Key Points
This report describes a multidisciplinary study characterizing the largest series of cyclin D1− MCL patients.
CCND2 translocations are the most frequent genetic event (55%) in cyclin D1− MCL.
Abstract
Cyclin D1− mantle cell lymphomas (MCLs) are not well characterized, in part because of the difficulties in their recognition. SOX11 has been identified recently as a reliable biomarker of MCL that is also expressed in the cyclin D1− variant. We investigated 40 lymphomas with MCL morphology and immunophenotype that were negative for cyclin D1 expression/t(11;14)(q13;q32) but positive for SOX11. These tumors presented clinically with generalized lymphadenopathy, advanced stage, and poor outcome (5-year overall survival, 48%). Chromosomal rearrangements of the CCND2 locus were detected in 55% of the cases, with an IG gene as partner in 18 of 22, in particular with light chains (10 IGK@ and 5 IGL@). No mutations in the phosphorylation motifs of CCND1, CCND2, or CCND3 were detected. The global genomic profile and the high complexity of the 32 cyclin D1− SOX11+ MCL patients analyzed by copy number arrays were similar to the conventional cyclin D1+/SOX11+ MCL. 17p deletions and high Ki67 expression conferred a significantly worse outcome for the patients. This comprehensive characterization of a large series of cyclin D1− MCL patients indicates that these tumors are clinically and biologically similar to the conventional cyclin D1+ MCL and provides a basis for the proper identification and clinical management of these patients.
Introduction
Mantle cell lymphoma (MCL) is genetically characterized by the t(11;14)(q13;q32) translocation that leads to the juxtaposition of the CCND1 gene to the immunoglobulin heavy chain (IGH@) gene, resulting in cyclin D1 overexpression.1,2 In recent years, there has been an ongoing debate after pathologists and geneticists identified a small subset of lymphomas resembling conventional MCL both morphologically and phenotypically but lacking the t(11;14)/CCND1 breakpoint and cyclin D1 expression. This observation prompted discussion regarding whether “true” MCL− for cyclin D1 exists. A gene-expression study shed light on this controversy, reporting that 6 recognized cyclin D1− lymphomas with morphologic, pathologic, clinical, and molecular features similar to typical MCL also had a similar global gene-expression profile (GEP).3 In the absence of cyclin D1, these cases overexpressed cyclin D2 or D3 but without evidence of chromosomal aberrations involving these loci. Subsequent studies have identified isolated cyclin D1− MCL harboring a t(2;12)(p12;p13) fusing the CCND2 gene to the IGK@ locus,4 t(12;22)(p13;q22) with IGL-CCND2 fusion,5 cryptic t(12;14)(p13;q32)/IGH-CCND26 or CCND2 breaks with unidentified partner.7 In addition, a translocation of the CCND3 gene has been detected in a single MCL patient,8 although more frequently in other mature B-cell malignancies.9 Nevertheless, these reports corresponded to isolated cases and the real incidence of CCND2 or CCND3 translocations has not been well studied in this MCL variant.
Because they are rare and not easy to recognize and reliable criteria for their diagnosis are lacking, cyclin D1− MCLs are not well characterized. Initial studies suggested that these tumors may be biologically and clinically similar to cyclin D1+ MCL, however, the number of cases investigated is small.3,10 In current trials of MCL, cyclin D1− cases have not been included. Given the increasing number of new therapeutic protocols developed for MCL and the improved survival of these patients,11 it is important to clarify whether cyclin D1− MCL is a variant of the same disease, because these patients could benefit from advances in the management of MCL.
The use of the immunohistochemical detection of cyclin D2 or cyclin D3 in routine diagnosis is not helpful in recognizing these tumors because these cyclins are also expressed in other small B-cell lymphomas.12,13 The SOX11 transcription factor has been found to be a reliable and highly specific biomarker for conventional cyclin D1+ MCL. In addition, it is not expressed in other low-grade B-cell lymphomas, offering a valuable practical tool to properly recognize cases of cyclin D1− MCL12,14-19 that were previously only identified by GEP.3
We report herein a complete characterization of 40 cyclin D1− MCL patients investigated for SOX11 expression and FISH for CCND2 and CCND3 translocations. In addition, mutational analysis of the cyclin D genes and high-resolution copy number (CN) profiling were performed.
