Abstract
The biology and outcome of adult T-cell acute lymphoblastic leukemia are poorly understood. We present here the clinical and biologic features of 356 patients treated uniformly on the prospective trial (UKALL XII/ECOG 2993) with the aim of describing the outcome and identifying prognostic factors. Complete remission was obtained in 94% of patients, and 48% survived 5 years. Positivity of blasts for CD1a and lack of expression of CD13 were associated with better survival (P = .01 and < .001, respectively). NOTCH1 and CDKN2A mutations were seen in 61% and 42% of those tested. Complex cytogenetic abnormalities were associated with poorer survival (19% vs 51% at 5 years, P = .006). Central nervous system involvement at diagnosis did not affect survival (47% vs 48%, P = not significant). For 99 patients randomized between autograft and chemotherapy, 5-year survival was 51% in each arm. Patients with a matched sibling donor had superior 5-year survival to those without donors (61% vs 46%, χ2, P = .02); this was the result of less relapse (25% vs 51% at 5 years, P < .001). Only 8 of 123 relapsed patients survive. This study provides a baseline for trials of new drugs, such as nelarabine, and may allow risk-adapted therapy in patients with poor-prognosis T-cell ALL.
Introduction
The results of therapy of adult acute lymphoblastic leukemia (ALL) remain unsatisfactory.1 The relative rarity of T-cell ALL in adults has made it difficult to describe clinical and biologic factors determining outcome.2 Rowe et al3 reported the results of induction therapy from the Medical Research Council (MRC) UKALL XII/Eastern Cooperative Oncology Group (ECOG) 2993 study with more than 90% of patients achieving complete remission (CR). Diagnostic white cell count (WBC) was predictive of survival with less than 30 × 109/L for B-cell and less than 100 × 109/L for T-cell disease being predictive of better survival (P = .001). At the time of analysis, T-lineage was associated with a better 5-year survival than B-lineage disease (48% vs 41%, P = .001) and also affected survival on Cox regression analysis (P = .02). However, other major studies have not shown an independent effect of immunophenotype on survival.4
The full study report1 contained a donor versus no-donor analysis of the effect of allografting and a comparison of autografting in CR1 versus standard maintenance, but there were no separate analyses of the effects of these therapies on patients with T- or B-cell disease. In a further report, central nervous system (CNS) disease was found to be more common in patients with T-cell compared with those with pre–B-cell disease (9.6% vs 4.4%, P < .001),5 and CNS involvement was associated with poorer survival. Fielding et al6 showed that the outcome of relapsing patients was very poor and that patients with T-cell disease had a 5% 5-year survival compared with 8% in those with B-ALL (P = .06). Within this study was also the most extensive analysis of karyotypic abnormalities in adult ALL patients ever performed7 but not a specific analysis of abnormalities in T-cell ALL.
Our study describes the largest cohort of adults with T-lineage ALL so far treated uniformly on a prospective randomized trial. It will focus on clinical features, cytogenetics and molecular changes, immunophenotype, and outcome with each treatment modality and compare it with ALL of B-lineage.
Methods
Patient identification
A total of 1927 adult patients with ALL were registered on the UKALL XII/ECOG 2993 study in the United Kingdom or United States between 1993 and 2006. Investigators were asked to classify the cell lineage as B-cell or T-cell based on institutional immunophenotyping. Thirteen patients were found not to have ALL, one relapsed pretrial entry and one who was over the maximum trial entry age were excluded, as were the 269 patients with Philadelphia chromosome–positive ALL.
Immunophenotyping
A total of 108 patients had immunophenotyping studies in ECOG's Leukemia Translational Studies Laboratory and designated as T-lineage ALL. This was based on the detection of intracytoplasmic CD3 in the entire blast cell population. These studies were not performed in the United Kingdom, and detailed immunophenotypic data on the United Kingdom patients were not collected.
Between 1993 and 1998, flow cytometry was done on a FACScan flow cytometer (Lysys II software), and from 1999 on a FACSCalibur flow cytometer (CellQuest software; all from BD Biosciences). Lymphoblasts were gated by 3-color flow cytometry based on antigen expression of the leukemic cells. In addition to cytoplasmic CD3 (cCD3), T-lymphoid–affiliated antigens tested included CD1a, CD2, surface CD3, CD4, CD5, CD8, CD62L, CD57, and surface α/β and γ/δ. B-lymphoid antigens included intracytoplasmic and surface CD22, CD19, CD10, and CD20. Myeloid antigens included myeloperoxidase, CD117, CD33, CD13, CD65(s), CD15(s), CD11b, and CD14. Uncommitted antigens CD45, CD34, HLA-DR, and TdT were analyzed. Further details of immunophenotyping methods are provided as supplemental data (available on the Blood website; see the Supplemental Materials link at the top of the online article).
Cytogenetic, FISH, and molecular genetic investigations
Cytogenetic analysis of pretreatment bone marrow or peripheral blood samples was performed locally, reviewed centrally, and collated retrospectively using standard definitions.7 Fluorescence in situ hybridization (FISH) was performed using fixed-cell suspensions from pretreatment samples. Details of the probes used can be found in supplemental data.
Therapy and risk assignment
This has been described in detail in previous publications.3 Briefly, all patients from 15 to 59 years of age with newly diagnosed ALL received identical 4-drug induction therapy, including CNS prophylaxis and treatment of CNS disease, if present at diagnosis. In 2003, the upper age limit of the study was raised to 64 years and eligibility for allograft was raised from 49 to 54 years. Patients with a human leukocyte antigen (HLA)–matched sibling donor were assigned to receive an allograft. Those without a human leukocyte antigen-matched sibling donor, or over the age limit, were to be randomized to receive a single autologous transplantation or consolidation/maintenance therapy. Before receiving the assigned or randomized therapy, all patients received intensification with 3 doses of high-dose methotrexate and asparaginase. In this study, patients older than 35 years or those with a high WBC at presentation (≥ 100 × 109/L for T-lineage and ≥ 30 × 109/L for B-lineage), along with all patients with the Philadelphia chromosome, were subsequently shown to be high-risk. All others were classified as standard risk. The study was approved by the relevant Institutional Review Boards of each center, and informed consent was obtained in accordance with the Declaration of Helsinki.
Statistical methods
In comparisons of B- and T-lineage patients, B-lineage included null but excluded mature B immunophenotypes. Patient characteristics and remission rates were compared using χ2 tests for heterogeneity, Mantel-Haenszel test for trend, or Mann-Whitney U test. Patients with CNS involvement at diagnosis unreported (∼ 25%) were assumed for analyses to have no CNS involvement.
The comparison between those with versus those without a matched related donor was used as an unbiased assessment of the effect of matched related donor allograft. This comparison included only patients younger than 50 or 55 years, commensurate with the age limit for related donor allografts.
The primary outcome measure was overall survival (OS). Censoring was at the date of last contact, or October 31, 2008, if earlier. Relapse incidence/relapse-free survival excluded nonremitters and censored at nonrelapse mortality, whereas the risk of nonrelapse mortality censored at relapse. Time to relapse or nonrelapse mortality was measured from diagnosis except in the case of analysis by grade of acute graft-versus-host disease (GVHD), where they were measured from the date of allograft. The median follow-up in the 164 surviving T-cell patients was 7 years (range, 3 months to 16 years).
Kaplan-Meier curves were used and comparisons made using the log-rank method unless otherwise specified. Where the hazard is nonproportional and long-term survival was to be compared, the χ2P value for the difference in the survival percentages at 5 years was quoted. Cox regression analysis was used to investigate WBC as a continuous variable. All P values are 2-sided.
Forest plots8 were used to illustrate the effect on outcome of randomized allocation and of matched sibling donor availability within subgroups.
Results
Clinical characteristics by lineage
A total of 356 patients (of the total 1643 patients with confirmed Philadelphia chromosome–negative ALL) were classified as having T-cell disease, 1176 were B-lineage (including 101 pro-B and 29 mature B), and 111 had unconfirmed lineage. Patient characteristics are shown in Table 1. Forty-one percent of patients were registered with ECOG rather than the MRC, compared with 34% of the B-cell patients (P = .01). T-cell disease was more commonly seen in males: nearly three-quarters of T-cell patients were male compared with 59% of B-cell patients (P < .001). T-cell disease was more common in the 20- to 29-year and 30- to 39-year age groups (P < .001, Table 1); however, the median ages at diagnosis of B- and T-cell ALL patients were similar (30 and 29 years, respectively). There was an association between T-cell disease and a high WBC with 42% of T-cell patients having a WBC more than 50 × 109/L compared with 19% of B-cell patients (P < .001). Furthermore, 9% of T-cell patients had CNS involvement at diagnosis compared with 4% in B cell (P < .001).
Clinical characteristics of patients with T-cell compared with B-cell acute lymphoblastic leukemia and overall survival in T-cell patients
| . | T-cell ALL, n (%) . | B-cell ALL, n (%)* . | P . | 5-year overall survival in T-cell ALL, percentage (95% CI) . | P . |
|---|---|---|---|---|---|
| Group | |||||
| ECOG | 146 (41) | 389 (34) | 47 (39-56) | ||
| NRCI | 210 (59) | 758 (66) | 48 (41-55) | ||
| P | .01† | > .1‡ | |||
| Age, y | |||||
| Younger than 20 | 55 (15) | 256 (22) | 53 (40-67) | ||
| 20-29 | 136 (38) | 311 (27) | 52 (43-60) | ||
| 30-39 | 85 (24) | 216 (19) | 46 (35-57) | ||
| 40-49 | 46 (13) | 203 (18) | 47 (32-63) | ||
| 50 or older | 34 (10) | 161 (14) | 27 (11-43) | ||
| P | < .001†/.06§ | .009‖ | |||
| 35 or younger | 244 (69) | 704 (61) | 52 (46-58) | ||
| Older than 35 | 112 (31) | 443 (39) | 38 (29-48) | ||
| P | .01† | .004‡ | |||
| Sex | |||||
| Male | 260 (73) | 680 (59) | 50 (44-57) | ||
| Female | 96 (27) | 467 (41) | 41 (31-51) | ||
| P | < .001† | .05‡ | |||
| Diagnostic WBC, ×109/L | |||||
| Less than 50 | 203 (58) | 920 (81) | 49 (42-56) | ||
| 50-99 | 54 (15) | 96 (8) | 56 (42-69) | ||
| 100 or more | 96 (27) | 125 (11) | 41 (31-51) | ||
| P | < .001§ | .09‖ (< 100 vs ≥ 100, P = .03‡) | |||
| CNS involvement | |||||
| Yes | 32 (9) | 46 (4) | 47 (29-64) | ||
| No/unknown | 324 (91) | 1101 (96) | 48 (42-54) | ||
| P | < .001†/ < .001¶ | > .1‡/> .1 |
| . | T-cell ALL, n (%) . | B-cell ALL, n (%)* . | P . | 5-year overall survival in T-cell ALL, percentage (95% CI) . | P . |
|---|---|---|---|---|---|
| Group | |||||
| ECOG | 146 (41) | 389 (34) | 47 (39-56) | ||
| NRCI | 210 (59) | 758 (66) | 48 (41-55) | ||
| P | .01† | > .1‡ | |||
| Age, y | |||||
| Younger than 20 | 55 (15) | 256 (22) | 53 (40-67) | ||
| 20-29 | 136 (38) | 311 (27) | 52 (43-60) | ||
| 30-39 | 85 (24) | 216 (19) | 46 (35-57) | ||
| 40-49 | 46 (13) | 203 (18) | 47 (32-63) | ||
| 50 or older | 34 (10) | 161 (14) | 27 (11-43) | ||
| P | < .001†/.06§ | .009‖ | |||
| 35 or younger | 244 (69) | 704 (61) | 52 (46-58) | ||
| Older than 35 | 112 (31) | 443 (39) | 38 (29-48) | ||
| P | .01† | .004‡ | |||
| Sex | |||||
| Male | 260 (73) | 680 (59) | 50 (44-57) | ||
| Female | 96 (27) | 467 (41) | 41 (31-51) | ||
| P | < .001† | .05‡ | |||
| Diagnostic WBC, ×109/L | |||||
| Less than 50 | 203 (58) | 920 (81) | 49 (42-56) | ||
| 50-99 | 54 (15) | 96 (8) | 56 (42-69) | ||
| 100 or more | 96 (27) | 125 (11) | 41 (31-51) | ||
| P | < .001§ | .09‖ (< 100 vs ≥ 100, P = .03‡) | |||
| CNS involvement | |||||
| Yes | 32 (9) | 46 (4) | 47 (29-64) | ||
| No/unknown | 324 (91) | 1101 (96) | 48 (42-54) | ||
| P | < .001†/ < .001¶ | > .1‡/> .1 |
NCRI indicates National Cancer Research Institute.
Excludes 29 mature B patients.
χ2 test for heterogeneity.
Log-rank test for heterogeneity.
χ2 test for trend.
Log-rank test for trend.
After controlling for WBC.
Immunophenotypic subclassification in ECOG T-cell ALL patients
T-lineage ALL was diagnosed in all centrally reviewed ECOG patients based on the presence of intracytoplasmic CD3 in all blast cells.9 CD7 was the only surface T-cell antigen expressed by all T lymphoblasts in all cases. Attempts to stratify patients according to their maturation stage, as suggested by the WHO classification,9 failed to provide subsets that were associated with outcome, with the exception of the cortical thymocyte stage, characterized by expression of CD1a. With respect to myeloid antigens, CD13 was found in 51% and CD33 in 30% of patients, whereas the carbohydrate antigens, CD65(s) and CD15(s), were present in only 4% and 12% of patients, respectively. By including the distribution profile of CD13, 2 major subclasses of T-ALL with prognostic significance and minimal immunophenotypic overlap became apparent, which accounted for 84 of 100 patients tested for both CD13 and CD1a: CD1a+ T-ALL lacking CD13 (31% of ECOG patients) and CD1a−CD13+ T-ALL (46% of ECOG patients). Seven patients coexpressed CD1a and CD13 (7%). The remaining 16 patients failed to present as a distinct group based on their antigen profile or clinical response.
The demographics of patients with detailed immunophenotyping were similar to those of the remaining patient population. There was no difference in gender or diagnostic WBC; patients with central immunophenotyping were slightly older (median age, 31 vs 28 years, P = .05).
Genetic classification
Table 2 outlines the frequency of abnormalities detected by cytogenetics, FISH, and/or molecular genetic studies. NOTCH1/FBXW7 mutations were the most frequent abnormalities (61% and 18%, respectively) and often occurred together (n = 11). TLX1 or TLX3 translocations were detected in 16% patients, whereas translocations affecting LMO1 or LMO2 were much rarer (2%). Up-regulation of TAL1 via del(1)(p32) or t(1;14) was detected in 13% of patients. CDKN2A deletions were detected by FISH in 42% of cases and were more prevalent than deletions of 9p (10%) detected by conventional cytogenetic studies.10 CDKN2A deletions were biallelic (n = 13), monoallelic (n = 10), or both (n = 4). G-banding analysis revealed deletions/abnormalities of 6q, 11q, 13q, and 17p in 5% to 11% cases, with 8% having a complex karyotype. There were few significant correlations between genetic abnormalities and sex, age, WBC, or immunophenotype. The exceptions were: (1) TLX1 patients were older (median age, 34 vs 27 years, P = .005); (2) t(11;14)(p13;q11) and del(6q) patients had a higher median WBC (144 vs 38 × 109/L, P = .02; 92 vs 28 × 109/L, P = .002, respectively); and (3) patients expressing CD2 rarely had a complex karyotype (1 of 48, 2% vs 4 of 15, 27%, P < .01).
Incidence and outcome of cytogenetic subgroups and genetic abnormalities in adult T-ALL
| Cytogenetic/genetic subgroup . | No. of cases (positive/tested)* . | Frequency, percentage . | CR (%) . | 5-year survival, percentage (95% CI) . |
|---|---|---|---|---|
| Cytogenetics attempted | 303/356 | 85 | 287 (95) | 49 (44-55) |
| Successful cytogenetics† | 204/303 | 67 | 194 (95) | 49 (42-56) |
| Failed cytogenetics† | 99/303 | 33 | 93 (94) | 51 (41-61) |
| Abnormal karyotype† | 146/204 | 72 | 138 (95) | 46 (38-55) |
| Normal karyotype† | 58/204 | 28 | 56 (97) | 54 (41-67) |
| SIL-TAL1‡ | 8/62 | 13 | 8 (100) | 50 (15-85) |
| TLX3-BCL11B | 4/62 | 6 | 4 (100) | 75 (32-100) |
| t(10;14)(q24;q11)/TLX1-TRA/D@§ | 21/215 | 10 | 20 (95) | 47 (26-69) |
| t(10;11)(p13;q14-q21)/AF10-CALM | 6/214 | 3 | 6 (100) | 67 (13-100) |
| t(11;14)(p15;q11)/LMO1-TRA/D@‖ | 0/215 | 0 | — | — |
| t(11;14)(p13;q11)/LMO2-TRA/D@ | 4/215 | 2 | 4 (100) | 0 |
| MLL translocations¶ | 3/216 | 1 | 3 (100) | 67 (13-100) |
| CDKN2A deletion** | 28/67 | 42 | 28 (100) | 52 (33-71) |
| NUP214-ABL1 | 2/104 | 2 | 2 (100) | 50 (0-100) |
| NOTCH1 mutation†† | 55/90 | 61 | 54 (98) | 53 (39-67) |
| FBXW7 mutation‡‡ | 16/88 | 18 | 16 (100) | 63 (39-86) |
| del(6q) | 22/204 | 11 | 21 (95) | 46 (25-66) |
| del(9p) | 21/204 | 10 | 21 (100) | 48 (26-69) |
| Abnormality of 11q | 13/204 | 6 | 12 (92) | 39 (12-65) |
| del(13q) | 13/204 | 6 | 13 (100) | 46 (19-73) |
| del(17p) | 10/204 | 5 | 9 (90) | 20 (0-45) |
| Complex karyotype§§ | 17/204 | 8 | 15 (88) | 19 (0-38) |
| Cytogenetic/genetic subgroup . | No. of cases (positive/tested)* . | Frequency, percentage . | CR (%) . | 5-year survival, percentage (95% CI) . |
|---|---|---|---|---|
| Cytogenetics attempted | 303/356 | 85 | 287 (95) | 49 (44-55) |
| Successful cytogenetics† | 204/303 | 67 | 194 (95) | 49 (42-56) |
| Failed cytogenetics† | 99/303 | 33 | 93 (94) | 51 (41-61) |
| Abnormal karyotype† | 146/204 | 72 | 138 (95) | 46 (38-55) |
| Normal karyotype† | 58/204 | 28 | 56 (97) | 54 (41-67) |
| SIL-TAL1‡ | 8/62 | 13 | 8 (100) | 50 (15-85) |
| TLX3-BCL11B | 4/62 | 6 | 4 (100) | 75 (32-100) |
| t(10;14)(q24;q11)/TLX1-TRA/D@§ | 21/215 | 10 | 20 (95) | 47 (26-69) |
| t(10;11)(p13;q14-q21)/AF10-CALM | 6/214 | 3 | 6 (100) | 67 (13-100) |
| t(11;14)(p15;q11)/LMO1-TRA/D@‖ | 0/215 | 0 | — | — |
| t(11;14)(p13;q11)/LMO2-TRA/D@ | 4/215 | 2 | 4 (100) | 0 |
| MLL translocations¶ | 3/216 | 1 | 3 (100) | 67 (13-100) |
| CDKN2A deletion** | 28/67 | 42 | 28 (100) | 52 (33-71) |
| NUP214-ABL1 | 2/104 | 2 | 2 (100) | 50 (0-100) |
| NOTCH1 mutation†† | 55/90 | 61 | 54 (98) | 53 (39-67) |
| FBXW7 mutation‡‡ | 16/88 | 18 | 16 (100) | 63 (39-86) |
| del(6q) | 22/204 | 11 | 21 (95) | 46 (25-66) |
| del(9p) | 21/204 | 10 | 21 (100) | 48 (26-69) |
| Abnormality of 11q | 13/204 | 6 | 12 (92) | 39 (12-65) |
| del(13q) | 13/204 | 6 | 13 (100) | 46 (19-73) |
| del(17p) | 10/204 | 5 | 9 (90) | 20 (0-45) |
| Complex karyotype§§ | 17/204 | 8 | 15 (88) | 19 (0-38) |
— indicates not applicable.
Tested by cytogenetics, FISH, reverse-transcribed polymerase chain reaction, or a combination thereof.
An abnormal karyotype was defined as one with a clonal chromosomal abnormality. In the absence of a clonal abnormality, cases with 20 or more analyzable metaphases were classified as having a normal karyotype whereas those with fewer than 20 cells were classified as having failed cytogenetics. All cases with a normal or abnormal karyotype were classified as having successful cytogenetics.
Includes 2 cases with t(1;14)(p32;q11).
Includes 2 cases with t(7;10)(q34-36;q24).
Includes 2 cases with t(7;11)(q34-36;p13).
Includes 2 cases with t(11;19)(q23;p13.3).
Types of deletion: biallelic (n = 13), monoallelic (n = 10), and both (n = 4).
Includes 2 cases with t(7;9)(q34;q34).
Eleven of 16 patients with a FBXW7 mutation also had a NOTCH1 mutation.
Five or more chromosomal abnormalities in the absence of an established subgroup.
Treatment
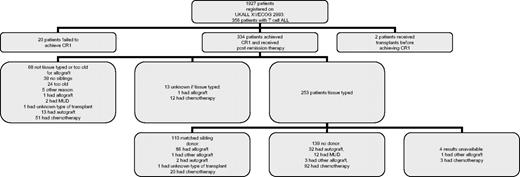
Twenty (5.6%) of the 356 T-ALL patients failed to achieve CR (16 died during induction and 4 did not enter remission) and 2 patients had a transplantation without remission. Thus, 334 evaluable patients (94%) achieved CR (Figure 1). Subsequent therapy was sibling allograft (n = 88), autologous stem cell transplantation (n = 47), unrelated donor allograft (n = 14), other type of allograft (4 mismatched and 1 allograft with reduced intensity conditioning), unknown type of transplantation (n = 2), and chemotherapy (n = 178). Ninety-nine patients were randomized between chemotherapy and autograft. Of the 45 patients randomized to chemotherapy, none was transplanted. The role of sibling donor allograft was assessed by a comparison of those with those without a sibling donor. A total of 253 patients were tissue typed of whom 110 had a related donor and 139 did not (and 4 are unknown). Of the 54 patients randomized to autograft, 33 had an autograft, 19 had chemotherapy, one had an unrelated donor allograft, and one patient received a transplantation from an unknown donor (assumed to be unrelated).
Outcome
The remission rate in T-cell patients was 94% compared with 93% in B-cell patients (P = .5). Similar to the results found in B-lineage patients, remission rates were higher in younger patients (98% at ages 15-19 and 20-29, 93% at ages 30-39 and 40-49, and 79% in those 50 years of age and older, P < .001). No significant differences in remission rates were found by sex, diagnostic WBC, or CNS involvement at diagnosis. Patients with detailed immunophenotyping had similar CR rates to the remaining T-cell patient population (95% vs 94%, P = .6). In ECOG T-ALL patients with central immunophenotyping, no antigen was found to be significantly related to remission rate (P < .01). There was no evidence that presence of CD1a+ or CD13+ affected remission (P > .1 in both cases). Similarly, none of the specific genetic abnormalities analyzed affected the likelihood of achieving CR (Table 2).
A total of 123 patients relapsed (37%). The date of relapse is unknown for 2 patients who died; these were scored as relapse one day before the date of death. Relapse occurred at a median of 12 months, with the majority of relapses occurring within 2 years. The actuarial incidence of relapse at 2 years was 35% (95% confidence interval [CI], 30%-40%) and at 5 years was 42% (36%-47%). Therapy after relapse was: 9 sibling donor allografts (2 reduced intensity conditioning), 17 matched unrelated donor allografts, 6 autografts, one mismatched unrelated donor allograft, and 90 no transplantation. Thus, only 27 of 123 patients had a potentially curative allograft. Only 8 survived at a median of 5.2 years (1.1-13.3 years), including 6 who were treated with an allograft (4 unrelated donor).
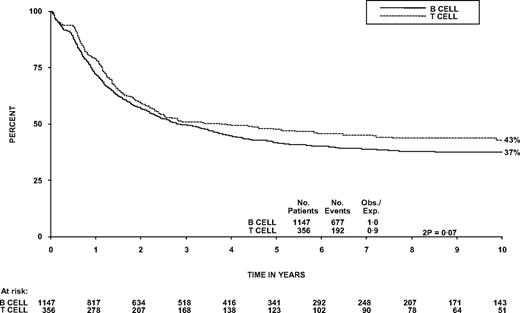
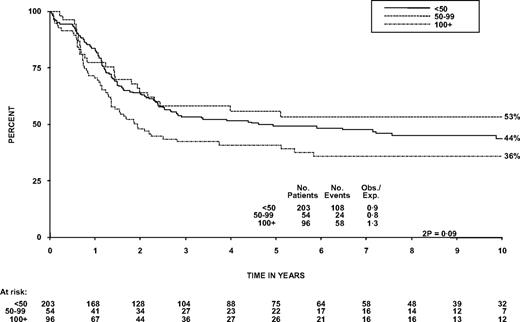
The OS at 5 years was 48% (95% CI, 42%-53%) compared with 42% (39%-45%) in patients with B-ALL (Figure 2, P = .07). The survival of patients who received chemotherapy only was also similar for T- and B-cell disease (40% in each arm, P = 1.0). Within T-cell patients, there was not a significant trend for diagnostic WBC to affect OS, nor was it significant as a continuous variable in Cox regression, although the 96 patients (27%) with a WBC more than 100 × 109/L did have poorer OS at 5 years than patients with a WBC less than 100 × 109/L (P = .03, Table 1; Figure 3). This is different from B-cell disease where there was a highly significant trend (P < .001; heterogeneity in the effect of WBC by lineage P = .007). T-cell patients over the age of 35 years and females had significantly worse OS (P = .004 and .05, respectively, Table 1). Thirty-two patients had CNS involvement at diagnosis (9%); this did not affect 5-year OS significantly (46% vs 48%, P = .8, Table 1). Sex of the patient was not significant in a model including age. In the 55 patients 15 to 19 years of age at entry, OS at 5 years was 53% (40%-69%).
Overall survival from diagnosis of patients with B- versus T- lineage disease.
Overall survival from diagnosis by WBC in patients with T-lineage ALL.
Looking at the first of the 2 major subtypes of T-cell leukemia identified, OS in CD1a+ patients at 5 years was 64% (95% CI, 48%-80%) versus 39% (26%-52%) in CD1a− patients (P = .01). This appears to be the result of a higher risk of relapse in CD1a− patients (50%; 36%-65%) at 5 years compared with 23% (8%-38%) in CD1a+ patients (P = .02). The majority of CD1a+ cases lacked CD34 (P < .001), both CD13 and CD33 (P < .001), as well as CD11b, a member of the integrin α chain family (P < .001). Typical for this cortical T-ALL stage,9,11 73% of patients expressed the leukocyte selectin CD62L, and 50% expressed CD10, an antigen commonly found in B-lymphoid precursor cells; associations between CD62L or CD10 and CD1a were not significant at P < .01. Although 61% of CD1a+ cases expressed both CD4 and CD8 (P < .001), only half of these cases lacked CD3; on the other hand, half of single CD4+ or CD8+ cases lacked surface CD3.
The second major immunophenotypic subset of T-ALL with prognostic significance was CD13+CD1a− T-ALL, which was present in 46 of 100 ECOG patients tested for both antigens. Five-year OS in CD13+ T-ALL was inferior with 35% (95% CI, 22%-48%), compared with CD13− patients (61%, 48%-75%, P < .001) mainly the result of an increased risk of TRM in CD13+ T-ALL (30%, 15%-45%) at 5 years compared with 13% (2%-23%) in CD13− patients (P = .008). There may be an increase in relapses in those with CD13+. T-ALL (50%, 34%-66%) at 5 years compared with 33% (19%-47%) in CD13− patients, but this does not reach statistical significance (P = .1). However, CD13 status did not significantly affect relapse risk (P = .1). The presence of CD33 did not add to the adverse prognostic effect of CD13 whether CD33 was considered in CD13+ or CD13− patients, suggesting that CD13+ T-ALL represents a novel prognostic leukemia subtype. Despite the obvious driving role of CD13 in determining the outcome of T-ALL, the dual CD13/CD33+ subset (21 patients) presented with a unique antigen profile. CD34 expression levels (P = .06) and the incidence of CD11b+ (P = .006), triple CD3−CD4−CD8− blasts (P < .001) were higher in CD13/CD33+ than CD13+CD33− populations, consistent with an early T maturation stage. Patients with detailed immunophenotyping had identical survival at 5 years to the remaining T-cell patient population (P = 1.0).
