic mouse line bearing Bim−/− B cells. (A) Schematic of the cyclin D1 transgene, floxed Bim locus, and CD19CRE locus. (B) Reverse transcriptase-PCR of spleen RNA for the transgenic cyclin D1 locus in wild-type (WT) and EμCycD1 mice. (C) Western blot of cyclin D1 in spleens and thymi of WT and EμCycD1 mice. (D) Western blot of cyclin D1 in B220-sorted splenocytes stimulated with lipopolysaccharide. (E) CD19CREBimfl/fl and EμCycD1CD19CREBimfl/fl mice manifest similarly elevated WBC counts at 20 to 25 weeks of age compared with EμCycD1 mice. (F) CD19CREBimfl/fl and EμCycD1CD19CREBimfl/fl mice exhibit a similar degree of splenomegaly compared with EμCycD1 mice. *P < .05; **P < .01; ***P < .001; 2-tailed Student t test. mRNA, messenger RNA.
Rare EμCycD1CD19CREBimfl/fl mice develop cyclin D1–positive malignancies
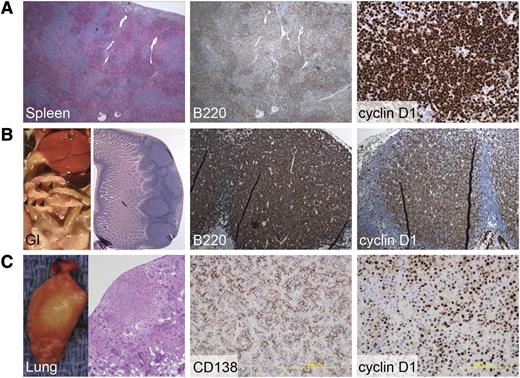
CD19CREBimfl/fl mice, which delete Bim only in the B-cell lineage, were crossed to cyclin D1 transgenic mice to investigate the potential collaborative role of apoptotic deregulation that derives from Bim loss in promoting human cyclin D1–positive MCL. CD19CREBimfl/fl and EμCycD1CD19CREBimfl/fl mice manifested similar and statistically significant increases in both total WBC count and spleen weight as compared with EμCycD1 mice (Figure 1E-F). Among the 25 EμCycD1CD19CREBimfl/fl mice subjected to complete histopathologic evaluation, 3 (12%) developed a cyclin D1–positive hematopoietic neoplasm (Figure 2). Two of the cases were small B-cell neoplasms occurring in the spleen and gastrointestinal tract, with morphology similar to MCL (Figure 2A-B). The third case was a cyclin D1–positive plasma cell neoplasm of the lung (Figure 2C). Of note, approximately 15% of multiple myeloma cases harbor the t(11;14) immunoglobulin H; cyclin D1 translocation, which is present in MCL.30 In contrast, none of the 22 EμCycD1 mice subjected to histopathologic evaluation developed any form of B-cell lymphoma, although 1 mouse developed a cyclin D1–positive histiocytic neoplasm. As expected, CD19CREBimfl/fl mice, which do not overexpress cyclin D1, did not develop cyclin D1–positive tumors, although we did observe organ infiltration by B cells in a subset of animals, as reported for mice with global Bim deletion31 (supplemental Figure 2).
Rare cyclin D1–positive malignancies develop in EμCycD1CD19CREBimfl/flmice. (A) Expanded spleen follicles that are diffusely positive for the B-cell marker B220 and for cyclin D1 were observed in an affected EμCycD1CD19CREBimfl/fl mouse. (B) Multiple gastrointestinal polyps were observed in a mouse with lymphomatoid polyposis; sheets of B220 and cyclin D1–positive B cells were evident. (C) One mouse developed lung nodules that were positive for the plasma cell marker CD138 and for cyclin D1. Original magnification (left to right): (A) ×40, ×40, ×400; (B) ×40, ×200, ×200; (C) ×40, ×400, ×400.
Rare cyclin D1–positive malignancies develop in EμCycD1CD19CREBimfl/flmice. (A) Expanded spleen follicles that are diffusely positive for the B-cell marker B220 and for cyclin D1 were observed in an affected EμCycD1CD19CREBimfl/fl mouse. (B) Multiple gastrointestinal polyps were observed in a mouse with lymphomatoid polyposis; sheets of B220 and cyclin D1–positive B cells were evident. (C) One mouse developed lung nodules that were positive for the plasma cell marker CD138 and for cyclin D1. Original magnification (left to right): (A) ×40, ×40, ×400; (B) ×40, ×200, ×200; (C) ×40, ×400, ×400.
No MCL elicited by pristane treatment of EμCycD1CD19CREBimfl/fl mice
A prior study indicated that pristane treatment of elderly EμCycD1 mice produced MCL.22 Thus, we tested whether pristane would have the same effect on our independently derived cyclin D1 transgenic mice and if the loss of Bim in B cells aggravated the phenotype. We subjected EμCycD1 and EμCycD1CD19CREBimfl/fl mice to monthly IP injections of pristane (400 mg) for 3 months. Because, in the prior study, pristane did not produce lymphomas in mice younger than 9 months,22 we tested 2 cohorts, an older group aged 13 to 17 months and a younger group aged 3 to 6 months. The pristane treatment resulted in a statistically significant increase in WBC and spleen weight among young EμCycD1 mice, but not in any of the other subgroups (supplemental Figure 3A-B). This difference was mostly due to 2 young pristane-treated EμCycD1 mice, 1 of which had both an elevated WBC count and enlarged spleen. However, neither of these 2 mice developed lymphoma, as evaluated by whole body necropsy. For each subgroup, treatment with pristane resulted in increased mediastinal lymphadenopathy (supplemental Figure 3C). Histopathologic analysis of 21 EμCycD1 mice and 20 EμCycD1CD19CREBimfl/fl mice, however, identified only 2 mice of each genotype with extensive lymphoproliferation, which manifested as multiple prominent gastrointestinal-associated B-cell lymphoid nodules (supplemental Figure 4). Notably, cyclin D1 expression was not a prominent feature of these proliferations, as documented by cyclin D1 immunohistochemistry.
Upregulation of mantle cells in EμCycD1CD19CREBimfl/fl mice upon SRBC injection
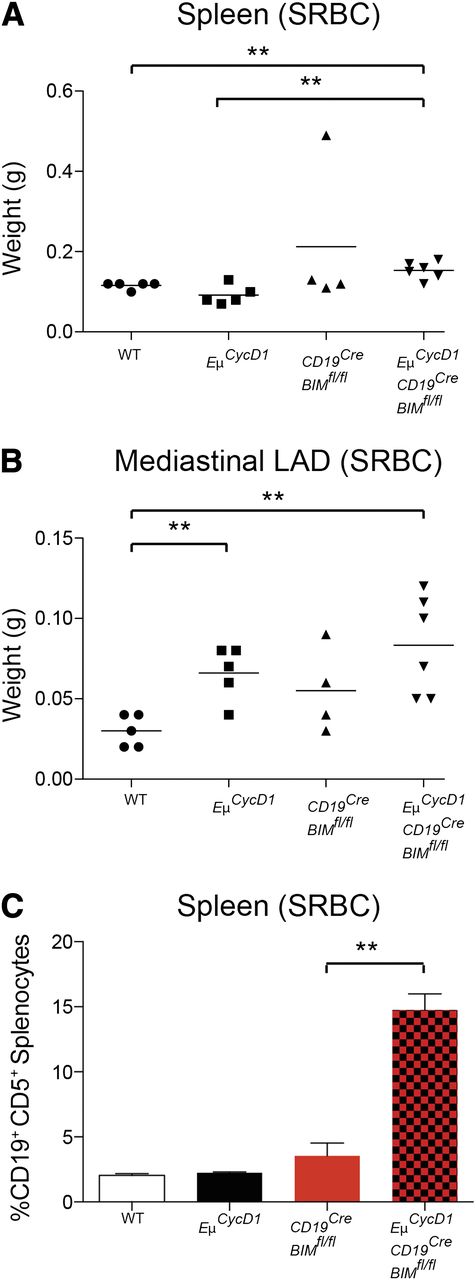
Polyclonal antigenic stimulation by immunization with SRBC stimulates germinal center formation and plasma cell differentiation, with a concomitant increase in apoptosis of B cells undergoing negative selection during the process of B-cell maturation.32 To determine if SRBC stimulation could induce MCL in the context of a Bim-null–mediated apoptotic blockade and cyclin D1–dependent proliferative drive, we subjected WT, EμCycD1, CD19CREBimfl/fl, and EμCycD1CD19CREBimfl/fl mice to 2 IP injections of SRBC (0.3 mL of a 3.3% suspension), administered 2 months apart, and then killed the mice for histopathologic evaluation 2 months after the second injection. As expected, all mice injected with SRBC manifested induction of plasma cells by 12 days after the first injection. By 2 months after the second injection, EμCycD1CD19CREBimfl/fl mice showed a small but statistically significant increase in spleen weight compared with WT and EμCycD1 mice, whereas both the EμCycD1CD19CREBimfl/fl and EμCycD1 mice displayed a statistically significant increase in mediastinal lymphadenopathy compared with WT mice (Figure 3A-B). Immunohistochemical staining of spleens from SRBC-injected mice of each genotype revealed a similar, focal pattern of cyclin D1–positive cells aligned along the mantle zone (supplemental Figure 5). However, flow cytometry detected a dramatic increase in CD5-positive B cells in the spleens of SRBC-treated EμCycD1CD19CREBimfl/fl mice, with no elevations observed in WT, EμCycD1, or CD19CREBimfl/fl mice (Figure 3C). Whereas under homeostatic conditions the CD5-positive mantle B-cell population comprises a very small subset of total B cells, clonal expansion of B cells that express CD5 is characteristic of MCL and chronic lymphocytic leukemia.1
CD5+B-cell expansions in the spleens of EμCycD1CD19CREBimfl/flmice subjected to SRBC injection. Mice of the indicated genotypes were injected with SRBC and evaluated by gross examination and flow cytometry. (A) EμCycD1CD19CREBimfl/fl mice developed larger spleens than WT and EμCycD1 mice, but all other groups were statistically similar. (B) EμCycD1 and EμCycD1CD19CREBimfl/fl mice manifested larger mediastinal lymphadenopathy (LAD) compared with WT mice; no other statistically significant differences were observed among the other groups. (C) Flow cytometry determined the percentage of splenic mononuclear cells that were positive for CD5 and either CD19 or B220. WT (n = 5), EμCycD1 (n = 5), CD19CREBimfl/fl (n = 4), and EμCycD1CD19CREBimfl/fl (n = 5). Data are mean and standard error of the mean. *P < .05; **P < .01; ***P < .001; 2-tailed Student t test.
CD5+B-cell expansions in the spleens of EμCycD1CD19CREBimfl/flmice subjected to SRBC injection. Mice of the indicated genotypes were injected with SRBC and evaluated by gross examination and flow cytometry. (A) EμCycD1CD19CREBimfl/fl mice developed larger spleens than WT and EμCycD1 mice, but all other groups were statistically similar. (B) EμCycD1 and EμCycD1CD19CREBimfl/fl mice manifested larger mediastinal lymphadenopathy (LAD) compared with WT mice; no other statistically significant differences were observed among the other groups. (C) Flow cytometry determined the percentage of splenic mononuclear cells that were positive for CD5 and either CD19 or B220. WT (n = 5), EμCycD1 (n = 5), CD19CREBimfl/fl (n = 4), and EμCycD1CD19CREBimfl/fl (n = 5). Data are mean and standard error of the mean. *P < .05; **P < .01; ***P < .001; 2-tailed Student t test.
Selective induction of MCL in EμCycD1CD19CREBimfl/fl mice
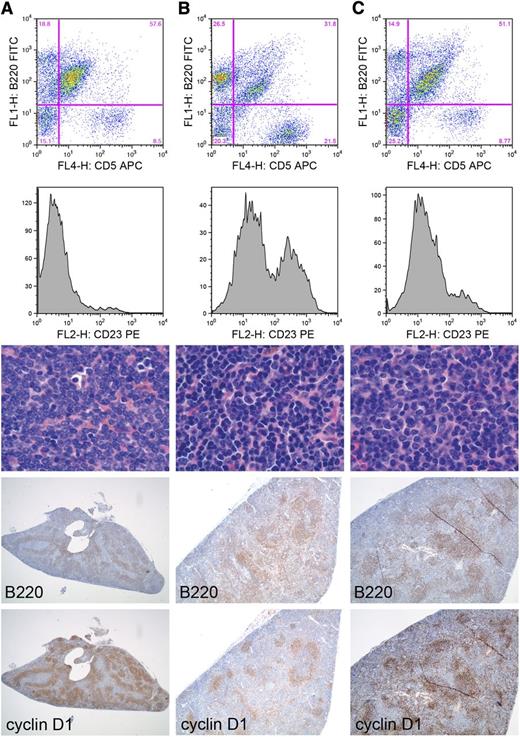
To determine if the observed selective expansion of CD5-positive B cells upon SRBC injection of EμCycD1CD19CREBimfl/fl mice could lead to MCL, we subjected the mice to combined SRBC and pristane treatments. Nine EμCycD1CD19CREBimfl/fl mice aged 13 to 15 months were treated with a single IP injection of SRBC followed by pristane injection 1 week later. Three of 9 injected mice (33%) developed lymphoma by the 4-month posttreatment evaluation time point (Figure 4). All 3 animals contained an extensive population of cyclin D1–positive B cells in the spleen, as demonstrated by immunohistochemistry. Of the B220-positive splenocytes analyzed by flow cytometry, 54% to 78% were also CD5-positive. As with human MCL, the B220+CD5+ B cells were mostly negative for CD23, a germinal center B-cell marker absent in resting cells of the surrounding mantle zone. Large, infiltrating populations of cyclin D1–positive B cells were also detected in the lymph nodes of 2 mice, the lung of 1 mouse, and the gastrointestinal tract of another, highlighting the systemic involvement of the disease (supplemental Figure 6). In all cases, the lymphoma comprised small-sized, mature lymphocytes with condensed chromatin and indistinct nucleoli, reflecting the characteristic morphology of common variant MCL (supplemental Figure 6).
Development of MCL in EμCycD1CD19CREBimfl/flmice treated with SRBC and pristane. (A-C) Disease manifestations of distinct EμCycD1CD19CREBimfl/fl mice. First row: Flow cytometry of splenic mononuclear cells (y-axis B220, x-axis CD5). Second row: Flow cytometric analysis of the B220+CD5+ cells for expression of CD23, plotted as a histogram. Third row: Hematoxylin and eosin staining of spleens (original magnification: ×1000). Fourth row: B220 immunohistochemistry (original magnification, left to right: ×20, ×40, ×40). Fifth row: cyclin D1 immunohistochemistry (original magnification, left to right: ×20, ×40, ×40). APC, allophycocyanin; FITC, fluorescein isothiocyanate; PE, phycoerythrin.
Development of MCL in EμCycD1CD19CREBimfl/flmice treated with SRBC and pristane. (A-C) Disease manifestations of distinct EμCycD1CD19CREBimfl/fl mice. First row: Flow cytometry of splenic mononuclear cells (y-axis B220, x-axis CD5). Second row: Flow cytometric analysis of the B220+CD5+ cells for expression of CD23, plotted as a histogram. Third row: Hematoxylin and eosin staining of spleens (original magnification: ×1000). Fourth row: B220 immunohistochemistry (original magnification, left to right: ×20, ×40, ×40). Fifth row: cyclin D1 immunohistochemistry (original magnification, left to right: ×20, ×40, ×40). APC, allophycocyanin; FITC, fluorescein isothiocyanate; PE, phycoerythrin.
Importantly, the use of pristane as a co-stimulus was not essential, as similar findings were elicited in EμCycD1CD19CREBimfl/fl mice treated with single-dose SRBC injection followed 1 week later by 400 cGy irradiation. Three of 9 mice in this cohort also developed the characteristic features of MCL described previously by 4 months posttreatment (Figure 5A). Even without pristane or irradiation co-stimuli, an intensive weekly SRBC injection regimen for 4 months induced MCL features in 3 of 6 EμCycD1CD19CREBimfl/fl mice (Figure 5B). These alternate stimuli also resulted in systemic involvement by MCL as shown in the liver (Figure 5C), lymph node (Figure 5D), and gastrointestinal tract (Figure 5E). As in human MCL, EμCycD1CD19CREBimfl/fl mice that developed MCL after SRBC exposure manifested a significant increase in immunoglobulin M–positive B cells (supplemental Figure 7). Comparing EμCycD1CD19CREBimfl/fl mice stimulated with SRBC and irradiation revealed that only the diseased animal bearing the aberrant CD5+CD23− B-cell population was clonal for an immunoglobulin heavy chain gene rearrangement (Figure 6). In each case, the lymphomas that arose in the differentially treated cohorts of EμCycD1CD19CREBimfl/fl mice were diffusely positive for B220 and cyclin D1 by immunohistochemistry, CD5 positive and typically CD23 negative by flow cytometry, and exhibited the characteristic histopathologic appearance of MCL (Figures 4 and 5).
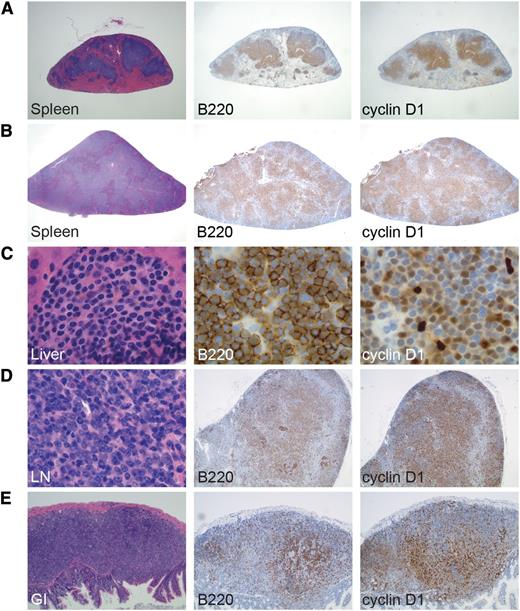
Development of MCL in EμCycD1CD19CREBimfl/flmice exposed to SRBC/irradiation or weekly SRBC. Spleens from mice treated with (A) SRBC and irradiation or (B) weekly SRBC show diffuse infiltration by cyclin D1–positive B cells (original magnification: ×20). Cyclin D1–positive B cells are shown invading (C) the liver of a mouse treated with weekly SRBC injections (original magnification: ×1000), (D) the lymph node (LN) of a mouse treated with SRBC and irradiation (original magnification: ×1000, ×40, ×40), and (E) the gastrointestinal (GI) tract of a mouse treated with SRBC and irradiation (original magnification, ×100).
Development of MCL in EμCycD1CD19CREBimfl/flmice exposed to SRBC/irradiation or weekly SRBC. Spleens from mice treated with (A) SRBC and irradiation or (B) weekly SRBC show diffuse infiltration by cyclin D1–positive B cells (original magnification: ×20). Cyclin D1–positive B cells are shown invading (C) the liver of a mouse treated with weekly SRBC injections (original magnification: ×1000), (D) the lymph node (LN) of a mouse treated with SRBC and irradiation (original magnification: ×1000, ×40, ×40), and (E) the gastrointestinal (GI) tract of a mouse treated with SRBC and irradiation (original magnification, ×100).
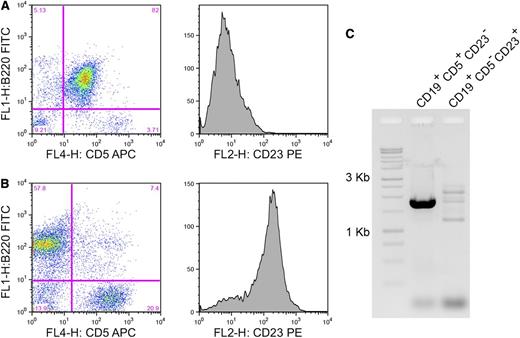
Clonality of splenic tissue from EμCycD1CD19CREBimfl/flmice. Spleens from 2 exemplary EμCycD1CD19CREBimfl/fl mice, (A) 1 that developed MCL and (B) 1 that did not are compared for CD5 expression, CD23 expression, and (C) immunoglobulin H clonality by framework 3 PCR.
Clonality of splenic tissue from EμCycD1CD19CREBimfl/flmice. Spleens from 2 exemplary EμCycD1CD19CREBimfl/fl mice, (A) 1 that developed MCL and (B) 1 that did not are compared for CD5 expression, CD23 expression, and (C) immunoglobulin H clonality by framework 3 PCR.
Cell-cycle activation in EμCycD1CD19CREBimfl/fl mice that develop MCL
Immunohistochemical and microarray gene expression analyses were used to further characterize the MCLs that arose in the immunostimulated EμCycD1CD19CREBimfl/fl mice. Elevated Ki-67 proliferation indices were observed in EμCycD1CD19CREBimfl/fl mice that developed MCL upon immunostimulation (Figure 7A; supplemental Figure 8B). Scattered cellular positivity for the phosphorylated form of the retinoblastoma protein, a marker of cyclin D1 activity, was also observed in all mice that developed MCL (Figure 7A; supplemental Figure 8C). As expected, BIM was not expressed in the lymphocytes of stimulated EμCycD1CD19CREBimfl/fl mice that developed MCL, with only scattered myeloid cells showing BIM immunoreactivity (Figure 7A). The fidelity of the BIM antibody was further confirmed by the presence of immunoreactivity in lymphoid tissues from WT but not globally deleted Bim−/− mice (supplemental Figure 8A).
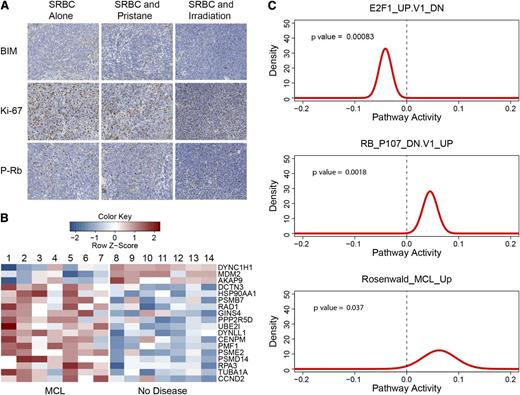
Cell-cycle analysis of EμCycD1CD19CREBimfl/flmice that developed MCL. (A) Spleens from mice treated with weekly SRBC alone (first column), SRBC and pristane (second column), or SRBC and irradiation (third column) all show absence of BIM expression (first row), high Ki-67 staining (second row), and scattered phosphorylated-retinoblastoma protein expression (third row). (B) Gene expression profiling was performed on spleens from EμCycD1CD19CREBimfl/fl mice stimulated with SRBC alone (2, 5, and 6), SRBC with irradiation (1, 3, and 7), or SRBC with pristane (4) that developed MCL (n = 7) and compared with EμCycD1CD19CREBimfl/fl mice stimulated with SRBC alone (8 and 9), SRBC with irradiation (12-14), or SRBC with pristane (10 and 11) that did not develop disease (n = 7). Shown are gene expression intensities (row normalized) of cell cycle–related genes that manifested statistically significant differential regulation. (C) Probability densities for gene set activity estimated by QuSAGE of MCL-bearing vs nondiseased EμCycD1CD19CREBimfl/fl mouse spleens for genes downregulated in E2F1 overexpressing cells (top), genes upregulated in retinoblastoma protein–deficient cells (middle), and genes that comprise a human MCL signature (bottom).
Cell-cycle analysis of EμCycD1CD19CREBimfl/flmice that developed MCL. (A) Spleens from mice treated with weekly SRBC alone (first column), SRBC and pristane (second column), or SRBC and irradiation (third column) all show absence of BIM expression (first row), high Ki-67 staining (second row), and scattered phosphorylated-retinoblastoma protein expression (third row). (B) Gene expression profiling was performed on spleens from EμCycD1CD19CREBimfl/fl mice stimulated with SRBC alone (2, 5, and 6), SRBC with irradiation (1, 3, and 7), or SRBC with pristane (4) that developed MCL (n = 7) and compared with EμCycD1CD19CREBimfl/fl mice stimulated with SRBC alone (8 and 9), SRBC with irradiation (12-14), or SRBC with pristane (10 and 11) that did not develop disease (n = 7). Shown are gene expression intensities (row normalized) of cell cycle–related genes that manifested statistically significant differential regulation. (C) Probability densities for gene set activity estimated by QuSAGE of MCL-bearing vs nondiseased EμCycD1CD19CREBimfl/fl mouse spleens for genes downregulated in E2F1 overexpressing cells (top), genes upregulated in retinoblastoma protein–deficient cells (middle), and genes that comprise a human MCL signature (bottom).
Principal component analysis of microarray gene expression profiles among the immunostimulated EμCycD1CD19CREBimfl/fl cohorts also revealed a clear distinction between EμCycD1CD19CREBimfl/fl mice that did and did not develop MCL, regardless of which immunostimulatory regimen they received (supplemental Figure 9A). QuSAGE analysis27 on REACTOME gene sets revealed that tissues harboring MCL were enriched for numerous cell-cycle gene sets compared with the corresponding tissues from the unaffected cohort, with the most significant set being REACTOME_CELL_CYCLE (supplemental Figure 9B). These expression profiling results are consistent with the immunohistochemical findings. Eighteen cell-cycle regulatory genes from the REACTOME_CELL_CYCLE gene set were differentially expressed between EμCycD1CD19CREBimfl/fl mice that did and did not develop MCL (Figure 7B). Of note, cyclin D2 was included in this group, whereas cyclin D3, CDK2, CDK4, and CDK6 did not show statistically different expression between the diseased and nondiseased mice. Moreover, activation of cyclin D1 in MCL-bearing mice would be predicted to manifest phosphorylation and inactivation of the retinoblastoma protein and activation of E2F.33-35 Indeed, based on QuSAGE analysis on MSigDB C6 oncogenic gene signatures, mice that developed MCL manifested (1) upregulation of a gene set that is turned on when retinoblastoma protein is inactivated (false discovery rate = 0.007), and (2) downregulation of a gene set that is turned off when E2F1 is overexpressed (false discovery rate = 0.005) (Figure 7C). Most striking, a gene signature defined for human MCL by Rosenwald et al28 was enriched in the disease-bearing mice (Figure 7C). The 25 human genes within this signature that could be mapped in the mouse were sufficient to differentiate the diseased vs nondiseased EμCycD1CD19CREBimfl/fl mice by principal component analysis (supplemental Figure 9C). Differences in BCL-2 family member expression among the affected vs unaffected cohorts of EμCycD1CD19CREBimfl/fl mice were not seen (supplemental Figure 9D). Thus, we find that the lymphoma arising in immunostimulated EμCycD1CD19CREBimfl/fl mice not only manifests the histopathologic and immunophenotypic hallmarks of classical MCL, but also bears the genetic signatures of cell-cycle activation and human MCL.
Discussion
In normal lymph nodes, naive B cells surround the germinal center to form a small rim known as the mantle zone.36 As these cells enter the germinal center, they are exposed to antigen, expand rapidly, and then the majority of them die by apoptosis during the process of negative selection.36 The majority of MCL cases are thought to derive from naive B cells in the mantle zone, based on the presence of a restricted immunoglobulin variable heavy chain,37 evidence of somatic mutations,37 and genetic profiling.38 Furthermore, the restricted use of variable heavy chains implies that the malignant transformation may be antigen-driven.37 Within these aberrant cells, a translocation places cyclin D1 under the regulation of the immunoglobulin heavy chain locus, resulting in the overexpression of this cell-cycle proliferation gene in B cells. However, the proliferative drive alone may be insufficient to produce MCL, because generating this translocation in mouse models fails to induce the disease. Among the numerous secondary mutations that have been identified in MCL, frequent biallelic deletion of the proapoptotic BH3-only protein BIM5,6 suggests that antigenic stimulation in the context of both an apoptotic blockade (Bim loss) and proliferative drive (cyclin D1 overexpression) could effectively collaborate to produce MCL (supplemental Figure 10).
Biochemical, genetic, and clinical evidence have implicated the BH3-only protein BIM as a candidate tumor suppressor, with loss of Bim expression observed in a wide variety of human cancers.5,39-42 Indeed, this central BCL-2 family regulator of apoptosis would be predicted to have robust tumor suppressor activity based on its breadth of protein interactions. BIM not only binds and inhibits all known antiapoptotic proteins with high affinity,43,44 but also directly interacts with and activates the proapoptotic effectors BAX and BAK.45-47 Biallelic deletion of Bim has been reported in up to 40% of MCLs.6 A second study confirmed the frequent deletion of Bim in MCL and extended the analysis to reveal silencing of the Bim promoter through hypermethylation of CpG islands in 50% to 80% of Burkitt lymphomas.5 Nearly 80% of renal cell carcinomas show loss of Bim expression, which is associated with metastasis and B symptoms.42 Reduced Bim expression in melanoma correlates with poor 5-year survival,39 whereas elevated Bim expression in colorectal cancer is associated with more favorable disease-free and overall survival.41 Finally, BIM has recently been shown to play an essential role in tyrosine kinase inhibitor–induced apoptosis. For example, inhibition of the MEK-ERK pathway relies on BIM induction for apoptosis of EGFR-mutant non–small cell lung cancer (NSCLC),48 HER2-amplified breast cancer,48,49 and BRAF-mutant colorectal cancer and melanoma.50 Therefore, high Bim expression is associated with longer progression-free survival in EGFR-mutant NSCLC and HER2-positive breast cancer patients treated with tyrosine kinase inhibitors.48 Conversely, patients with a germline polymorphism in Bim that deletes its BH3 domain exhibit relative tyrosine kinase inhibitor resistance in chronic myelogenous leukemia as well as EGFR-positive NSCLC.40
Bim-null mouse models have yielded a mixture of autoimmune and cancer phenotypes. A subset of Bim−/− mice dies during the embryonic period, whereas the remaining mice develop a progressive lymphadenopathy and a fatal systemic autoimmune disease.31 Similarly, transgenic overexpression of the miR-17-92 microRNA cluster, which decreases Bim expression, results in lymphoproliferative disease with autoimmune pathology.51 Hematopoietic tumors, including diffuse large B-cell lymphoma and histiocytic sarcomas, have been reported in 20% of Bim−/− and 60% of Bim−/−Puma−/− mice.52 In addition, heterozygous loss of Bim potentiates Myc-driven B-cell leukemias,53 whereas homozygous loss of Bim potentiates Ras-driven subcutaneous growth of immortalized epithelial cells.54
Consistent with the genomic analyses of human MCL, here we find that a murine lymphoma exhibiting the key genetic, phenotypic, and morphologic characteristics of human MCL can be elicited in cyclin D1 transgenic mice bearing Bim-deleted B cells. Even within a germ-free environment, rare EμCycD1CD19CREBimfl/fl mice develop disease with striking similarity to MCL. To drive antigenic stimulation, we treated mice with either SRBC or pristane. Although pristane was previously reported to induce MCL in older EμCycD1 mice, we did not observe this outcome in our colony, likely because of different transgene expression levels and/or background strain of mice. In contrast, 2 SRBC injections led to a striking expansion of mantle cells only in EμCycD1CD19CREBimfl/fl mice, as reflected by B cells coexpressing CD5. Indeed, the expansion of this cell population within the spleen may represent incipient disease, much like the CD5 expansion seen in monoclonal B-cell lymphocytosis before the development of chronic lymphocytic leukemia.55 More intensive stimulation with either serial SRBC injections alone or a single SRBC injection complemented by pristane exposure or irradiation produced murine lymphomas in a subset of mice with classic characteristics of common variant human MCL.
The elicited lymphomas in EμCycD1CD19CREBimfl/fl mice may represent a bona fide model of MCL, reflecting the pathologic consequences of established genetic lesions in the human disease. In contrast to other models with different genetic backgrounds that generated lymphomas with blastoid features, the disease that arose in EμCycD1CD19CREBimfl/fl mice has the classic morphologic appearance of human MCL. Thus, this model may be especially useful for studying mantle cell biology, the progression to MCL, and the response to experimental therapeutics. Indeed, the pathogenic role of Bim loss in this model highlights the potential pharmacologic utility of BIM mimetics that “replace” the critical function of the BIM BH3 domain. Early studies using the BH3-mimetic ABT-737 and ABT-263 demonstrated efficacy in cultured MCL cells,10,56,57 an MCL xenograft model,57 and in a cyclin D1–driven mouse lymphoma model.12 Given the potential importance of the BIM-BCL-2 complex, rather than BCL-2 expression alone,58 in mediating the indirect activation of apoptosis by such BCL-2 inhibitors, the efficacy of this mechanism may be thwarted in the context of Bim-null cancers. Indeed, beyond MCL, the loss of Bim or its expression occurs in Burkitt lymphoma,5 renal cell carcinoma,42 melanoma,39 colorectal cancer,41 and tyrosine kinase inhibitor–resistant chronic myelogenous leukemia and EGFR–positive NSCLC.40 Taken together, these findings underscore the importance of advancing therapeutics that can replace the lost functionality of the BIM protein, such as stapled BIM BH3 peptides16 or other direct activators of the proapoptotic effectors BAX59 and BAK.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The Specialized Histopathology Core of the Dana-Farber/Harvard Cancer Center is funded in part by a National Cancer Institute Cancer Center Support Grant (National Institutes of Health 5 P30CA06516). This work was supported by a Leukemia and Lymphoma Society (LLS) Fellow Award and a Lymphoma Research Foundation Mantle Cell Lymphoma grant to S.G.K. and an LLS/Marshall A. Lichtman Specialized Center of Research project grant to L.D.W.
Authorship
Contribution: S.G.K. designed and performed experiments, analyzed results, generated the figures and wrote the paper; J.L.L. designed and performed experiments and analyzed results; H.M. analyzed results and generated figures; R.P.V and J.K.F. generated mice and performed experiments; H.S. performed experiments; S.J.R. analyzed results; S.H.K. analyzed results; and L.D.W. designed experiments, analyzed results, and wrote the paper.
Conflict-of-interest disclosure: L.D.W. is a consultant and scientific advisory board member for Aileron Therapeutics. The remaining authors declare no competing financial interests.
Correspondence: Loren D. Walensky, Dana-Farber Cancer Institute, 450 Brookline Ave, Mayer 664, Boston, MA 02215; e-mail: loren_walensky@dfci.harvard.edu.