Methods
Lymphoma samples and clinical data
The inclusion criteria were lymphomas with MCL morphology, MCL phenotype consistent with the disease, SOX11 expression, and negativity for cyclin D1 expression and/or t(11;14). A total of 41 patients were recruited, but 1 patient (ID45) with morphology and phenotype consistent with MCL but negative for SOX11 was excluded from further studies. Twenty-three patients were included in previous studies (cases 1-23).3,4,6-8,12 The morphology, growth pattern, and cyclin D1, cyclin D2, cyclin D3, cyclin E1, SOX11, p27, Ki67, CD5, CD23, and CD10 protein expression were evaluated in formalin-fixed paraffin-embedded (FFPE) tissue sections by immunohistochemistry (IHC) as described previously.12 Flow cytometry was performed in patient ID36 with leukemic involvement and no tissue available. Morphologic and clinical data for all patients included in the study are summarized in Tables 1 through 3 and supplemental Table 1 (available on the Blood Web site; see the Supplemental Materials link at the top of the online article). This study was approved by the institutional review board of the participating institutions where required and was conducted in accordance with the Declaration of Helsinki.
FISH analyses
Interphase FISH analysis was performed on FFPE sections, frozen tissue sections, or cells left over from prior cytogenetic analyses. Complete methods and probes used for the detection of breakpoints or gene fusions affecting the CCND1, CCND2, CCND3, IGH@, IGL@, and IGK@ loci are described in supplemental Methods and shown in supplemental Table 2.
DNA extraction and aCGH
Material from 27 samples was obtained for DNA extraction. DNA of 21 samples was extracted from FFPE tissue blocks using the phenol-chloroform extraction method according to previously published protocols with minor modifications.20 In addition, DNA of 6 cases was extracted from frozen tissue (3 cases), cells in fixative (2 cases), and DMSO-cryopreserved cells (1 case) using standard protocols. Tumor cell content was > 70% in all samples. Details on DNA extraction and array-comparative genomic hybridization (aCGH) methods are described in supplemental Methods. SurePrint G3 Human aCGH Microarray 1M (Agilent Technologies) hybridization was outsourced to qGenomics (www.qgenomics.com; Gene Expression Omnibus [GEO] accession no. GSE42854.) Detection of CN alterations (CNAs) was performed using the Aberration Detection Method-2 algorithm by Agilent Technologies and the Nexus 6.0 Discovery Edition (Biodiscovery). A noise-reduction method “waves aCGH correction algorithm” based on GC content and fragment size adjustment was applied.21 Previously published CN data (CGH and Affymetrix SNP-500K array) of 6 cyclin D1− MCL patients were included.22,23 Data from 92 previously reported cyclin D1+/SOX11+ MCL patients analyzed using SNP-100K and 500K arrays were used for the comparison of genetic profiles.23,24
Molecular analyses
In patients ID17 and ID36, from whom no tissue for IHC analysis of SOX11 was available, quantitative PCR (qPCR) of SOX11 expression was performed. In patients with available RNA, we analyzed cyclin D1, cyclin D2, cyclin D3, and cyclin E1 expression by qPCR. Detailed procedures are specified in supplemental Methods.
Mutational analysis of TP53 gene (exons 4-10) in patients with 17p alterations was performed as described previously.25 Mutations altering the phosphorylation motif of the CCND1, CCND2, and CCND3 genes were investigated in cases without cyclin D gene rearrangements. The primers used are described in supplemental Table 3. Sanger sequencing was performed on amplified products for TP53 and cyclin D genes using an ABI Prism 3100 (Applied Biosystems).
Statistical analyses
The χ2 test was used to compare morphologic and clinical characteristics between cyclin D1− and cyclin D1+ MCL patients. A probe-wise comparison of CNAs in cyclin D1− and cyclin D1+ MCL patients using the Fisher exact test was performed. Overall survival (OS) was defined as the time from MCL diagnosis to the date of the last follow-up or death as a result of any cause. Survival was estimated with the Kaplan-Meier method and survival curves were compared using the log-rank test. The Cox proportional hazards model was used to analyze prognostic factors. P < .05 was considered statistically significant.
Results
Histopathology and immunohistochemistry
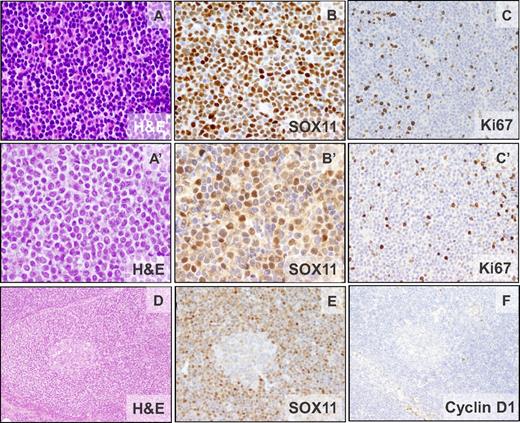
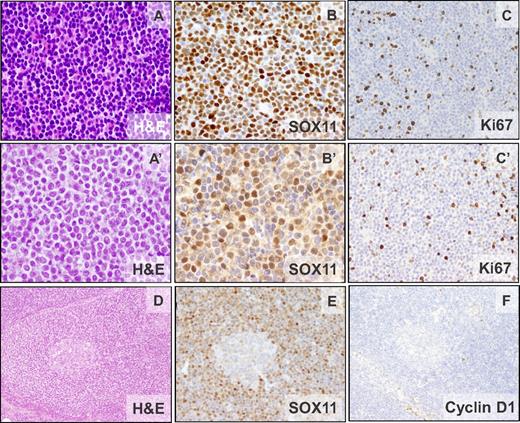
The pathologic features of 40 patients are summarized in Table 1 and supplemental Table 1. IHC analysis revealed strong positivity of SOX11 in 38 patients (Figure 1) and in 2 additional patients with only blood cells available, high SOX11 expression was demonstrated by qPCR. The morphology was classic in 38 patients, blastoid in 1 patient, and 1 patient progressed from classic morphology to pleomorphic 4 years after diagnosis. All patients had a diffuse/nodular growth pattern except 1 patient with a restricted mantle zone pattern (Figure 1D-F). Neoplastic cells were monomorphic and small to medium-sized with hyperchromatic indented nuclei. All patients were CD20+ and cyclin D1−. CD5 was positive in 39 of 40 (98%) patients, whereas CD10 was negative in 26 of 27 (96%) patients analyzed and positive in a subset of cells in 1 patient. CD23 was negative in 34 of 35 (97%) patients, with the only positive patient showing weak staining. p27 was negative or showed a weaker expression than the associated T cells in 29 of 31 (93%) patients. Nine of 24 (38%) patients had a high (≥ 35%) Ki67-proliferative index (Figure 1, Table 1, and supplemental Table 1). Six of 7 and 6 of 8 evaluated patients showed nuclear positivity for cyclin D2 or cyclin D3, respectively, but the staining was undistinguishable from that observed in other B-cell lymphomas (supplemental Table 4).12
Morphologic and IHC features of cyclin D1− MCL. Patients ID38 (CCND2 translocation–negative) and ID44 (IGK-CCND2 translocation–positive). (A-A′) Monomorphic small- to medium-sized cells with slightly irregular hyperchromatic nuclei (H&E; 400×). (B-B′) Nuclear positivity for SOX11. (C-C′) The cellular proliferation marker Ki67 shows positivity in 10%-20% of the neoplastic cells (IHC; 200×). (D) Cyclin D1− MCL with mantle zone growth pattern (ID42). Note the expanded mantle zone (H&E; 100×). (E-F) SOX11 is strongly positive (E) and cyclin D1 is not expressed in the neoplastic cells (F; IHC; 100×).
Morphologic and IHC features of cyclin D1− MCL. Patients ID38 (CCND2 translocation–negative) and ID44 (IGK-CCND2 translocation–positive). (A-A′) Monomorphic small- to medium-sized cells with slightly irregular hyperchromatic nuclei (H&E; 400×). (B-B′) Nuclear positivity for SOX11. (C-C′) The cellular proliferation marker Ki67 shows positivity in 10%-20% of the neoplastic cells (IHC; 200×). (D) Cyclin D1− MCL with mantle zone growth pattern (ID42). Note the expanded mantle zone (H&E; 100×). (E-F) SOX11 is strongly positive (E) and cyclin D1 is not expressed in the neoplastic cells (F; IHC; 100×).