There was no significant difference in OS according to whether cytogenetic analysis was performed, successful or detected an abnormality. Although none of the abnormalities was associated with an improved OS, it is of interest that only one of 4 TLX3 rearrranged patients died. Patients with a complex karyotype had a significantly lower OS at 5 years compared with patients with simple or normal karyotypes (19% vs 51%, P = .006, Table 2), and this effect was not mediated by a higher WBC or age. In addition, all 4 patients with t(11;14)(p13;q11) died within 5 years and those with del(17p) had a somewhat lower OS (20% vs 50% at 5 years, P = .07). It may be noteworthy that OS of patients with a CDKN2A deletion was identical to those who retained the gene (52% vs 52% at 5 years, P = 1.0). As we have recently reported,12 patients with a mutation in the NOTCH pathway (either NOTCH-1 and/or FBXW7) had higher event-free survival versus those without 51% (± 14%) versus 27% (± 19%), but this was not significant (P = .1), in keeping with other reports in adults13 with T-cell ALL.
Survival and treatment
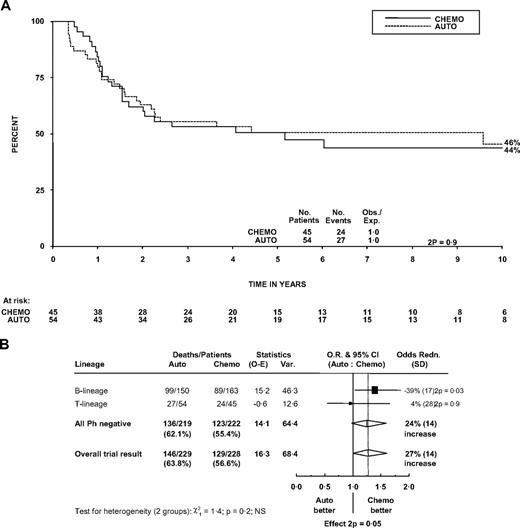
The 5-year OS in the 99 patients randomized between autograft and chemotherapy was 51% (36%-65%) in the chemotherapy arm and 51% (37%-64%) in the patients who were assigned to autograft (P = .9). Figure 4A shows survival in the 2 groups followed out to 10 years. Autografting was not shown to be inferior to chemotherapy in patients with T-cell disease, but there is no evidence that the effect of autograft in T-cell patients is different from that in B-cell patients (P for heterogeneity = .2; Figure 4B). The overall trial result showed inferior survival with autograft (P = .05). Larger numbers are needed to establish whether the effects are truly different between T- and B-lineage ALL.
Effect of randomized treatment on overall survival in patients. (A) Survival curve in patients with T lineage. (B) Forest plot within lineage subgroups. Survival was measured from randomization. The forest plot represents the treatment effect (odds ratio) and its 95% CI by a square and horizontal line (within subgroups) and the center and width of a diamond (overall).
Effect of randomized treatment on overall survival in patients. (A) Survival curve in patients with T lineage. (B) Forest plot within lineage subgroups. Survival was measured from randomization. The forest plot represents the treatment effect (odds ratio) and its 95% CI by a square and horizontal line (within subgroups) and the center and width of a diamond (overall).
Of the 19 patients randomized to autograft but who received chemotherapy, only 3 survived. We do not know whether stem cells were obtained in these 19 patients.
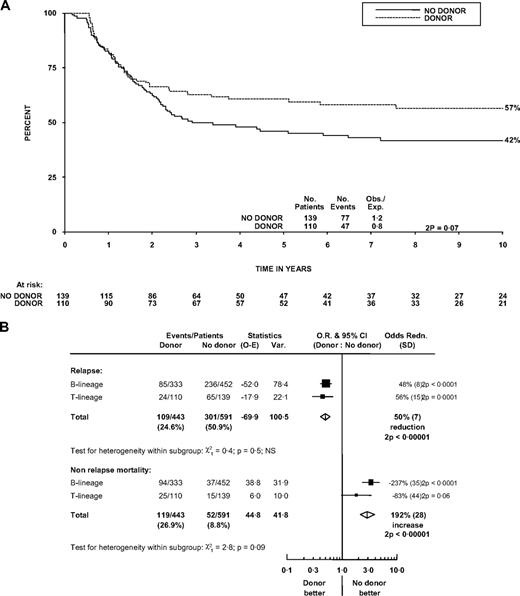
The role of sibling donor allograft was assessed by a comparison of those with versus those without a sibling donor. OS at 5 years was 46% (38%-55%) for the no donor group and 61% (51%-70%) for the donor group (log-rank P = .07, χ2 test of difference at 5 years, P = .02), a difference maintained at 10 years (Figure 5A).
Effect of matched sibling donor availability on outcome. (A) Survival curve in patients with T lineage. (B) Forest plot of effects on relapse and on nonrelapse mortality within lineage subgroups. Survival was measured from diagnosis. Forest plot format is as in Figure 4.
Effect of matched sibling donor availability on outcome. (A) Survival curve in patients with T lineage. (B) Forest plot of effects on relapse and on nonrelapse mortality within lineage subgroups. Survival was measured from diagnosis. Forest plot format is as in Figure 4.
