Key Points
Integrated genomic profiling identifies high-risk adult T-ALL patients with poor response to intensified chemotherapy.
Abstract
Adult T-cell acute lymphoblastic leukemia (T-ALL) is an aggressive hematologic tumor associated with poor outcome. In this study, we analyzed the prognostic relevance of genetic alterations, immunophenotypic markers, and microarray gene expression signatures in a panel of 53 adult T-ALL patients treated in the Eastern Cooperative Oncology Group E2993 clinical trial. An early immature gene expression signature, the absence of bi-allelic TCRG deletion, CD13 surface expression, heterozygous deletions of the short arm of chromosome 17, and mutations in IDH1/IDH2 and DNMT3A genes are associated with poor prognosis in this series. In contrast, expression of CD8 or CD62L, homozygous deletion of CDKN2A/CDKN2B, NOTCH1 and/or FBXW7 mutations, and mutations or deletions in the BCL11B tumor suppressor gene were associated with improved overall survival. Importantly, the prognostic relevance of CD13 expression and homozygous CDKN2A/CDKN2B deletions was restricted to cortical and mature T-ALLs. Conversely, mutations in IDH1/IDH2 and DNMT3A were specifically associated with poor outcome in early immature adult T-ALLs. This trial was registered at www.clinicaltrials.gov as #NCT00002514.
Introduction
T-cell acute lymphoblastic leukemias (T-ALLs) account for 25% of adult ALL cases.1 Clinically, T-ALL is characteristically 3 times more frequent in men than women and typically presents with hematopoietic failure resulting from bone marrow infiltration by an immature lymphoblast with a T-cell immunophenotype and high white blood cell counts. In addition, T-ALL patients frequently show mediastinal masses and leukemic infiltration of the central nervous system at diagnosis. T-ALL arises as result of a multistep oncogenic process in which different genetic alterations induce aberrant cell proliferation, increased cell survival, and blocked differentiation of immature T-cell progenitors. In this context, deletion of the CDKN2A locus encompassing the p16/INK4A and p14/ARF suppressor genes in chromosome band 9p21 is the most frequent genetic alteration in T-ALL, present in >70% of all T-ALL cases.1,2 In addition, constitutive activation of NOTCH1 signaling as a result of activating mutations in the NOTCH1 gene and mutations in the FBXW7 tumor suppressor are found in 50% to 60% of T-ALLs.3 Moreover, and most uniquely, T-ALLs also show aberrant expression of oncogenic transcription factors as a result of chromosomal translocations involving strong regulatory elements associated with the TCR genes. T-ALL transcription factor oncogenes include basic helix-loop-helix factors such as TAL1; LIM-only domain (LMO) genes such as LMO1 and LMO2; the TLX1/HOX11, TLX3/HOX11L2, and HOXA homeobox genes; MYC; and MYB and TAN1, a truncated and constitutively activated form of the NOTCH1 receptor.4 Furthermore, recent studies have uncovered a growing number of mutations and deletions disrupting the function of key transcriptional regulators such as WT1, LEF1, BCL11B, GATA3, RUNX1, and ETV6 and epigenetic factors including EP300, PHF6, SETD2, EZH2, EED, and SUZ12.4 The mutation landscape of T-ALL also includes chromosomal rearrangements and mutations driving increased proliferation, including translocations involving CCND2 and NUP214-ABL1 and mutations in IL7R, FLT3, PTEN, and NRAS.4 Finally, the genetic landscape of T-ALL is completed with mutations in genes such as the ribosomal protein RPL105 and the dynamin DNM2 gene,6 whose functional role in T-cell transformation is yet to be fully understood.
Still, and despite this diversity of molecular and cytogenetic lesions, microarray gene expression studies have shown that T-ALLs can be classified in a limited number of transcriptional groups, which are related to the activation of specific transcription factor oncogenes and show distinct gene expression signatures related to their arrest at different stages of T-cell differentiation.1,7-9 Thus, early immature T-ALLs, which are arrested early at the double-negative stage of thymocyte development,1,10,11 show a transcriptional program related to hematopoietic stem cells and myeloid progenitors,6,11 higher levels of LYL1 and LMO2 expression,1 and mutations in acute myeloid leukemia genes,6,11 together with inactivation of important transcription factors such as RUNX1, GATA3, and ETV6.6,11 Notably, these early immature tumors frequently show the absence of bi-allelic TCRG deletions12 and are associated with very poor prognosis.10,12 In contrast, CD1a+ T-ALLs show an early cortical gene expression signature, frequent activation of TLX1, and a favorable prognosis.1,13 Finally, leukemias with aberrant expression of TAL1 together with LMO1 and LMO2 are characterized by a gene expression program related to those of late cortical thymocytes.1
To gain insight in the clinical relevance of different genetic alterations and oncogenic pathways involved in T-cell transformation, we analyzed the prognostic significance of transcriptional, cytogenetic, immunophenotypic, and molecular features of T-ALLs treated in the Eastern Cooperative Oncology Group (ECOG) E2993 clinical trial.14
Methods
Patient samples
Bone marrow lymphoblast samples from 53 T-ALL patients treated in the E2993 ECOG clinical trial14 were included in this study. All samples were collected with informed consent at trial entry according to the declaration of Helsinki and analyzed under the supervision of the Columbia University Medical Center Institutional Review Board.