FISH screening for CCND2 and CCND3 translocations, mRNA expression, and gene mutations
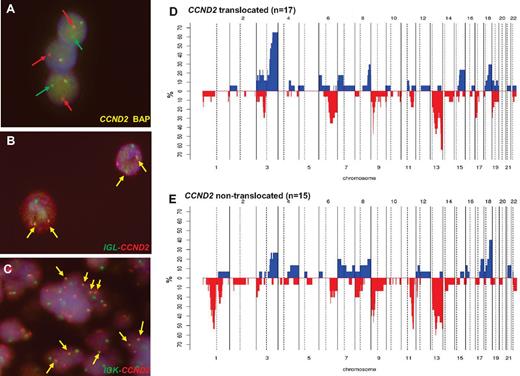
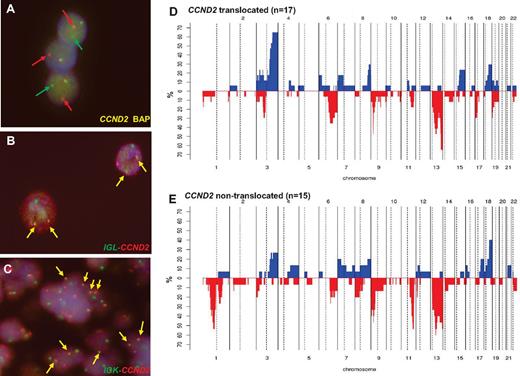
FISH information was gathered from contributors in 18 cases3,4,6-8,12 and 22 new patients were studied for CCND2 and CCND3 translocations and IG breaks (including 4 previously published cases). By a first FISH screening using break-apart probes, CCND2 breakpoints were recognized in 22 of the 40 patients (55%; Table 2): 10 patients had IGK-CCND2, 5 had IGL-CCND2, and 3 had IGH-CCND2 (Figure 2A-C). In the 4 additional patients, the partner of CCND2 remained undisclosed. In 2 of these 4 patients, the 3 IG genes were excluded as possible partners; in a third patient, the IGL@ probe failed repeatedly, and in the remaining patient, no additional material was available for further studies. In 18 patients (45%) in which no CCND2 breaks were identified, a CCND3 break-apart FISH test was performed. No CCND3 translocations were detected in any case. Complete FISH results are detailed in supplemental Table 5.
Genetic features of cyclin D1− MCL. (A) FISH analysis shows CCND2 breaks in patient ID36. (B) FISH image shows an IGL-CCND2 fusion in patient ID36. (C) FISH image shows and IGK-CCND2 fusion in patient ID28. Frequency of genetic alterations in CCND2 translocation–positive (D; n = 17) and CCND2 translocation–negative (E; n = 15). On the x-axis, the chromosomes are represented horizontally from 1-22; on the y-axis, the percentages of patients with CNAs are shown. Gains are represented on the positive y-axis and colored in blue and losses are represented on the negative y-axis in red.
Genetic features of cyclin D1− MCL. (A) FISH analysis shows CCND2 breaks in patient ID36. (B) FISH image shows an IGL-CCND2 fusion in patient ID36. (C) FISH image shows and IGK-CCND2 fusion in patient ID28. Frequency of genetic alterations in CCND2 translocation–positive (D; n = 17) and CCND2 translocation–negative (E; n = 15). On the x-axis, the chromosomes are represented horizontally from 1-22; on the y-axis, the percentages of patients with CNAs are shown. Gains are represented on the positive y-axis and colored in blue and losses are represented on the negative y-axis in red.
The levels of cyclin D1, D2, D3, and E1 expression were evaluated using qPCR in cyclin D1− MCL with and without CCND2 rearrangements. In addition, for comparison, conventional cyclin D1+ MCL and other B-cell neoplasms (diffuse large B-cell lymphoma [DLBCL], nodal marginal zone lymphoma, follicular lymphoma, chronic lymphocytic leukemia [CLL], and Burkitt lymphoma) were analyzed. We observed that CCND2-translocated MCL (n = 8) had high expression levels of cyclin D2 mRNA (mean 160 relative units) compared with the other groups (mean, 14.7 relative units; P = .003; supplemental Figure 1A). In addition, cyclin D3 and cyclin E1 mRNA levels were low in all samples analyzed (supplemental Figure 1B). CCND1, CCND2, and CCND3 mutational analysis performed in patients without rearrangements of these genes showed no mutations of the phosphorylation site (supplemental Table 6).