We then compared the positive (protection from relapse) and negative (nonrelapse mortality) effects of allograft in patients with T- and B-ALL (Figure 5B). Having a donor had a similar effect on relapse protection in patients with T-cell disease (25% vs 51%, P < .001) to that in B-cell disease (30% vs 55% at 5 years, P < .001; P heterogeneity = .5). Nonrelapse mortality was more modestly increased in the T-cell donor group compared with the no-donor group (22% vs 12% at 5 years, P = .06) than it was in the B-cell cohort (32% vs 10% at 5 years, P < .001; P heterogeneity = .09).
Data on GVHD grade were available in 76 of 88 T-cell patients who had allografts. Sixteen patients had grade 2-IV GVHD. There was no significant difference between these and those with grade 0-I GVHD (relapse-free survival 83% at 5 years vs 80%; P = .8 and nonrelapse mortality 33% vs 19%, P = .2). Of the 14 patients who had an unrelated donor transplantation in CR1, 9 survive. Seven of these patients had a WBC more than 100 × 109/L.
Subgroup analyses of the effect of different treatments (donor vs no donor and autogaft vs chemotherapy) on outcome show no evidence that the treatment effect is affected by the presence or absence of any of CD1a, CD13, or complex cytogenetics. This is not surprising considering the small patient numbers in these subgroups.
Discussion
We present a detailed description of 356 uniformly treated patients with T-lineage ALL and, for the first time, describe biologic characteristics that affect outcome. The overall CR rate (in a large multicenter trial) was high and nearly half the patients survived 5 years, but (as in all adult ALL) there is considerable room for improvement. Older patients (> 35 years) and females had a poorer outcome and may require different strategies. Twenty-seven percent of patients presented with a WBC at diagnosis more than 100 × 109/L; these patients had slightly inferior survival. CNS disease at diagnosis did not affect outcome, although it did in the complete patient cohort.5 This may be a chance effect as there was no evidence of CNS disease having a different effect in T- compared with B-lineage patients (heterogeneity P = .1).
In 2006, Vitale et al2 presented the results of 90 adult patients with T-ALL treated with the GIMEMA LAL 0496 protocol. Similar to our study, they found a predominance of males (68% vs 73% in our study). In their study, males had a higher CR rate (84% vs 52%, P = .004). Although males in our study had a superior survival at 5 years, we found that gender had no impact on the chance of achieving remission. Our study had nearly twice the incidence of abnormal karyotypes (72% vs 36.5%), enabling us to explore the prognostic significance of these abnormalities in more detail. A partial deletion of 6q was the most common abnormality seen in the Italian cohort, and no cytogenetic lesion significantly impacted on outcome. We confirmed that associations between immunophenotypic characteristics and cytogenetic abnormalities are much less pronounced in T- than B-ALL14 and only detected an association between CD2 positivity and complex karyotypes.
This is one of the largest genetic studies of adult T-ALL and one of the few to assess the prognostic relevance of several abnormalities simultaneously in the context of a single trial. Still, our analysis was limited by the number of patients with specific abnormalities. We were able to establish that patients with a complex karyotype have a significantly inferior survival.7 In contrast, we were unable to confirm previous observations15,16 that patients with TLX1 overexpression/translocations (21 patients) have a superior outcome. Although this discrepancy could be the result of differential response to varied protocols, our data suggest that involvement of the TLX1 gene does not necessarily confer a good prognosis. Finally, our data provide strong evidence that deletion of CDKN2A is not associated with outcome in T-ALL, probably because it is a secondary abnormality and hence does not define a distinct biologic subgroup.
We made several novel observations regarding antigen profiles and their clinical significance in adult T-ALL. Although only performed in ECOG patients, our cohort represents the largest series of adult T-ALL for which multiparameter flow-cytometric data are available. We defined the diagnosis of T-ALL by the presence of cytoplasmic CD3, the only antigen specific for the T-cell lineage, together with surface CD7, the antigen universally expressed by but not specific for T-lineage ALL.11,17-19 Contrary to earlier studies,2,20 we did not base antigen positivity on arbitrary cut-off levels, given that antibody binding data were restricted to lymphoblasts gated with a blast cell marker, the preferred method for blast identification.18,21 Doubts regarding the clinical relevance of subgrouping T-lineage ALL according to thymocyte maturational stages have been raised since the early days of immunophenotyping.22 Our data finally settle this controversy and confirm that, with the exception of the CD1a+ cortical stage, the maturation level of T lymphoblasts is not associated with prognostic significance. In agreement with previous studies,11 an increased number of our CD1a+ patients survived for at least 5 years. However, CD1a expression did not affect CR rate. Although Vitale et al2 reported that cortical and mature T-ALL patients had a higher CR rate than patients with more immature T-ALL stages, implying a beneficial effect of CD1a on CR achievement, their definition of cortical ALL lacked the assessment of CD1a expression. The favorable outcome occasionally reported for dual CD4/CD8 or CD10 expression in T-ALL11,18 most probably mirrors the advantageous effect of CD1a, an antigen frequently associated with these markers.