Flow cytometry
Immunophenotypic analysis was performed at the ECOG leukemia reference laboratory as previously described14 (supplemental Table 1 on the Blood website). Briefly, lymphoblasts were gated by 3-color flow cytometry based on antigen expression of the leukemic cells. In addition to cytoplasmic CD3 (cCD3), which established the diagnosis of T-ALL, T-lymphoid–affiliated antigens tested included CD1a, CD2, surface CD3, CD4, CD5, CD8, CD62L, CD57, and surface α/β and γ/δ. The intensity of CD5 staining was determined as mean fluorescent intensity as previously described.15 Myeloid antigens included myeloperoxidase, CD117, CD33, CD13, CD65(s), CD15(s), CD11b, and CD14 and B-lymphoid–affiliated antigens included CD19 and CD10. Finally, uncommitted antigens included CD45, CD34, HLA-DR, and TdT.
Microarray gene expression profiling of primary adult T-ALL samples
RNA was isolated using the RNeasy plus mini kit (Qiagen) according to the manufacturer's protocol. Next, 500 ng of RNA was amplified and biotinylated using the TotalPrep RNA Amplification Kit (Ambion) and hybridized to the HumanHT-12 v4 Expression BeadChip (Illumina). Gene expression profiling data were normalized using log2 transformation and quantile normalization. Unsupervised consensus clustering was performed using the GenePattern application.16 Enrichment of the gene set associated with early T-cell precursor (ETP) T-ALL was analyzed by Gene Set Enrichment Analysis using the Student t test metric and 10 000 permutations of the genes. Microarray gene expression data are available in Gene Expression Omnibus (GEO accession code GSE42328).
Microarray-based array-CGH
Comparative genomic hybridization (array-CGH) analysis was performed using the SurePrint G3 Human 1 × 1M oligonucleotide array platform according to the manufacturer’s instructions (Agilent). Slides were scanned in a 2565AA DNA microarray scanner (Agilent). Microarray images were analyzed using feature extraction software (Agilent), and the data were subsequently imported into array-CGH analytics software (Agilent).
Mutation analysis
Exon sequences from NOTCH1, FBXW7, PTEN, IL7R, DNM2, PHF6, BCL11B, WT1, EZH2, ETV6, IDH1, IDH2, FLT3, NRAS, DNMT3A, GATA3, TP53, and RUNX1 were amplified from genomic DNA by polymerase chain reaction and analyzed by direct dideoxynucleotide sequencing. Mutational hotspot regions were sequenced for NRAS, PTEN, FLT3, DNMT3A, IDH1, IDH2, NOTCH1, IL7R, and FBXW7. Primer sequences used for FLT3,17 DNMT3A,18 IDH1,19 IDH2,19 NOTCH1,3 IL7R,20 FBXW7,21 DNM2,6 PHF6,22 BCL11B,23 WT1,24 EZH2,25 ETV6,11 GATA3,6 RUNX1,26 TP53,27 and PTEN28 have been previously described.
Statistical analyses
Survival time was measured from the date of randomization to date of death for patients who died and to the date of the last follow-up for those who were alive at the time of the analysis. The prognostic value of the different covariates was evaluated with the use of the Kaplan-Meier estimate of survival function. Differences between the survival functions were assessed with the log-rank test. The covariates included transcriptional gene signature status (early immature vs typical/mature), genetic characteristics (gene mutations and genomic deletions/amplifications), and expression of immunophenotypic markers. Any antibody binding by a distinct subset of gated T-lymphoblasts was considered a positive finding. Weak intensity of antibody binding by the entire blast cell population above background staining was interpreted as positivity of all blasts for the respective antigen, albeit at low density.
Multivariate survival analyses were performed with the Cox proportional-hazards model. Covariates were selected using the least absolute shrinkage and selection operator (lasso)29 as this approach is particularly well suited when the number of explanatory variables is high with respect to the sample size. We checked the proportional-hazards assumption by testing for a nonzero slope in a regression of the scaled Schoenfeld residuals on functions of time. The Cox model was cross-validated using the leave-one-out cross-validation method proposed by Verweij and Van Houwelingen.30 The variables included in the final model appeared in >90% of the leave-one-out lasso–selected sets. We used the libraries survival and glmnet in R2.15.0. All analyses were performed with the use of Stata version 11.0 (www.stata.com) and R statistical software 2.15.0 (www.r-project.org).
Results
Prognostic value of transcriptionally defined early immature and cortical-mature adult T-ALL groups
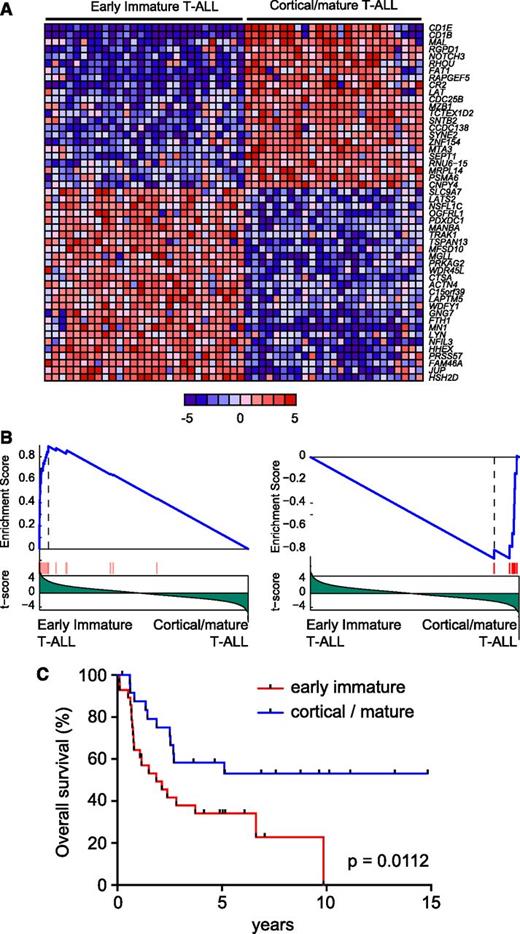
To analyze the prognostic relevance of molecular groups defined by gene expression profiling in adult T-ALL, we analyzed a series of 53 primary leukemia samples using gene expression oligonucleotide microarrays. As previously described,11 unsupervised clustering analysis of microarray gene expression data in this series revealed the presence of 2 robust clusters of samples (Figure 1A). The first of these clusters encompassed 28 samples and corresponded to early immature leukemias (Figure 1A-B), a group characterized by a gene expression signature related to that of hematopoietic stem cells and myeloid progenitors.11 In contrast, the second of these clusters, referred here as cortical/mature leukemias, contained 28 samples (Figure 1A) whose gene expression signatures were related to those of cortical and mature thymocytes11 (Figure 1B). Notably, univariate analysis in our patient series showed that early immature adult T-ALLs are associated with poor prognosis and reduced overall survival compared with cortical/mature adult T-ALL (5-year survival: early immature 34% [95% confidence interval: 17%-52%] vs cortical/mature 62% [95% confidence interval: 40%-78%]; P = .0112; Figure 1C).
Prognostic value of immature adult T-ALL. (A) Top 50 differentially expressed genes between early immature and cortical/mature adult T-ALL. Genes in the heat map are shown in rows, and each individual sample is shown in 1 column. The scale bar shows color-coded differential expression from the mean in standard deviation units, with red indicating higher levels and blue lower levels of expression. (B) Gene Set Enrichment Analysis of genes associated with pediatric ETP leukemias in early immature vs cortical/mature adult T-ALLs. (C) Kaplan-Meier survival curves in adult T-ALL patients treated in ECOG clinical trial ECOG2993 with early immature vs cortical/mature gene expression signatures.
Prognostic value of immature adult T-ALL. (A) Top 50 differentially expressed genes between early immature and cortical/mature adult T-ALL. Genes in the heat map are shown in rows, and each individual sample is shown in 1 column. The scale bar shows color-coded differential expression from the mean in standard deviation units, with red indicating higher levels and blue lower levels of expression. (B) Gene Set Enrichment Analysis of genes associated with pediatric ETP leukemias in early immature vs cortical/mature adult T-ALLs. (C) Kaplan-Meier survival curves in adult T-ALL patients treated in ECOG clinical trial ECOG2993 with early immature vs cortical/mature gene expression signatures.
Prognostic value of copy number alterations
Array-CGH analysis in our adult T-ALL series revealed on average 6 copy number alterations (5 deletions and 1 amplification) per sample (supplemental Tables 2 and 3). Recurrent deletions of the short arm of chromosome 9 centered in 9p22 and encompassing the CDKN2A/CDKN2B tumor suppressor genes were present in 22 samples (Figure 2A). Additional recurrent copy number aberrations included 1p33 deletions generating the SIL-TAL1 fusion transcript (n = 3); 4q25 deletions targeting LEF1 (n = 2); 4q31.3 deletions targeting the FBXW7 tumor suppressor gene (n = 3); duplications of the MYB oncogene (n = 10) at 6q23.3; 9q34 deletions generating the SET-NUP214 fusion oncogene (n = 2); 11p13 deletions targeting the WT1 tumor suppressor (n = 3); 12p13 deletions targeting the ETV6 and CDKN1B (n = 5); 13q deletions targeting RB1 (n = 9); 14q32 deletions targeting the BCL11B gene (n = 2); TP53 deletions at 17p (n = 5); and 17q11 deletions encompassing the NF1 and SUZ12 tumor suppressors (n = 3; Figure 2A; supplemental Table 3). Finally, array-CGH analysis also revealed the rearrangement status of the T-cell receptor (TCR) loci evidenced as highly recurrent deletions in chromosome bands 7p14 (TCRG), 7q34 (TCRB), and 14q11 (TCRA/D).
Prognostic value of copy number defects in adult T-ALL. (A) Human chromosomal ideograms showing the areas of genetic gain and loss identified by aCGH in adult T-ALL. Red bars represent areas of gain. Light blue bars represent areas of heterozygous copy number loss, and dark blue bars indicate homozygous deletions. (B-D) Kaplan-Meier survival curves in adult T-ALL patients (B) with or without the absence of bi-allelic deletion of the TCRG locus (ABD); (C) with or without homozygous CDKN2A/CDKN2B deletion; and (D) with or without heterozygous TP53 (17q) deletion, treated in the ECOG2993 clinical trial.
Prognostic value of copy number defects in adult T-ALL. (A) Human chromosomal ideograms showing the areas of genetic gain and loss identified by aCGH in adult T-ALL. Red bars represent areas of gain. Light blue bars represent areas of heterozygous copy number loss, and dark blue bars indicate homozygous deletions. (B-D) Kaplan-Meier survival curves in adult T-ALL patients (B) with or without the absence of bi-allelic deletion of the TCRG locus (ABD); (C) with or without homozygous CDKN2A/CDKN2B deletion; and (D) with or without heterozygous TP53 (17q) deletion, treated in the ECOG2993 clinical trial.
Pediatric T-ALL patients with the absence of the bi-allelic TCRG deletion (ABD), a feature linked with developmental arrest at the earliest stages of thymocyte development, exhibit early induction failure and inferior overall survival rates.12 Array-CGH analysis of TCRG copy number in our series showed ABD of the TCRG locus in 27 of 53 (51%) adult T-ALL samples. In addition, 22 of 27 (81%) ABD adult T-ALL samples showed an early immature gene expression signature, suggesting biological overlap between the ABD and the early immature subtype in adult T-ALL. Notably, as in the case of pediatric T-ALLs, ABD adult T-ALL patients showed a worse overall survival (ABD 35% [95% confidence interval: 18%-53%] vs bi-allelic TCRG deletion 59% [95% confidence interval: 38%-76%]; P = .0217; Figure 2B). In contrast, homozygous deletion of CDKN2A/CDKN2B (n = 15) was associated with favorable outcome (5-year survival: no bi-allelic CDKN2A/CDKN2B deletion 38% [95% confidence interval: 23%-53%] vs bi-allelic CDKN2A/CDKN2B deletion 71% [95% confidence interval: 41%-88%]; P = .0119; Figure 2C), whereas heterozygous deletions of the short arm of chromosome 17 covering the TP53 tumor suppressor gene (n = 15) predicted worse clinical outcome (2-year survival: TP53 deletion 20% [95% confidence interval: 9%-58%] vs no TP53 deletion 66% [95% confidence interval: 50%-77%]; P = .0005; Figure 2D). Within these TP53-deleted leukemia samples, 4 of 5 (80%) patients were ABD positive and 3 of 5 (60%) showed an early immature gene expression signature (Figure 3).
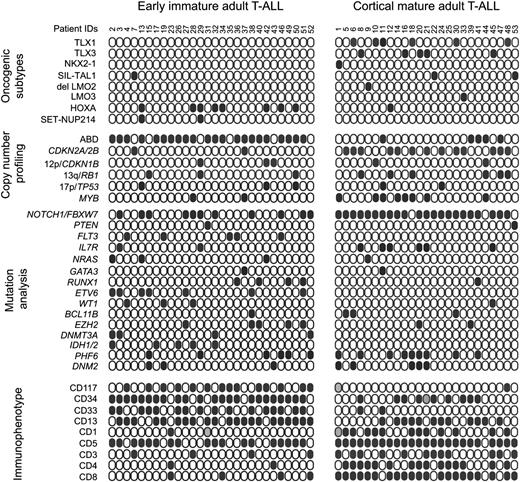
Genetic and immunophenotypic characteristics of adult T-ALL. Schematic comparison of copy number lesions, genetic mutations, and surface marker expression between early immature and mature/cortical adult T-ALL. Solid circles represent positive leukemia samples. Data were not available for leukemia patients represented by gray filled circles. T-ALL oncogenic subtypes are based on aCGH alterations and microarray expression of T-ALL transcription factor oncogenes.
Genetic and immunophenotypic characteristics of adult T-ALL. Schematic comparison of copy number lesions, genetic mutations, and surface marker expression between early immature and mature/cortical adult T-ALL. Solid circles represent positive leukemia samples. Data were not available for leukemia patients represented by gray filled circles. T-ALL oncogenic subtypes are based on aCGH alterations and microarray expression of T-ALL transcription factor oncogenes.
Prognostic value of somatic gene mutations
Mutation analysis of T-ALL oncogenes and tumor suppressor genes (Figure 3; supplemental Table 4) identified NOTCH1 and FBXW7 mutations, resulting in activation of NOTCH signaling, in 62% (33/53) of adult T-ALL patients. In addition, 49% (27/53) of adult T-ALL cases showed mutations targeting epigenetic and/or chromatin remodeling factors including DNMT3A, IDH1, IDH2, EZH2, and PHF6. Moreover, mutations targeting transcription factors including GATA3, RUNX1, WT1, BCL11B, and ETV6 were identified in 38% (20/53) of adult T-ALL patients. Finally, mutations in signaling pathways (FLT3, NRAS, IL7R, and PTEN) were found in 36% (19/53) of T-ALL patients, whereas mutations targeting the endocytosis and membrane trafficking factor gene DNM2 were present in 8 of 53 T-cell leukemias. No mutations were identified in TP53.
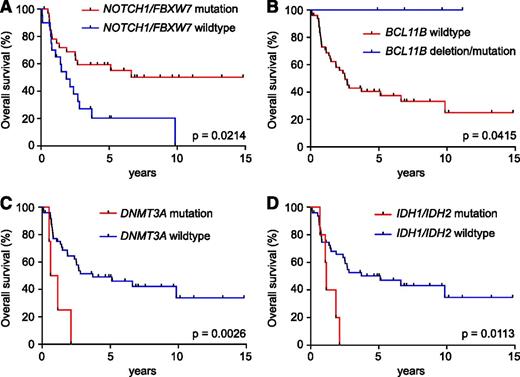
Favorable outcome was observed in adult T-ALL patient samples with NOTCH1 and/or FBXW7 mutations (5-year survival: wild type 24% [95% confidence interval: 7%-46%] vs mutant 59% [95% confidence interval: 41%-74%]; P = .0214; Figure 4A); and in cases with heterozygous inactivating mutations or deletions in the BCL11B tumor suppressor gene (5-year survival: wild type 43% [95% confidence interval: 28%-56%] vs mutation/deletion 100%; P = .0415; Figure 4B). In contrast, somatic mutations in genes targeting the epigenetic regulators DNMT3A (1-year survival: mutant 50% [95% confidence interval: 6%-84%] vs wild type 77% [95% confidence interval: 63%-87%]; P = .0026; Figure 4C) and IDH1/2 (2-year survival: mutant 20% [95% confidence interval: 8%-58%] vs wild type 66% [95% confidence interval: 50%-77%]; P = .0113; Figure 4D) were associated with worse prognosis. Consistent with previous reports,11 alterations in DNMT3A and IDH1/IDH2 were uniquely present in the early immature adult T-ALL group.
Prognostic value of somatic gene mutations in adult T-ALL. (A) Kaplan-Meier survival curve in adult T-ALLs treated in the ECOG2993 clinical trial with or without NOTCH1/FBXW7 mutations. (B) Kaplan-Meier survival curve in adult T-ALLs treated in the ECOG2993 clinical trial with or without BCL11B mutations/deletions. (C) Kaplan-Meier survival curve in adult T-ALLs treated in the ECOG2993 clinical trial with or without DNMT3A mutations. (D) Kaplan-Meier survival curves in adult T-ALLs treated in the ECOG2993 clinical trial with or without IDH1/IDH2 mutations, treated in the ECOG2993 clinical trial.
Prognostic value of somatic gene mutations in adult T-ALL. (A) Kaplan-Meier survival curve in adult T-ALLs treated in the ECOG2993 clinical trial with or without NOTCH1/FBXW7 mutations. (B) Kaplan-Meier survival curve in adult T-ALLs treated in the ECOG2993 clinical trial with or without BCL11B mutations/deletions. (C) Kaplan-Meier survival curve in adult T-ALLs treated in the ECOG2993 clinical trial with or without DNMT3A mutations. (D) Kaplan-Meier survival curves in adult T-ALLs treated in the ECOG2993 clinical trial with or without IDH1/IDH2 mutations, treated in the ECOG2993 clinical trial.
Prognostic value of cell surface markers
Immunophenotype analysis showed that our 53 adult T-ALL series encompasses 25 immature (CD4−CD8−CD1a−), 13 early cortical (CD1a+), 6 late cortical/mature (CD8+CD3+) leukemias, and 9 cases with overlapping patterns of antigen expression. Notably, leukemic blasts from 39 of 53 adult T-ALL patients (73%) showed expression of the myeloid antigens CD13 and/or CD33 (Figure 3). In addition, expression of the stem cell marker c-KIT (CD117) was identified in 15 of 53 (28%) leukemia samples (Figure 3; supplemental Table 1).
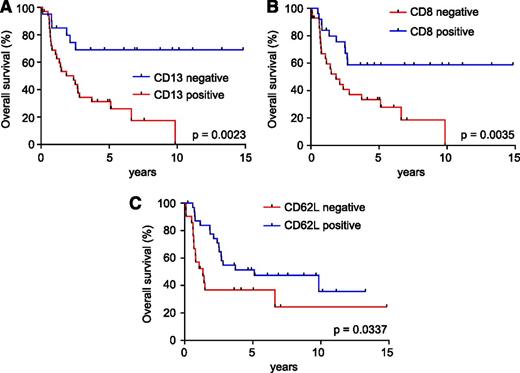
Prognosis analysis of these immunophenotypic markers revealed that expression of CD13 was associated with poor survival (5-year survival: CD13+ 34% [95% confidence interval: 18%-50%] vs CD13− 69% [95% confidence interval: 44%-85%]; P = .0023; Figure 5A), as previously described.14 In addition, expression of CD8 (5-year survival: CD8− 33% [95% confidence interval: 17%-51%] vs CD8+ 63% [95% confidence interval: 41%-79%]; P = .0035; Figure 5B) and CD62L (5-year survival: CD62L− 37% [95% confidence interval: 17%-57%] vs CD62L+ 54% [95% confidence interval: 35%-70%]; P = .0337; Figure 5C) conferred better prognosis.
Prognostic value of cell surface markers in adult T-ALL. (A) Kaplan-Meier survival curves in adult T-ALLs treated in the ECOG2993 clinical trial according to CD13 expression. (B) Kaplan-Meier survival curves in adult T-ALLs treated in the ECOG2993 clinical trial according to CD8 expression. (C) Kaplan-Meier survival curves in adult T-ALLs treated in the ECOG2993 clinical trial according to CD62L antigen expression.
Prognostic value of cell surface markers in adult T-ALL. (A) Kaplan-Meier survival curves in adult T-ALLs treated in the ECOG2993 clinical trial according to CD13 expression. (B) Kaplan-Meier survival curves in adult T-ALLs treated in the ECOG2993 clinical trial according to CD8 expression. (C) Kaplan-Meier survival curves in adult T-ALLs treated in the ECOG2993 clinical trial according to CD62L antigen expression.
Multivariate analysis
Next, multivariate Cox regression analysis revealed that CD13 and CD62L antigen expression, heterozygous 17p deletions, and mutations in NOTCH1/FBXW7 and DNMT3A are independent prognostic markers in adult T-ALL treated on the E2993 protocol (supplemental Table 5).
Risk stratification in early immature and cortical/mature adult T-cell leukemias
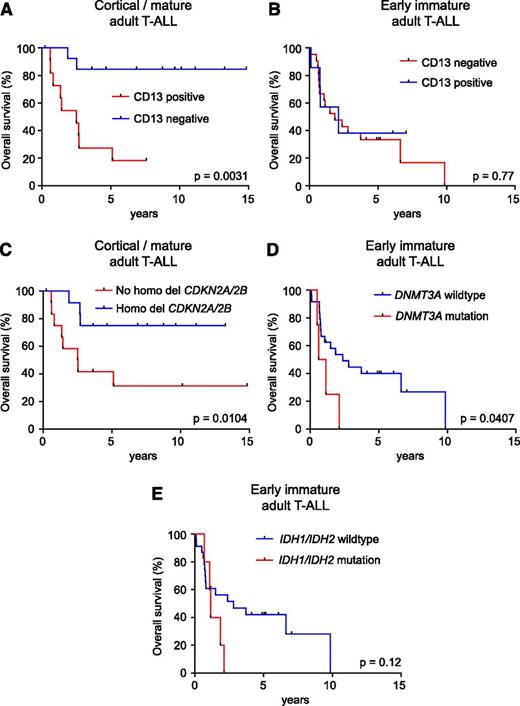
Given the broad gene expression, immunophenotypic, and genetic differences between early immature and cortical/mature adult T-cell leukemias, we tested the significance of different prognostic markers in these 2 groups. This analysis revealed that the poor prognostic effect of CD13 expression was restricted to the otherwise favorable clinical group of mature/cortical leukemias (5-year survival: CD13+ 36% [95% confidence interval: 11%-63%] vs CD13− 85% [95% confidence interval: 51%-96%]; P = .0031; Figure 6A) and was not observed in T-ALLs with an early immature gene expression signature (5-year survival: CD13+ 33% [95% confidence interval: 15%-53%] vs CD13− 38% [95% confidence interval: 6%-72%]; P = .77; Figure 6B). In addition, the overall favorable prognostic effect of homozygous CDKN2A/CDKN2B deletions was confirmed within the mature/cortical gene expression profile group (5-year survival: no bi-allelic CDKN2A/CDKN2B deletion 42% [95% confidence interval: 15%-67%] vs bi-allelic CDKN2A/CDKN2B deletion 83% [95% confidence interval: 48%-96%]; P = .0104; Figure 6C), and the overall poor prognosis of DNMT3A mutations was confirmed within the high-risk group of T-ALLs with an early immature gene expression signature (1-year survival: mutant 25% [95% confidence interval: 9%-67%] vs wild type 62% [95% confidence interval: 40%-78%]; P = .0407; Figure 6D). This is in contrast with IDH1/IDH2 mutations, which are also restricted to early immature adult T-ALLs, but only showed a trend toward worse clinical outcome (2-year survival: mutant 20% [95% confidence interval: 8%-58%] vs wild type 56% [95% confidence interval: 34%-74%]; P = .12; Figure 6E) in this group.
Risk stratification in early immature and cortical/mature adult T-ALL. (A) Kaplan-Meier survival curve of early immature adult T-ALLs treated in the ECOG2993 clinical trial according to CD13 expression. (B) Kaplan-Meier survival curve of cortical/mature adult T-ALLs treated in the ECOG2993 clinical trial according to CD13 expression. (C) Kaplan-Meier survival curve of cortical/mature adult T-ALLs treated in the ECOG2993 clinical trial according to the presence or absence of homozygous CDKN2A/CDKN2B deletion. (D) Kaplan-Meier survival curve of early immature adult T-ALLs treated in the ECOG2993 clinical trial according to the presence or absence of DNMT3A mutations. (E) Kaplan-Meier survival curve of early immature adult T-ALLs treated in the ECOG2993 clinical trial according to the presence or absence of IDH1/IDH2 mutations.
Risk stratification in early immature and cortical/mature adult T-ALL. (A) Kaplan-Meier survival curve of early immature adult T-ALLs treated in the ECOG2993 clinical trial according to CD13 expression. (B) Kaplan-Meier survival curve of cortical/mature adult T-ALLs treated in the ECOG2993 clinical trial according to CD13 expression. (C) Kaplan-Meier survival curve of cortical/mature adult T-ALLs treated in the ECOG2993 clinical trial according to the presence or absence of homozygous CDKN2A/CDKN2B deletion. (D) Kaplan-Meier survival curve of early immature adult T-ALLs treated in the ECOG2993 clinical trial according to the presence or absence of DNMT3A mutations. (E) Kaplan-Meier survival curve of early immature adult T-ALLs treated in the ECOG2993 clinical trial according to the presence or absence of IDH1/IDH2 mutations.
Discussion
ETP leukemias are characterized by a cell surface antigen profile consisting of lack of CD1 and CD8, weak CD5, and the expression of at least one myeloid- or stem cell–related marker.10 These leukemias are transcriptionally related to early T-cell progenitors, a subtype of thymocytes that retain multilineage potential, and have been associated with poor prognosis in different clinical series including the Total Therapy Studies XIII, XIV, and XV at St. Jude Children’s Research Hospital,10 the ALL-2000 protocol of the Associazione Italiana Ematologia Oncologia Pediatrica,10 and the L99-15 study of the Tokyo Children's Cancer Study Group.31 Our microarray gene expression signature analyses demonstrate a high prevalence of early immature leukemias closely related to pediatric ETP-ALLs associated with reduced overall survival in adult T-ALL. This observation is in line with previous immunophenotype-based correlative analyses showing that adult T-ALLs with immature immunophenotypic features are associated with poor outcome.14,32 Of note, the German Multicenter Study Group for Adult ALL study protocols currently applies an immunophenotype-based risk stratification in which early adult T-ALLs (sCD3−, CD1a−) are selected for a more intensive treatment regimen including allogeneic stem cell transplantation.33
Absence of a bi-allelic TCRG deletion, a molecular marker linked with a very early block in T-cell differentiation, has been associated with poor response to induction therapy and dismal 5-year event-free and overall survival in pediatric T-ALL patients treated on COG P9404 or DFCI 00-01 protocols.12 More recently, the prognostic relevance of ABD was confirmed in pediatric T-ALL patients treated according to the Taiwan Pediatric Oncology Group protocols34 and pediatric T-cell lymphoblastic lymphoma patients treated following a Berlin-Frankfurt-Munich ALL-type strategy.35 In our patient series, ABD was identified in about half of adult T-ALL samples and found to confer with poor overall survival. In addition, 22 of 27 ABD adult T-ALL samples showed an early immature gene expression signature, suggesting biological overlap between the ABD and early immature subgroups in adult T-ALL. Altogether, these data show that early immature leukemias (defined by their transcriptional profile or TCRG status) are associated with poor prognosis in adult T-ALL. In addition, and as previously documented,14 CD13 expression was strongly associated with poor survival in our patient series, whereas other early immature or myeloid antigens including CD34 and CD33 showed no clinical impact. Importantly, our results suggest that CD13 expression could identify patients at increased risk of therapeutic failure in the otherwise good prognostic subtype of adult T-ALLs with a mature/cortical gene expression profile. However, given the small number of patients in the different subgroups, these results should be interpreted with caution and await validation in an independent series.
T-ALL is a cytogenetically stable disease with a limited number of chromosomal alterations per sample. Comparison of our aCGH analysis results with those of previously reported pediatric series showed major overlap and some differences. Among these, the frequency of MYB duplications (10/53; 19%) and RB1 deletions (9/53; 17%) in our adult patient series was higher than previously documented in pediatric studies.36,37 In addition, we failed to identify genomic deletions at the 6q14-6q22 locus, which occur in 20% to 30% of pediatric T-ALLs.38 Finally, the presence of multiple amplifications at 13q and 17q, and recurrent genomic deletions at 1p36 and 5q, suggest that these chromosomal regions could harbor novel T-ALL oncogenes and tumor suppressors, respectively.
Importantly, TP53 mutations have been previously associated with poor survival in pediatric T-ALL patients treated on the Pediatric Oncology Group protocol POG8862.39 Moreover, TP53 deletions and mutations at relapse are associated with chemotherapy resistance, failure to achieve a second complete remission, and poor survival.40 In line with this notion, heterozygous deletions of the short arm of chromosome 17 encompassing the TP53 tumor suppressor gene predicted for worse clinical outcome in adult T-ALL. In contrast, homozygous deletions of the CDKN2A/CDKN2B locus on the short arm of chromosome 9 conferred better clinical outcome in our series. Consistently, loss of heterozygosity at the short arm of chromosome 9 has been associated with a favorable initial treatment response in pediatric T-ALL patients treated in the Berlin-Frankfurt-Munster ALL-95 study protocol.41
Activation of NOTCH signaling through NOTCH1 and/or FBXW7 mutations has been associated with a favorable prognosis in several, but not all, pediatric study protocols.42-45 Similarly, controversial results on the prognostic effect of NOTCH activation have been obtained in adult T-ALL.46-49 These inconsistent results might at least in part be explained by the prognostic relevance of functional interplay between specific genetic lesions in T-ALL, as recently shown for NOTCH1 activation and inactivation of PTEN in children with T-ALL treated according to Berlin-Frankfurt-Munster study protocols.50 In our study, both univariate and multivariate analyses revealed that patients whose leukemias showed mutations in NOTCH1 and/or FBXW7 had a favorable outcome.
Recently, we documented a high prevalence of epigenetic and signaling mutations targeting FLT3, NRAS, DNMT3A, IDH1, and IDH2, typically found in myeloid leukemias, in adult early immature T-ALLs.11 Notably, activating internal tandem duplication mutations in the FLT3 tyrosine kinase gene correlated with poor overall survival and an increased risk of relapse in AML.51 Similarly, mutations in DNMT3A occur in ∼20% of AML patients and seem to confer poor overall survival in this disease.18,52 In contrast, mutations in IDH1 and IDH2 are associated with a favorable clinical outcome in AML, provided they co-occur together with NPM1 mutations.53 In our series, mutations targeting the epigenetic regulators DNMT3A, IDH1, and IDH2 conferred worse prognosis in adult T-ALL, and multivariate analysis pointed to DNMT3A mutations as an independent prognostic factor. Importantly, these findings confirm the recent association of DNMT3A mutations with poor outcome in a panel of 90 adult T-ALL patients treated according to the German Multicenter Study Group for Adult ALL protocols.54
Overall, the comprehensive analysis presented here shows that an early immature gene expression signature and the absence of bi-allelic TCRG deletion are associated with poor prognosis in adult T-ALL. In addition, we show that CD13 expression and homozygous CDKN2A/CDKN2B deletions might serve as prognostic markers to stratify low-risk cortical/mature adult T-ALLs, whereas DNMT3A mutations may be useful for risk stratification within high-risk early immature adult T-ALLs. Together, these analyses identified a subset high risk adult T-ALLs who may benefit from new emerging targeted therapies or alternative chemotherapy approaches.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This study was supported by the Research Foundation Flanders (postdoc fellowship and Odysseus type 2 grant to P.V.V. and postdoc fellowship to K.D.K.), the Eastern Cooperative Oncology Group tumor bank, National Institutes of Health grants R01CA120196 (A.A.F.), U24 CA114737, and U10 CA21115 (E.P.) provided by the National Cancer Institute, the Stand Up To Cancer Innovative Research Award (A.A.F.), the Chemotherapy Foundation (A.A.F.), and the Swim Across America Foundation (A.A.F.).
Authorship
Contribution: P.V.V. performed experiments and wrote the manuscript; A.A.-I. performed bioinformatic analyses; K.D.K. performed aCGH analysis; M.H. performed sequencing analysis; M.R. and C.F. performed statistical analysis; E.P., J.M.R., M.S.T., and J.M.R provided samples and correlative clinical and immunophenotypic data from ECOG; and A.A.F. designed the studies, directed research, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Adolfo A. Ferrando, Institute for Cancer Genetics, Columbia University, Irving Cancer Research Center, 4-402A, 1130 St. Nicholas Ave, New York, NY 10032; e-mail: af2196@columbia.edu.