Genomic profiles of cyclin D1− MCL patients
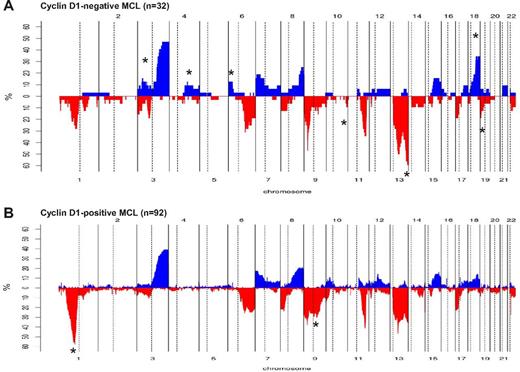
The genomic profile of 32 cyclin D1−/SOX11+ MCL patients (17 CCND2-translocated and 15 CCND2-nontranslocated cases) was analyzed with high-resolution aCGH and SNP array (Figures 2D-E and 3). CNAs were observed in all 32 patients, with a total of 225 losses, 102 gains, 9 homozygous deletions, and 6 high-level amplifications (supplemental Table 7). Both the global profile and the number of alterations per patient were similar to the previously reported cyclin D1+/SOX11+ MCL patients.2,22-24,26 Frequent gains were found at 3q26-q29 (47%), 8q24 (25%), and 18q21-q23 (31%). The only recurrent minimal region of amplification was 3p21.31-p22.3 (1 patient with and 1 without CCND2 translocation). Frequent losses were located at 1p22-p31 (22%), 1p13-p22 (22%), 6q21 (31%), 6q22 (28%), 9p21 (47%), 11q22-q23 (34%), 13q33-q34 (56%), 13q14-q21 (47%), and 17p13 (22%). Recurrent homozygous deletions were detected at 9p21.3 involving CDKN2A (7 patients), whereas biallelic losses in single cases were found at 13q14.3 and 17p13.1 (partially involving the TP53 gene).
Comparison of chromosomal imbalances detected by high-resolution CN array in cyclin D1− and cyclin D1+ MCL patients. (A) Relative frequency of genomic imbalances in cyclin D1− MCL patients (n = 32). (B) Previously reported cyclin D1+ MCL (n = 92). In both panels, the statistically significant gains and losses between both groups are indicated with an asterisk.
Comparison of chromosomal imbalances detected by high-resolution CN array in cyclin D1− and cyclin D1+ MCL patients. (A) Relative frequency of genomic imbalances in cyclin D1− MCL patients (n = 32). (B) Previously reported cyclin D1+ MCL (n = 92). In both panels, the statistically significant gains and losses between both groups are indicated with an asterisk.
Cyclin D1− MCL patients with and without CCND2 rearrangements had similar CN profiles (Figure 2D-E and supplemental Table 8), with median number of CNAs similar between the groups (10 vs 12 CNAs/case, respectively). In addition, no significant differences in terms of CNAs were detected between CCND2 translocation–positive patients with the IGK@ (n = 7) and IGL@ (n = 4) genes as partners (supplemental Figure 2).
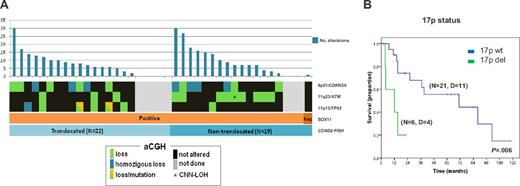
Similar to cyclin D1+ MCL patients, a high proportion (15 of 32, 47%) of cyclin D1−/SOX11+ MCL patients had 9p21 deletions of different sizes and half were homozygous. All 9p21 deletions spanned the CDKN2A, CDKN2B, and MTAP loci (supplemental Figure 3 and supplemental Table 6). The 17p deletions, including the TP53 gene, were observed in 7 of 32 patients (22%). Patient ID35 had a small homozygous deletion (68.2 kb) affecting the promoter region and exon 1 (detected by aCGH). Mutational analysis of the TP53 gene was performed in 4 patients with available material. Two patients carried mutations of the remaining allele (ID18 and ID40). In patient ID38, exons 7 and 10 were unmutated and no material was available for additional analyses and ID34 was found to be unmutated (supplemental Table 6). All 12 patients with 11q alterations involved the target gene ATM. Interestingly, CCND2-translocated MCL patients had more frequent 17p deletions than nontranslocated patients (35% vs 7%, P = .05) whereas nontranslocated MCL had more frequent losses of 11q (60% vs 18%, P = .01) and 1p (67% vs. 12%, P = .011). The frequency of 9p deletions was extremely high and equal in the subgroups (47%; Figure 4A).
Characterization of cyclin D1− MCL patients according to their genetic and molecular features. (A) The genomic complexity is illustrated with bar plots at the top of the panels, reflecting the number of alterations for each patient. The plots below indicate the SOX11 expression and the presence of CCND2 translocations. Orange, SOX11+ MCL; brown: SOX11− MCL; light blue, CCND2 translocation–positive MCL; and dark blue, CCND2 translocation–negative MCL. The status of CDKN2A, ATM, and TP53 genes according to aCGH and SNP-array data are indicated by color (black, not altered; green, loss; blue, homozygous loss; green/orange, loss and mutation; and gray, unknown). (B) Kaplan-Meier estimates of OS for MCL cyclin D1− patients according to 17p status (17p altered vs 17p not altered). D indicates dead.
Characterization of cyclin D1− MCL patients according to their genetic and molecular features. (A) The genomic complexity is illustrated with bar plots at the top of the panels, reflecting the number of alterations for each patient. The plots below indicate the SOX11 expression and the presence of CCND2 translocations. Orange, SOX11+ MCL; brown: SOX11− MCL; light blue, CCND2 translocation–positive MCL; and dark blue, CCND2 translocation–negative MCL. The status of CDKN2A, ATM, and TP53 genes according to aCGH and SNP-array data are indicated by color (black, not altered; green, loss; blue, homozygous loss; green/orange, loss and mutation; and gray, unknown). (B) Kaplan-Meier estimates of OS for MCL cyclin D1− patients according to 17p status (17p altered vs 17p not altered). D indicates dead.
We next compared the profile of CNAs with 92 cyclin D1+/SOX11+ MCL patients from our previously published studies.20,22-24 Cyclin D1− MCL patients (n = 32) had more frequent deletions of 10q26, 13q32-q34, and 19p13 and gains of 3p24-p22, 4q25-q28, 4q31, 6p25-p22, and 18q21-q23, but a low incidence of 1p32-p21 and 9q21-q22 deletions (Figure 3A-B and supplemental Table 8).
Clinical features of cyclin D1−/SOX11+ MCL patients
The clinical characteristics of the patients are summarized in Table 2. There were 29 men and 11 women with a median age of 63 years (range, 33-90). Lymphadenopathy (mainly cervical and inguinal) was reported in 38 of 39 (97%) patients with available information. Extranodal involvement was identified in 37 of 39 (95%) patients. Two patients had multiple intestinal lymphomatous polyposis, an uncommon but highly characteristic presentation of conventional MCL. High serum levels of lactate dehydrogenase and B-symptoms were detected in 8 of 23 (35%) and 5 of 19 (26%) patients, respectively.
All but 2 patients with follow-up information received treatment, including Adriamycin-containing chemotherapy in 5 patients, Adriamycin-containing immunochemotherapy in 14, and other treatments in 8. The 5-year OS of the patients was 48% (95% confidence interval, 28%-68%; supplemental Figure 4A). High serum lactate dehydrogenase and high expression of Ki67 (≥ 35%) significantly predicted poor OS (P < .001 and P = .03, respectively; supplemental Figure 4B-C). Regarding genetic alterations, patients carrying 17p deletions had a significant worse prognosis than those without this aberration (median survival: 12 vs 71 months, respectively, P = .006; Figure 4B). Neither the genetic complexity nor any other loss or gain had a significant impact on OS. In a bivariate analysis including Ki67 expression and 17p deletion, only the latter remained as an independent prognostic parameter for OS (P = .047; relative risk = 13; 95% confidence interval, 1.0-160). Finally, the main clinical features and outcome were similar for CCND2-translocated and CCND2-nontranslocated patients.
Discussion
MCL is a well-characterized lymphoid neoplasm with the t(11;14) as the primary genetic event leading to overexpression of cyclin D1 and cell-cycle deregulation. Several findings have suggested the existence of a particular subset of tumors with the same morphology and phenotype as conventional MCL but lacking cyclin D1 expression and the t(11;14) translocation.3-8 The transcription factor SOX11 has been demonstrated to be a reliable marker for MCL in both cyclin D1+ and cyclin D1− patients.12,14,18 We report herein the largest series of well-defined cyclin D1− MCL patients with a molecular and genetic comparative study to the conventional cyclin D1+ MCL. The suitability of adapted high-resolution CN arrays and FISH analyses for the examination of FFPE material made it possible to characterize the genomic profile of these tumors at high resolution.
For a well-defined series of MCL patients, only those with consistent morphology, phenotype, and SOX11 positivity were selected for further analysis. Based on these criteria, only 1 patient with consistent morphology/phenotype was excluded because of a lack of SOX11 expression. This case presented a very simple CN profile with only 1 small deletion at 17q12 that was concordant with its indolent clinical behavior. The morphology of all patients was indistinguishable from cyclin D1+ MCL. Only 1 patient in the present series had blastoid morphology at diagnosis (3%), a frequency lower than in most series of conventional cyclin D1+ MCL patients. This suggests that cyclin D1− MCL with blastoid or pleomorphic morphology might be underappreciated and probably classified in other categories such as DLBCL.14 All patients had diffuse/nodular growth pattern except 1 with a mantle zone pattern. As would be expected for MCL, almost all patients were positive for CD5 and negative for CD23, CD10, and p27.
FISH screening demonstrated that more than half of the cyclin D1− MCL patients carried CCND2 rearrangements with IG genes. Interestingly, in 2 patients, interphase FISH confirmed a clear CCND2 break with normal IGH@, IGK@, and IGL@ genes, indicating the probability of a novel translocation partner in addition to the described translocations. As expected and observed in previous studies, high levels of cyclin D2 mRNA were detected only in patients with CCND2 translocation.7 In MCL patients without CCND2 rearrangement, we cannot exclude CCND2 breakpoints with far 5′ or 3′ breaks as well as insertions of CCND2 into another locus and vice versa. However, this seems unlikely because these patients, in contrast to the CCND2-translocated patients, have very low levels of cyclin D2, suggesting that other genetic mechanism(s) must be involved in the pathogenesis of these lymphomas. A previous report identifying a CCND3 translocation in 1 MCL patient8 could not be substantiated in this large series.
Mutations in the phosphorylation site of cyclin D genes have been observed in DLBCL, CLL, and Burkitt lymphoma.27-29 These mutations eliminate the phosphorylated residue needed for the nuclear export of the cyclins and subsequent degradation by the proteasome. Therefore, these mutations generate a more stable protein with high levels of expression. No cyclin D mutations were found in the nonrearranged CCND2 patients, excluding these mutations as an alternative mechanism to cyclin D translocations in these tumors. Further molecular studies will be needed to determine the primary genetic alteration of the MCL patients without cyclin D translocation or mutations that in the present series represented up to 45% of the cases.
The overall genetic complexity and the genomic profile of the 32 cyclin D1−/SOX11+ patients was similar to those described in cyclin D1+ MCL.2,22-24,26 The frequency of deletions targeting specific loci (eg TP53, CDKN2A, and ATM deletions) was similar between cyclin D1− and cyclin D1+ MCL. However, in the CCND2-translocated MCL patients, the high-risk-conferring 17p loss was observed frequently, whereas 11q loss was more frequently observed in patients without CCND2 translocation.
The proportion of cyclin D1− MCL patients with 9p losses was higher in both the CCND2 translocation–positive and –negative subgroups (47%) compared with cyclin D1+ MCL patients (7%-30% by conventional CGH, 10%-36% by array platforms).2 Moreover, the pattern of chromosome 9 losses, and specifically 9p in cyclin D1− MCL, was complex; the repertoire included patients with single CDKN2A deletions (focal, larger, or spanning the whole 9p arm), narrow homozygous deletions, monosomy of whole chromosome 9, patients with multiple independent losses in 9p, and patients with independent losses of 9q. The mechanism triggering 9p21 deletions has been well documented in lymphoblastic leukemias,30 in which the deletions are formed at a few defined regions by illegitimate activity of the RAG protein complex in V(D)J recombination; conversely, the double strand breaks in solid tumors with 9p21 deletions are formed at unspecific regions by microhomology-mediated mechanisms. To determine the exact mechanism operating in MCL, a detailed analysis of sequences of the breakpoints would be needed.
Interestingly, 2 patients had gains of the mir17-92 cluster at 13q31.3. This cluster has been demonstrated experimentally to act as an oncogene in cooperation with MYC. Its amplification and overexpression are present in different types of lymphomas, including MCL.31-33
The existence of cyclin D1− MCL was confirmed initially by GEP analysis of patients with the suggested pathologic features. However, this technique is not applicable in clinical practice. In contrast, the simple detection of SOX11 expression by IHC may help in the identification of these tumors, and this prompted us to perform FISH analysis looking for CCND2 rearrangements or using RT-PCR to quantify the CCND2 mRNA levels, all routine techniques that can be applied in FFPE material.
One important question is whether patients lacking both CCND1 and CCND2 translocations represent “true” MCL and not misclassified cases of CD5+ marginal zone lymphoma or CLL. The differential diagnosis is challenging because these entities may mimic MCL morphologically. However, the strong expression of SOX11 in the tumor cells, together with the lack of expression of p27 and CD23, makes this possibility very unlikely. Indeed, the differential diagnosis is important and relevant for patient treatment and prognosis.
Although clinically similar, the cyclin D1− MCL patients presented extranodal involvement more frequently than cyclin D1+ MCL patients (Table 3). Despite the low number of patients, the 17p deletion was shown to have an adverse impact on outcome.
In conclusion, to our knowledge, this is the largest series to date of this peculiar variant of MCL. From a clinical point of view, the majority of the patients diagnosed with cyclin D1− MCL have similar pathologic and genetic characteristics compared with cyclin D1+ MCL patients. The clinical presentation and evolution are also similar to cyclin D1+ patients. These observations suggest that these patients may benefit from the same therapeutic protocols used for the management of patients with conventional MCL. The frequent CCND2 translocations together with SOX11 expression may be very valuable diagnostic tools with which to identify these patients. Understanding of the alternative molecular mechanisms involved in cyclin D2− and cyclin D1− MCL will require further studies.
There is an Inside Blood commentary on this article in this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Miriam Prieto, Reina Zühlke-Jenisch, Magret Ratjen, and Manel García-Aragonés for excellent technical assistance and Frédéric Leprêtre and Lluís Armengol for assistance in the application and adaptation of the aCGH correction algorithm to the high-resolution aCGH data.
This study was supported by the Instituto de Salud Carlos III, Fondo de Investigaciones Sanitarias (PI08/0077; PI11/01177 to S.B.), Alexander von Humboldt Foundation (to I.S.) and Subprograma Juan de la Cierva Ministerio de Ciencia e Innovación (JCI-2011-10232 to I.S.), Instituto de Salud Carlos III, Beca Predoctoral de Formación en Investigación en Salud (FI08/00347 to C.R.) Comisión Interministerial de Ciencia y Tecnología Española (SAF08/3630 to E.C.), Red Temática de Investigación Cooperativa del Cáncer (RD06/0020/0039 ISCIII to E.C.), and the Spanish Ministry of Science and Innovation and European Regional Development Fund (Unió Europea “Una manera de fer Europa”).
Authorship
Contribution: I.S. and C.R. performed the molecular analysis, analyzed the data, and wrote the manuscript; A.C.-C., J.Y.S., R.W., G.R., W.K., E.M.H., P.S., I.W., J.A.F., P.G., G.O., A.R., A.L-G., L.Q-M., N.L.H., E.S.J., and R.S. provided samples and clinical data and performed the pathology or molecular review; G.C. performed the statistical analysis; A.N. and A.V. performed the molecular analysis; and I.S., E.C. and S.B. conceived the project, analyzed the data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Sílvia Beà, Centre Esther Koplowitz, 2nd Floor, Rosselló, 153, 08036-Barcelona, Spain; e-mail: sbea@clinic.cat.
References
Author notes
I.S. and C.R. contributed equally to this work.