We found that, among myeloid antigens, CD13 was expressed most frequently (51% of all patients, irrespective of CD1a expression), often together with CD33, an incidence much higher than reported previously.2,20,23 On the other hand, CD65(s) and CD15(s) were rarely detected, independent of T-blast maturation, a finding different from that in B-lineage ALL, where CD13/CD33 versus CD65(s)/CD15(s) expression is tied to B-lymphoid blast maturation.11,24 Except for 2 patients with CD117+ disease who expressed myeloperoxidase in 3% to 5% of cCD3+ T lymphoblasts,25 none of our myeloid antigen-positive patients fits the correct definition of biphenotypic leukemia, requiring dual expression of distinct lineage-specific antigens.11,17,18 Only 7 CD1a+ cases expressed CD13, consistent with minor overlap between the 2 prognostic antigen subgroups. Significantly fewer CD13+ than CD13− patients survived 5 years, whereas CD33 did not add to this inferior outcome. CD13, and particularly CD13/CD33, dual positivity was associated with a CD34+, triple CD3/CD4/CD8 negative, immature phenotype that resembles the adult T-ALL molecular risk group recently defined by high BAALC and ERG expression.26 These data suggest that CD13+ T-ALL is derived from the earliest thymic precursors, which possess dual T and myeloid potential.27
In our cohort, none of the antigens tested affected CR rate. Vitale et al2 had found that CD34 positivity lowered the CR rate from 84% to 54%. In our patients, the CR rate was 100% in patients lacking CD34 expression, compared with 93% in patients with 1% to 79% CD34+ blasts, compared with 91% in patients with more than 79% CD34+ blasts (not significant). Although disparities in flow cytometric technology and lack of centralized immunophenotyping in the GIMEMA study could contribute to this different finding, the most probable explanation is a superior treatment response in UKALL XII/E2993, abolishing any prognostic impact of CD34 on achievement of CR.
The outcome of relapse remains dismal, with 8 of 123 relapsed patients surviving (6 patients had allogeneic transplantation), as in the entire study population.6 Only 27 of 123 (23%) relapsed patients had a potentially curative allograft. We did not routinely collect data about therapy after relapse, nor do we know how many patients achieved a second CR. Strategies designed to achieve a second remission safely and to increase the number of patients proceeding to allografting should be the focus of future studies. Nelarabine may have a role in patients with relapsed T-cell disease, but large-scale efficacy and toxicity data are lacking.
This study has some limitations despite being the largest reported series of uniformly treated and prospectively followed adult T-ALL patients. Detailed immunophenotyping was only performed in the ECOG patients, representing 30% of the entire cohort. Nonetheless, after a comparison of demographic and prognostic factors, these patients appear to be representative. The spectrum of genetic abnormalities in T-ALL includes a high proportion requiring FISH or polymerase chain reaction for accurate detection (eg, cryptic chromosomal translocations and gene mutations). Moreover, this is a rapidly evolving field, and many abnormalities have only recently been characterized.16 As a result, only a minority of cases in this study were adequately screened; thus, only 41% could be classified into one of the genetic subgroups listed in Table 2. Although cytogenetic analysis was attempted in 85% of cases, it failed in 33% and yielded only normal metaphases in another 28%. These rates are significantly higher than those observed among B-lineage cases in this trial. The high incidence of normal karyotypes is the result of the high proportion of cryptic abnormalities in this disease, whereas the high cytogenetic failure rate is probably the result of unsatisfactory sample type (blood instead of marrow) and transit time to the cytogenetic laboratory,28 both factors that complicate multicenter trials conducted over long periods.
This study has many important new findings. The donor versus no donor comparison suggests that matched sibling allografting in CR1 remains a viable strategy in this patient subgroup. The outcome of autografting in CR1 (compared with chemotherapy) may be different from that in B-ALL, although the reason for this finding is uncertain.29 More evidence on treatment effects within subgroups will be provided eventually by a meta-analysis of similar trials. Finally, and most importantly, this study identified biologic factors associated with a poor outcome, namely, complex cytogenetics, CD13 positivity, and CD1a negativity. Achieving remission was not an issue in these poorer prognosis patients; relapse was the problem for CD1a− patients and death in remission for CD13+ patients. These patients are candidates for trials of aggressive therapy, such as alternative donor allografting in CR1 or the use of nelarabine to consolidate remissions. Future trials should consider individualizing therapy in high-risk patients, although minimal residual disease analyses may refine prognosis assignment and aid decision making.30-33 In the next study, patients who are minimal residual disease-positive after 2 cycles of induction will be eligible for unrelated donor allografts in CR1, although it remains uncertain whether high-dose chemoradiotherapy and cellular immunotherapy can cure these patients.33 Although our study identified biologic subgroups with a somewhat better outcome, we have not identified a group of patients whose outcome is good enough to contemplate de-escalation of therapy. Nonetheless, many questions remain and there are no standards of care for adults with T-cell ALL. To improve outcomes, investigators are urged to enter these rare patients into well-designed prospective clinical trials.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
A.V.M. thanks all member laboratories of the United Kingdom Cancer Cytogenetics Group for providing cytogenetic and FISH results and cell suspensions and Drs John Crolla and Fiona Ross and their teams at Wessex Regional Genetics Laboratory (Salisbury, United Kingdom) for the growing and preparation of the home-grown probes listed in “Cytogenetic, FISH, and molecular genetic investigations.”
This work was supported by the National Institutes of Health (R01CA120196; A.F.), the WOLF Foundation (A.F), and the Leukemia & Lymphoma Society (grants 1287-08 and 6237-08; A.F.). A.F. is a Leukemia & Lymphoma Society Scholar. A.V.M. was supported by Leukaemia Research United Kingdom. A Medical Research Council grant (G8223452) funded United Kingdom trial management.
National Institutes of Health
Authorship
Contribution: D.I.M., E.M.P., A.V.M., A.K.F., A.H.G., M.R.L., J.M.R., M.S.T., and H.M.L. designed research; D.I.M., E.M.P., A.V.M., G.D., A.F., R.P.K., and M.R.M. performed research; D.I.M., E.M.P., A.V.M., and H.M.L. wrote the paper; D.I.M., S.M.R., and G.B. analyzed data; and A.K.M. and S.M.L. critically reviewed the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: David I. Marks, Adult BMT Unit, University Hospitals Bristol NHS Foundation Trust, Upper Maudlin St, Bristol BS2 8BJ, United Kingdom; e-mail: David.Marks@ubht.nhs.uk.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal