In this issue of Blood, Salaverria et al report that more than half of Cyclin D1- (CCND1) negative SOX11-positive mantle cell lymphoma (MCL) had CCND2 gene rearrangement predominantly with immunoglobulin (IG) light chain genes.1
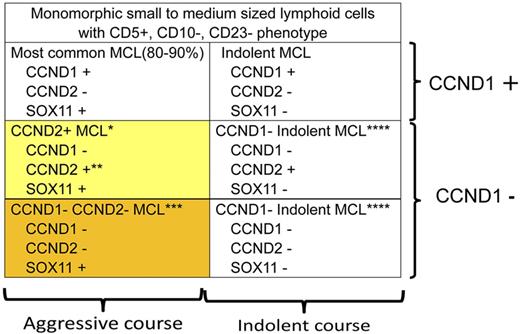
MCL is characterized by monomorphic small to medium-sized lymphoid cells with CD5+, CD10−, CD23− phenotype.2 CCND1 rearrangement and/or CCND1 immunostaining is a hallmark of diagnosis. However, it has been known that CCND1-negative MCL with a poor clinical outcome exists. Recently, SOX11 has been shown to be a good marker to identify such cases with poor outcome,5 although a controversy has been raised.7 Salaverria et al revealed that CCND2 gene rearrangement is frequently found (22 of 40 CCND1-negative MCL cases, 55%) and is a molecular basis for CCND1-negative SOX11-positive MCL (yellow box).1 Further study on molecular mechanisms of CCND1-negative CCND2-negative SOX 11-positive MCL with a poor clinical outcome (orange box) to provide appropriate therapy is needed. *More than half (22/40) of the CCND1-negative SOX11-positive MCL show CCND2 gene rearrangement. **Translocation of CCND2 takes place predominantly with IG light chain genes (15/22). ***Among CCND1-negative SOX11-positive MCL cases, 18 of 40 did not show CCND2 translocation. Because controversy exists on SOX11 staining with regard to clinical outcome, further study including identification of other diagnostic markers and molecular basis is warranted. ****CCND1-negative indolent MCL has not been well recognized and would be rare, but its understanding is important for selecting appropriate therapy and for differential diagnosis from the other types of lymphoma.
MCL is characterized by monomorphic small to medium-sized lymphoid cells with CD5+, CD10−, CD23− phenotype.2 CCND1 rearrangement and/or CCND1 immunostaining is a hallmark of diagnosis. However, it has been known that CCND1-negative MCL with a poor clinical outcome exists. Recently, SOX11 has been shown to be a good marker to identify such cases with poor outcome,5 although a controversy has been raised.7 Salaverria et al revealed that CCND2 gene rearrangement is frequently found (22 of 40 CCND1-negative MCL cases, 55%) and is a molecular basis for CCND1-negative SOX11-positive MCL (yellow box).1 Further study on molecular mechanisms of CCND1-negative CCND2-negative SOX 11-positive MCL with a poor clinical outcome (orange box) to provide appropriate therapy is needed. *More than half (22/40) of the CCND1-negative SOX11-positive MCL show CCND2 gene rearrangement. **Translocation of CCND2 takes place predominantly with IG light chain genes (15/22). ***Among CCND1-negative SOX11-positive MCL cases, 18 of 40 did not show CCND2 translocation. Because controversy exists on SOX11 staining with regard to clinical outcome, further study including identification of other diagnostic markers and molecular basis is warranted. ****CCND1-negative indolent MCL has not been well recognized and would be rare, but its understanding is important for selecting appropriate therapy and for differential diagnosis from the other types of lymphoma.
Mantle cell lymphoma (MCL) is a B-cell neoplasm composed of monomorphic small to medium-sized lymphoid cells. CCND1 translocation and CCND1 nuclear expression are used for diagnostic purposes and distinguish MCL from other types of lymphomas.2
Since the introduction of anti-CCND1 monoclonal antibody for immunostaining, it has been used to delineate MCL from the other types of lymphoma. CCND1-positive MCL has a poor prognosis. However, CCND1-negative MCL has remained controversial due to its variable clinicopathologic features.3,4
Recent study on indolent CCND1-positive MCL with the most common poor prognosis type MCL revealed that SOX11 is a good marker to distinguish the latter from the former.5
SOX11 is a neural transcription factor involved in central nervous system development and is over-expressed in gliomas compared with normal brain tissue, suggesting a role in malignant transformation. Immunostaining of SOX11 at nuclei in MCL was reported6 but the prognostic significance of this finding remains unclear.7 This point must be further clarified to establish appropriate therapies for each MCL subtype.
The molecular pathogenesis of CCND1-negative MCL with a poor clinical outcome has not been defined, but it was shown that more than one-half of such cases (22/40) had CCND2 translocation, most likely resulting in deregulation of CCND2 expression (see figure).1 Indeed, a preceding report of CCND1-negative MCL cases with CCND2 translocation has shown CCND2 overexpression in nuclei.8 Cyclin-D families (D1, D2, D3) are differentially expressed in various hematopoietic malignancies and are believed to play roles in malignancies.9 Therefore, it is possible that CCND3 plays a role in lymphoma development for CCND1-negative MCL, but Salaverria et al revealed no CCND3 rearrangement, suggesting mechanisms other than cyclin-D family pathways exist.1
The cases without CCND2 gene rearrangement (18/40) had a poor clinical outcome and are positive for SOX11 expression. This may indicate that the pathways involving SOX11 play a role in lymphoma development of CCND1- and D2-negative MCL. Exploration of both CCND1- and D2-negative SOX11-positive MCL (see figure) would lead to a clue underlying molecular mechanisms of MCL development including the most typical MCL.
It should be also noted that CCND2 predominantly translocates to IG light chain genes (IGK@ or IGL@; 15/22, 68%), and only 3 cases show IG heavy chain (IGH@) gene translocation. The remaining 4 of 22 cases are undisclosed.1 IG gene translocation with CCND1, BCL2, or BCL6 is a valuable diagnostic marker, but involvement of IGH@ gene is more frequent than the cases reported by Salaverria et al.
Non-Ig translocation of CCND2 is also noted,1 although approximately 40% cases with BCL6 translocation have been shown to possess non-IG gene translocation.10
High frequency of IG light chain gene translocation may indicate that the translocation takes place in a different stage of B-cell differentiation compared with the stages where CCND1, BCL2 or BCL6 translocation occurs. Alternatively, genomic and/or epigenomic configurations unique to CCND1-negative MCL may cause different environments for chromosome translocations.
CCND1-negative MCL is rare, but identification of CCND2 gene rearrangement in such cases provides a very robust marker indicating the need for intensive therapy. This report by Salaverria and colleagues surely opens a gate to better understanding the whole picture of molecular mechanisms of MCL development.
Conflict-of-interest disclosure: The author declares no competing financial interests. ■

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal