Key Points
CUX1 is a transcription factor encoded on a region of chromosome 7 that is frequently deleted in high-risk acute myeloid leukemia.
Haploinsufficiency of CUX1/cut promotes hematopoietic overgrowth in both Drosophila melanogaster and human xenograft mouse models in vivo.
Abstract
Loss of chromosome 7 and del(7q) [−7/del(7q)] are recurring cytogenetic abnormalities in hematologic malignancies, including acute myeloid leukemia and therapy-related myeloid neoplasms, and associated with an adverse prognosis. Despite intensive effort by many laboratories, the putative myeloid tumor suppressor(s) on chromosome 7 has not yet been identified. We performed transcriptome sequencing and SNP array analysis on de novo and therapy-related myeloid neoplasms, half with −7/del(7q). We identified a 2.17-Mb commonly deleted segment on chromosome band 7q22.1 containing CUX1, a gene encoding a homeodomain-containing transcription factor. In 1 case, CUX1 was disrupted by a translocation, resulting in a loss-of-function RNA fusion transcript. CUX1 was the most significantly differentially expressed gene within the commonly deleted segment and was expressed at haploinsufficient levels in −7/del(7q) leukemias. Haploinsufficiency of the highly conserved ortholog, cut, led to hemocyte overgrowth and tumor formation in Drosophila melanogaster. Similarly, haploinsufficiency of CUX1 gave human hematopoietic cells a significant engraftment advantage on transplantation into immunodeficient mice. Within the RNA-sequencing data, we identified a CUX1-associated cell cycle transcriptional gene signature, suggesting that CUX1 exerts tumor suppressor activity by regulating proliferative genes. These data identify CUX1 as a conserved, haploinsufficient tumor suppressor frequently deleted in myeloid neoplasms.
Introduction
Loss of chromosome 7 and del(7q) [−7/del(7q)] was first recognized as a frequent event in acute myeloid leukemia (AML) nearly 40 years ago.1 −7/del(7q) occurs in 8% of de novo AML2 and 50% of therapy-related myeloid neoplasms (t-MNs).3 −7/del(7q) is also found in myelodysplastic syndromes, AMLs arising from myeloproliferative neoplasms, the blast phase of chronic myelogenous leukemia, Ph+ acute lymphoblastic leukemia, and AMLs associated with inherited syndromes.4-10 −7/del(7q) is an adverse-risk prognostic indicator in myeloid disorders, and the long-term outcome for patients is typically poor. The median overall survival for patients with de novo AML or t-MNs with −7/del(7q) is ∼ 6 months.2,3
Loss of 1 or more tumor suppressor gene(s) (TSGs) is thought to contribute to leukemic growth in myeloid malignancies with −7/del(7q). Several groups have mapped a commonly deleted segment (CDS) of chromosome band 7q22 using polymorphic markers, conventional cytogenetic analysis, and FISH analysis.11-13 In one study of 81 patients with malignant myeloid disorders characterized by chromosome 7 abnormalities, the CDS was mapped to a 2.52-Mb region of 7q22 by FISH using YAC clones.11 However, deletion of a 2-Mb syntenic region in mice did not result in overt myeloid disease.14 Other studies have mapped rearrangements involving 7q22 and identified similar15 or slightly more centromeric intervals (Figure 1).12,13,16
CUX1 is within the 2.17-Mb CDS of 7q22.1. Copy number analysis of 7q derived from SNP arrays of 35 samples of de novo AML or t-MNs. Thirty-four samples are from primary leukemia samples, and UoCM1 is a cell line derived from the leukemia cells of a patient with de novo AML. Seventeen samples have −7/del(7q) detected by cytogenetic analysis as depicted above the sample identifiers. The percentage of cells exhibiting the abnormality is indicated in brackets. T12 has a cryptic deletion in 7q22.1 not visible by conventional cytogenetic analysis. The CDS spans 49 genes; however, only the genes bordering the CDS are shown for clarity. Data are visualized with Integrated Genome Viewer Version 2.0.50 Previously identified commonly deleted segments are shown on the right panel, and genomic coordinates are indicated in build hg19. LOH indicates loss of heterozygosity.
CUX1 is within the 2.17-Mb CDS of 7q22.1. Copy number analysis of 7q derived from SNP arrays of 35 samples of de novo AML or t-MNs. Thirty-four samples are from primary leukemia samples, and UoCM1 is a cell line derived from the leukemia cells of a patient with de novo AML. Seventeen samples have −7/del(7q) detected by cytogenetic analysis as depicted above the sample identifiers. The percentage of cells exhibiting the abnormality is indicated in brackets. T12 has a cryptic deletion in 7q22.1 not visible by conventional cytogenetic analysis. The CDS spans 49 genes; however, only the genes bordering the CDS are shown for clarity. Data are visualized with Integrated Genome Viewer Version 2.0.50 Previously identified commonly deleted segments are shown on the right panel, and genomic coordinates are indicated in build hg19. LOH indicates loss of heterozygosity.
In this study, we used single nucleotide polymorphism (SNP) arrays to map the 7q22 CDS with resolution of ∼ 1 kb. Overlaying transcriptome-sequencing with copy number aberrations, we identified the gene encoding the transcription factor, CUX1, to be within the CDS and expressed at haploinsufficient levels. Haploinsufficent levels of the CUX1 ortholog, cut (ct), in Drosophila melanogaster hemocytes led to hemocyte overproliferation and melanotic tumor formation in vivo. Similarly, decreased expression of CUX1 led to an engraftment advantage for human hematopoietic progenitors transplanted into immunodeficient mice. Thus, from invertebrates to humans, CUX1/cut is a conserved, haploinsufficient hematopoietic TSG.
Methods
Patient samples
This research was approved by the University of Chicago Institutional Review Board. Leukemia samples were obtained from the University of Chicago Hematopathology and Cancer Cytogenetics Laboratories (supplemental Table 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article) with informed consent per the Declaration of Helsinki.
SNP array
RNA sequencing library preparation
Paired-end libraries were prepared following the standard protocol recommended by Illumina. In brief, purified mRNA (MicroPoly(A) Purist kit, Ambion) was fragmented for 7 minutes at 85°C, and first-strand cDNA generated (Superscript II Reverse Transcriptase, Invitrogen) with random hexamers (Invitrogen), followed by second-strand synthesis with RNaseH and DNA Polymerase I (New England Biolabs). cDNA was repaired and polished with T4 DNA polymerase, Klenow, and T4 PolyNucleotide Kinase (New England Biolabs), followed by adenosine addition with Klenow Fragment 3′ → 5′ exo− (New England Biolabs). Paired-end adaptors (Illumina) were ligated with DNA T4 ligase, and libraries were amplified with p5 and p7 primers (Illumina) and Platinum Pfx taq polymerase (Invitrogen) for 18 PCR cycles; 450-bp fragments were gel-extracted using QIAquick Gel Extraction kit (QIAGEN).
Sequence analysis
Paired-end reads of lengths 36-100 bp were generated on the Illumina Genome Analyzer II. Reads were individually trimmed from the 3′ end, such that the trimmed 3′ bases had an average Phred-scaled quality score < 15. Reads were aligned to the human reference genome hg18 using TopHat (Version 1.3.1)18 and output in the BAM format.19 Further alignment manipulation was performed with SAMTools (Version 0.1.18),19 Picard (Version 1.52, http://picard.sourceforge.net), and custom perl scripts. Read alignments with mapping qualities < 10 were removed. Alignments were refined with the Genome Analysis Toolkit (Version 1.0.5) RealignerTargetCreator and IndelRealigner tools.20 Cufflinks (Version 1.3.0) was used to estimate transcript abundance using RefSeq gene models and output as Fragments Per Kilobase per Million mapped reads (FPKM) with upper-quartile normalization (supplemental Table 2).21 Statistical tests were performed on log2-transformed data after addition of 1 pseudocount. RNA-sequencing data are in the Short Read Archive SRA061655.
RT-PCR
cDNA was generated using the High Capacity RNA-to-cDNA Master Mix (Applied Biosystems). CUX1 was amplified with Power SYBR Green PCR Master Mix (Invitrogen) on the ABI Step One Plus (Applied Biosystems). cut was amplified from cDNA from hemocytes pooled from 6 larvae per group, using EXPRESS qPCR Supermix (Invitrogen) and detected with a dual-labeled probe (IDT). Primer sequences are provided (supplemental Table 3). Samples were run in triplicate, and expression levels relative to a housekeeping control gene were determined using the comparative CT method according to the manufacturer's software.
Fusion identification
deFuse22 (Version 0.5.0) was used to identify chimeric transcripts requiring > 7 split reads, > 7 spanning reads, splitr_span P value > 0.2, split position P value > 0.2, breakpoint homology < 11, and breakpoints in coding regions. To identify reads mapping to the fusion breakpoint junction, exon-exon junctions between the 2 genes were constructed and all reads were mapped to this junction library with BWA (Version 0.5.9).23 Reads with < 3 mismatches and > 14 bp anchor region around the junction were identified.
Cell culture
K562, Kasumi-1, KG-1, NB4, Mono7, and U-937 human AML cell lines, and HeLa adenocarcinoma cells were maintained in RPMI 1640-GlutaMAX (Invitrogen) supplemented with 10% fetal bovine serum (Invitrogen), 100 U/mL penicillin, 100 μg/mL streptomycin, and 0.25 μg/mL amphotericin B (Invitrogen). Cells were grown at 37°C in 5% CO2.
Western blot
A total of 2 million cells were lysed in RIPA with sodium orthovanadate, protease inhibitor cocktail, and phenylmethylsulfonyl fluoride (Santa Cruz Biotechnology). A total of 40 μg of supernatant, quantified by Micro BCA kit (Thermo Scientific), was electrophoresed on a 4%-15% Mini-PROTEAN TGX gradient gel (Bio-Rad) and transferred to nitrocellulose. Blots were blocked with Odyssey Blocking buffer (LI-COR) and probed with anti-CUX1 at 1:200 (SC-6327, Santa Cruz Biotechnology), followed by donkey anti–goat AlexaFluor-680 (Invitrogen). Mouse anti–β-actin (SC-4778, Santa Cruz Biotechnology) followed by goat anti–mouse IRDye 800CW (LI-COR) was used for normalization. Fluorescence was quantified by Odyssey imaging (LI-COR).
Fly stocks
Fly stocks were obtained from the Bloomington Drosophila Stock Center. To express the RNAi in larval hemocytes, hmlΔ-Gal424 or cg-Gal425 was used as the driver line to cross with the UAS-RNAi and UAS-2xEGFP lines. The following stocks were used: ctRNAi A, ctRNAi B, and luciferase RNAi (nonspecific control). RNAi-negative control larvae were from the crosses of hmlΔ-Gal4 or cg-Gal4,UAS-2xEGFP and w1118. All the crosses were set up at 25°C on standard cornmeal media.
Larval hemocyte counts
Circulating hemocytes from wandering third instar larvae were collected by opening the larval cuticles in 20 μL PBS. A total of 1 μL of hemocytes was fixed in 4% paraformaldehyde (Electron Microscopy Sciences), stained with 10 ng/mL Hoechst 33342 (Invitrogen), and counted using EBImage26 under a DM5000 B microscope (Leica).
Sanger sequencing
Sanger sequencing of CUX1 exons was performed on the 35 samples studied by SNP array plus an additional 109 leukemia samples consisting of 57 de novo AMLs, 47 t-MNs, and 5 AML cell lines (K562, Kasumi-1, KG-1, NB4, and U-937). Whole genome amplified DNA (REPLI-g, QIAGEN) was amplified using Taq 5X Master Mix (New England Biolabs) with primers for CUX1.27 PCR product was treated with 3.3 units Exonuclease I and 0.3 units Shrimp Alkaline Phosphatase (Affymetrix) for 30 minutes at 37°C followed by 15 minutes at 80°C. Sequencing traces were analyzed using MutationSurveyor Version 4.00 software (SoftGenetics).
Xenografts
Lentiviral GFP-IRES-shRNA pGIPZ vectors purchased from Thermo Scientific are as follows: CUX1 shRNA A (clone V2LHS_151077), CUX1 shRNA B (clone V2LHS_151078), and nonspecific shRNA control. Lentivirus was produced by transient transfection of pGIPZ, pCMV-dR8.74, and pMD2 VSVG in HEK293T cells and ultracentrifugation of supernatant. Human cord blood was collected at the University of Chicago. Hematopoietic stem cells were purified from Ficoll gradient-separated white blood cells by Lineage Cell Depletion Kit (Miltenyi Biotec) and cultured for 24 hours in growth media: Stemline II media (Sigma-Aldrich) supplemented with 50 ng/mL each of IL-3, IL-6, thrombopoietin, SCF, and Fms-like tyrosine kinase 3 ligand (R&D Systems). Transduction was performed by spinning cells on Retronectin-coated plates (Takara) with viral stock in Stemline II media at 2500 rpm for 90 minutes at 30°C on day 1 and repeated on day 2. After the spin, supernatant was removed and replaced with growth media. On day 3, cells were assayed for GFP expression by flow and 1 × 105 cells were injected via facial vein in 1- to 2-day-old NOD.Cg-PrkdcscidIL2rgtm1Wjl/SzJ (NOD scid gamma, NSG, The Jackson Laboratory) pups after irradiation with 115 cGy. Retro-orbital blood was collected and stained with the following antibodies (eBioscience): anti–mouse CD45-allophycocyanin-Cy7, anti–human CD45- peridinin chlorophyll protein, anti–human CD13-PE, anti–human CD33-allophycocyanin, and anti–human CD3-Pe-Cy7. Flow cytometry was performed on an LSR II (BD Biosciences) and analyzed by FlowJo (TreeStar). Sorting was performed on a FACSAria II (BD Biosciences).
K562 transfection
K562 cells were transfected with pGIPZ-shRNA by Nucleofector (Lonza) per the manufacturer's instructions and stably selected with 3 μg/mL puromycin (Invitrogen).
Statistical analysis
CUX1-associated genes were identified by regressing the expression level of each gene against the expression level of CUX1 using the function lm in the statistical package R. The significance of regression coefficients was quantified by t test. False discovery rates (FDR) were estimated by the Storey q-value method.28 Unless otherwise noted, ± indicates SD, and P values were calculated by t test.
Results
CUX1 resides within a commonly deleted segment of chromosome band 7q22
We performed SNP-array analysis of 35 de novo and t-MN leukemia samples and defined a 2.17-Mb CDS on chromosome band 7q22 (Figure 1). Cytogenetic analysis revealed clonal −7/del(7q) in 17 of these. None of the samples showed biallelic deletions of chromosome 7. As seen in previous reports,11-13 most samples had large deletions of chromosome 7 and were uninformative for defining the CDS of 7q22. One sample, T12, was found to have an interstitial deletion encompassing 7q22 not visible by cytogenetic analysis (Figure 1). Samples T12 and T20 (with a del(7)(q22q36)) had similar proximal breakpoints and defined the proximal boundary, and sample T12 defined the distal boundary of the 2.17-Mb CDS (Figure 1). Although this CDS is defined by a small number of patients, this region overlaps with prior studies.12,13,16 The CDS within the current study is only 500 kb centromeric to the CDS identified in Le Beau et al11 (Figure 1). The relatively lower resolution of the 500- to 1.5-Mb YAC probes used in this earlier study may have contributed to imprecision in defining the borders of the deleted segment.
CUX1 is expressed at haploinsufficient levels in −7/del(7q) leukemias
To identify genes that are differentially expressed in samples with −7/del(7q), RNA-sequencing was performed on 23 of the leukemia samples analyzed by SNP array. Eleven of the sequenced samples have loss of 7q22. Although sample A72 was reported to have−7[15%]/r(7)(p2?2q2?2)[75%] by cytogenetic analysis, we categorized it as having diploid 7q22 based on the SNP array (Figure 1). The percentage of blasts was similar in the 2 groups with 41.9% ± 30.1% and 49.7% ± 20.8% blasts in −7/del(7q) and other leukemias, respectively. Alignment statistics are provided in supplemental Table 4.
We performed an unbiased, genome-wide comparison of transcript levels in samples with −7/del(7q) versus other samples to define regions of haploinsufficient gene expression. Across all 20 139 expressed transcripts, 4 of the 5 most significantly, differentially expressed transcripts with > 1 fold change are located on chromosome band 7q22.1 and include CUX1 (Figure 2A). The RNA-sequencing dataset thereby provides an independent validation of the CDS defined by SNP-array (Figure 1). The 2.17-Mb CDS contains 49 RefSeq genes, 3 of which exhibited significantly decreased expression after Bonferroni correction (Figure 2B). CUX1 is the most significantly differentially expressed transcript within the CDS.
The most significantly decreased transcripts, genome-wide, in −7/del(7q) leukemias reside on chromosome band 7q22.1, and CUX1 is the most significantly decreased within the CDS. (A) Genes on chromosome band 7q22.1 are the most significantly altered in leukemias with −7/(del)7q. Gene expression levels quantified by FPKM from RNA-sequencing were compared between patient samples with loss of −7/del(7q) (n = 11) and all other leukemias (n = 12). Only genes from chromosome 7 are shown. Each circle represents 1 gene plotted on the x-axis by the 5′ genomic coordinate. The y-axis indicates the −log10(q value) of the comparison in gene expression levels between −7/del(7q) and other samples (Wilcoxon rank test). Solid circles indicate the top 5 most significant genes (genome-wide) with at least a 1-fold change in expression level. Hyphenated lines represent the CDS as defined by SNP-array analysis in Figure 1. (B) CUX1 transcripts are the most significantly decreased of the products of genes residing within the CDS. The expression levels of 49 RefSeq genes within chromosome 7:99,807,993-101,974,292 (hg19) are shown. Patient samples are divided into those with loss of 7q22 (gray bars) and others (white bars). Genes are ranked on the x-axis by the mean expression level of the gene in all samples. The graph is shown on a linear scale to illustrate relative expression differences between −7/del(7q) versus other leukemias. Error bars represent SEM. *Significant P value after Bonferroni correction (Wilcoxon rank test). The nominal P value is shown.
The most significantly decreased transcripts, genome-wide, in −7/del(7q) leukemias reside on chromosome band 7q22.1, and CUX1 is the most significantly decreased within the CDS. (A) Genes on chromosome band 7q22.1 are the most significantly altered in leukemias with −7/(del)7q. Gene expression levels quantified by FPKM from RNA-sequencing were compared between patient samples with loss of −7/del(7q) (n = 11) and all other leukemias (n = 12). Only genes from chromosome 7 are shown. Each circle represents 1 gene plotted on the x-axis by the 5′ genomic coordinate. The y-axis indicates the −log10(q value) of the comparison in gene expression levels between −7/del(7q) and other samples (Wilcoxon rank test). Solid circles indicate the top 5 most significant genes (genome-wide) with at least a 1-fold change in expression level. Hyphenated lines represent the CDS as defined by SNP-array analysis in Figure 1. (B) CUX1 transcripts are the most significantly decreased of the products of genes residing within the CDS. The expression levels of 49 RefSeq genes within chromosome 7:99,807,993-101,974,292 (hg19) are shown. Patient samples are divided into those with loss of 7q22 (gray bars) and others (white bars). Genes are ranked on the x-axis by the mean expression level of the gene in all samples. The graph is shown on a linear scale to illustrate relative expression differences between −7/del(7q) versus other leukemias. Error bars represent SEM. *Significant P value after Bonferroni correction (Wilcoxon rank test). The nominal P value is shown.
CUX1 transcript levels in samples with a single CUX1 allele are present at half the level (46.3% ± 20.2%) of samples with 2 copies of the gene (Figure 3A) consistent with a recent report.16 Transcript expression was confirmed by quantitative RT-PCR. Leukemia samples with loss of 7q22 express 45.8% ± 4.7% of the level of CUX1 expressed by other samples (Figure 3B). Loss of 1 copy of CUX1 is also associated with haploinsufficient protein levels in human AML cell lines (Figure 3C).
CUX1 RNA and protein levels are haploinsufficient in AML cells with loss of 7q22. (A) CUX1 expression levels in RNA-sequenced leukemia samples. Samples are divided into those with −7/del(7)(q22) (●) and other samples (○). Each circle represents an individual sample. P value calculated by Wilcoxon rank test. (B) Quantitative RT-PCR of CUX1 in RNA from primary patient samples. A19 and A35 have a normal karyotype, and A56 and T52 have −7/del(7q). One representative experiment of 3 independent experiments is shown. Samples with −7/del(7q) have 45.8% ± 2.4% of the level of CUX1 expressed by other samples (P = .045, comparing the mean within 1 experiment, A19 and A35 vs A56 and T52). (C) Protein lysates from human AML cell lines were probed for CUX1 protein by Western blot. UoCM1, Mono7, and KG-1 have −7/del(7q) karyotypes. HeLa, K562, and Kasumi-1 cell lines were used as controls. The full-length p180 protein isoform of CUX1 is shown. The same blot was probed for and normalized to β-actin. Error bars represent ± SEM from 3 independent experiments.
CUX1 RNA and protein levels are haploinsufficient in AML cells with loss of 7q22. (A) CUX1 expression levels in RNA-sequenced leukemia samples. Samples are divided into those with −7/del(7)(q22) (●) and other samples (○). Each circle represents an individual sample. P value calculated by Wilcoxon rank test. (B) Quantitative RT-PCR of CUX1 in RNA from primary patient samples. A19 and A35 have a normal karyotype, and A56 and T52 have −7/del(7q). One representative experiment of 3 independent experiments is shown. Samples with −7/del(7q) have 45.8% ± 2.4% of the level of CUX1 expressed by other samples (P = .045, comparing the mean within 1 experiment, A19 and A35 vs A56 and T52). (C) Protein lysates from human AML cell lines were probed for CUX1 protein by Western blot. UoCM1, Mono7, and KG-1 have −7/del(7q) karyotypes. HeLa, K562, and Kasumi-1 cell lines were used as controls. The full-length p180 protein isoform of CUX1 is shown. The same blot was probed for and normalized to β-actin. Error bars represent ± SEM from 3 independent experiments.
The CUX1 gene is disrupted by a translocation resulting in a chimeric transcript
As an orthogonal dataset to identify the putative TSG on chromosome 7, RNA-sequencing data were mined for chimeric fusion reads involving genes within the CDS. Nine high-confidence fusion events were identified: 4 validated by Sanger sequencing, 2 were false positives, and there was insufficient patient material to confirm the remaining (Table 1; supplemental Table 5). MLL-MLLT1 and USP42-RUNX1 fusions have been previously described in AML.29 Three expressed fusion events were identified in sample T20, 1 of which contains the first exon of CUX1 (7q22) upstream of exons 2-4 of CLND7 (chromosome band 17p13.1), in the same orientation (Figure 4). The resulting transcript is predicted to contain the codons for the first 21 amino acids of CUX1, followed by 8 amino acids in the wrong reading frame from CLDN7, followed by a premature stop codon, thus truncating 98.6% of the CUX1 transcript. The second allele of CUX1 in T20 probably remains intact based on RNA-sequencing (data not shown) and RT-PCR (Figure 4B). The level of CUX1 in T20 is 47.6% of the level in samples without loss of 7q22 as estimated by FPKM.
The CUX1 gene on 7q22 is disrupted by a fusion event involving CLDN7 on 17p13.1 resulting in a chimeric transcript in 1 sample. (A) Model of the chimeric fusion transcript. The fusion contains the first exon of CUX1 upstream of exons 2-4 of CLDN7. The fusion was identified by 35 RNA-sequencing reads wherein one end of the paired-end reads maps to one gene whereas the mate-pair maps to the other gene (indicated by black bars connected by hyphenated lines). In addition, 89 junction spanning reads (indicated as single bars) were identified that map across the exon-exon boundaries of the fused genes. Sanger sequencing of the PCR product confirmed the fusion. The CLDN7 exons are out of frame with respect to the CUX1 start site, resulting in a premature stop codon. (B) RT-PCR of RNA from t-MN sample T20 confirms the presence of both the CUX1-CLDN7 fusion transcript and native CUX1. K562 cells, which express both endogenous CUX1 and CLDN7, but not the fusion transcript, were used as a control.
The CUX1 gene on 7q22 is disrupted by a fusion event involving CLDN7 on 17p13.1 resulting in a chimeric transcript in 1 sample. (A) Model of the chimeric fusion transcript. The fusion contains the first exon of CUX1 upstream of exons 2-4 of CLDN7. The fusion was identified by 35 RNA-sequencing reads wherein one end of the paired-end reads maps to one gene whereas the mate-pair maps to the other gene (indicated by black bars connected by hyphenated lines). In addition, 89 junction spanning reads (indicated as single bars) were identified that map across the exon-exon boundaries of the fused genes. Sanger sequencing of the PCR product confirmed the fusion. The CLDN7 exons are out of frame with respect to the CUX1 start site, resulting in a premature stop codon. (B) RT-PCR of RNA from t-MN sample T20 confirms the presence of both the CUX1-CLDN7 fusion transcript and native CUX1. K562 cells, which express both endogenous CUX1 and CLDN7, but not the fusion transcript, were used as a control.
deFuse22 did not identify CUX1 in fusion events in any of the other RNA-sequenced samples. Sanger sequencing of CUX1 exons in 144 AML/t-MNs samples, 39 of which have −7/del(7q), did not reveal any somatic mutations (data not shown). Similarly, there are no mutations in CUX1 reported in 200 AML samples by the Cancer Genome Atlas (tcga-data.nci.nih.gov, Level 2 DNA sequencing somatic mutations, Version 2.2.0). There is 1 reported case of CUX1 mutation acquired during leukemic transformation of a myeloproliferative neoplasm.30 Thus, CUX1 function is disrupted primarily by chromosome loss or deletion (Figure 1) and infrequently by a translocation event (Figure 4) or mutation.30
CUX1 is associated with a promitotic transcriptional gene signature in leukemia
CUX1 is a compelling candidate TSG: it is a homeodomain-containing transcription factor that regulates cell cycle progression and apoptosis and has been implicated in tumorigenesis of solid tumors.31 Consistent with a TSG, we uncovered a CUX1-associated cell cycle program within the RNA-sequencing data. Of the 662 genes significantly associated with CUX1 expression (FDR < 0.20, P < .0083), there is a substantial enrichment for the mitotic cell cycle pathway (DAVID, GO:0000278, Benjamini P = 2.57 × 10−5).32
CUX1-associated genes replicate in an independent microarray dataset of 38 subtypes of normal human hematopoietic cells.33 There is a significant overlap (P = .0063, permutation test) between the top 1000 CUX1-associated genes (FDR < 1.58 × 10−10) in normal hematopoietic cells and CUX1-associated genes in leukemia cells (2300 genes at P < .05). 73.8% of the 126 overlapping genes have a concordant direction (Pearson χ2 test P = 6.24 × 10−7). CUX1-associated genes in normal cells are also enriched for cell cycle (GO:0007049, Benjamini P = .017). Thus, the CUX1-associated cell growth transcriptional profile is present in both leukemia and normal cells.
This transcriptional program is probably a result of direct regulation by CUX1, as many of these genes are bound by CUX1. We overlaid the CUX1-correlated leukemia gene set with previously reported CUX1 genome-wide promoter occupancy data from 8 human cancer cell lines, 5 of which are hematopoietic.34 There is a significant enrichment for direct targets of CUX1 (defined as binding with a P < .0005 in any cell type) within the leukemia CUX1-associated genes (P = .0016, permutation test). The 125 overlapping genes are also enriched for cell cycle genes (GO:0022403, Benjamini P = .05, supplemental Table 6). Nine of the 10 cell cycle genes in the overlapping gene set are inversely correlated with CUX1 expression, further implicating CUX1 as a negative regulator of hematopoietic proliferation.
The CUX1 ortholog, cut, is a conserved hematopoietic TSG in D melanogaster
The hematopoietic system in Drosophila is composed of hemocytes with functions similar to human myeloid cells.35 The development of hemocytes involves signaling molecules and transcription factors that are highly conserved in Drosophila and humans, such as GATA2, RUNX1, ZFPM1 (FOG-1), and NOTCH- and WNT-family orthologs.35 Several genetic lesions in human leukemia result in hemocyte hyperproliferation in Drosophila, including gain-of-function mutations of RAS- and JAK-family genes, and expression of the human RUNX1-RUNX1T1 fusion protein.35 Most human TSGs have Drosophila orthologs with evolutionarily conserved networks that regulate proliferation and differentiation; indeed, much of our understanding of TSGs comes from genetic screens in Drosophila. Ct is highly conserved between humans and Drosophila,36 and ectopic expression of human CUX1 can rescue ct-deficient phenotypes in Drosophila.37
To test the hypothesis that ct is a TSG in vivo, ct, which is expressed in hemocyte progenitors,38 was targeted by RNAi specifically in developing Drosophila hemocytes. Expression of ctRNAi A in developing hemocytes led to the development of melanotic pseudotumors35 in both third instar larvae (Figure 5A) and pupae (data not shown). In a small number of cases, tumors were seen as early as the second instar (data not shown). A total of 23.8% (10 of 42) of larvae developed tumors, whereas none (0 of 30) of the control larvae did. To confirm that tumor formation was not the result of an off-target effect of the ct shRNA, we tested a second RNAi construct. Expression of ctRNAi B in hemocytes also led to tumor formation in 16.1% (10 of 62) of larvae.
Haploinsufficiency of the D melanogaster CUX1 homologue, cut, leads to melanotic tumor formation and hemocyte hyperproliferation in vivo.ct knockdown was performed by crossing flies expressing ct RNAi in a construct containing a UAS promoter and GFP tag with flies expressing the GAL4 driver under the hemocyte-specific hml promoter. Two different ct targeting constructs were tested: ctRNAi A and ctRNAi B. (A) ctRNAi A but not wild-type (w1118;hml-GAL,UAS-GFP/+), Drosophila third instar larvae develop melanotic tumors (indicated by the arrow). (B) ct knockdown leads to increased numbers of hemocytes. Circulating hemocytes bled from wild-type, nonspecific RNAi, ctRNAi A, and ctRNAi B developing larvae were stained with Hoechst and quantified (mean ± SEM). (C) Haploinsufficient levels of ct are confirmed by quantitative RT-PCR of ct from hemocyte RNA. One representative experiment of 3 is shown.
Haploinsufficiency of the D melanogaster CUX1 homologue, cut, leads to melanotic tumor formation and hemocyte hyperproliferation in vivo.ct knockdown was performed by crossing flies expressing ct RNAi in a construct containing a UAS promoter and GFP tag with flies expressing the GAL4 driver under the hemocyte-specific hml promoter. Two different ct targeting constructs were tested: ctRNAi A and ctRNAi B. (A) ctRNAi A but not wild-type (w1118;hml-GAL,UAS-GFP/+), Drosophila third instar larvae develop melanotic tumors (indicated by the arrow). (B) ct knockdown leads to increased numbers of hemocytes. Circulating hemocytes bled from wild-type, nonspecific RNAi, ctRNAi A, and ctRNAi B developing larvae were stained with Hoechst and quantified (mean ± SEM). (C) Haploinsufficient levels of ct are confirmed by quantitative RT-PCR of ct from hemocyte RNA. One representative experiment of 3 is shown.
Melanotic tumors are a phenotype of hemocyte overgrowth35 ; however, we confirmed that there are increased numbers of circulating hemocytes. ctRNAi A and ctRNAi B knockdown larvae have significantly increased numbers of hemocytes compared with nonspecific shRNA-expressing larvae (Figure 5B). In addition, we tested a second hemocyte-specific GAL4-driver.39 Nine of 16 cg-GAL4/UAS-ctRNAi A larvae developed tumors and had significantly increased numbers of circulating hemocytes (33 737 ± 5719 SE in cg-GAL4/UAS-ctRNAi A, n = 7, vs 18 826 ± 1984 cg-GAL4 wild-type, n = 6, P = .042, data not shown). Residual expression of ct transcripts in hemocytes was 38.3% ± 3.8% (ctRNAi A, Figure 5C) and 28.9% ± 6.8% (ctRNAi B) of control, indicating that hemocyte overgrowth was a result of haploinsufficient levels of ct.
CUX1 is a haploinsufficient TSG in human hematopoietic stem cells
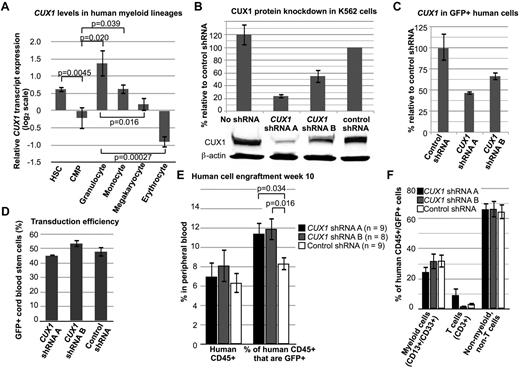
To determine whether CUX1 is a conserved TSG in humans, we targeted CUX1 for knockdown in human hematopoietic progenitors. CUX1 is normally expressed highly in multipotent hematopoietic stem cells (Figure 6A).33 CUX1 expression decreases in common myeloid progenitors and varies significantly in terminally differentiated myeloid cells (Figure 6A).
Knockdown of CUX1 leads to increased engraftment of human hematopoietic cells in immunodeficient mice. (A) CUX1 is highly expressed in normal human hematopoietic stem cells. The CUX1 transcript expression profile was extracted from published microarray data.33 Hematopoietic stem cells (HSC, lineage−, CD133+, CD34dim, n = 10), common myeloid progenitors (CMP, lin−, CD34+, CD38+, IL-3Rlo, CD45RA−, n = 4), granulocytes (FSChi, SSChi, CD16+ CD11b+, n = 4), monocytes (FSChi, SSClo, CD14+, CD45dim, n = 5), megakaryocytes (CD34−, CD41+, CD61+, CD45−, n = 6), and erythrocytes (CD34−, CD71−, GlyA+, n = 6). Data were normalized such that the mean of each gene, across all 38 hematopoietic cell types tested, is zero. (B) shRNA constructs targeting CUX1 lead to haploinsufficient CUX1 protein levels in K562 cells. Western blot was performed on lysates of K562 cells stably expressing CUX1 shRNA A, shRNA B, or control shRNA. Data represent the mean ± SD from 3 experiments. (C) Residual CUX1 transcript levels in human hematopoietic stem cells transduced with lentivirus expressing shRNA targeting CUX1. Lineage-depleted human cord blood cells were transduced with 2 different shRNA constructs targeting CUX1 (shRNA A or shRNA B), or a nonspecific control shRNA. RT-PCR was performed on sorted GFP+ cells on the day of transplantation. Each sample was performed in triplicate, and 1 of 2 experiments is shown. (D) Transduction efficiency was similar across the 3 groups of transplanted cord blood stem cells. Transduction efficiency was determined by flow cytometry for GFP+ cells before transplantation. (E) Knockdown of CUX1 leads to increased engraftment of human hematopoietic cells in NSG mice. Peripheral blood from mice transplanted with human xenografts was collected on week 10 after transplantation and tested for human CD45 expression (left bars) and coexpression of human dCD45 and GFP (right bars). The numbers of mice per group are indicated. Data are mean ± SEM. (F) Peripheral blood human CD45+/GFP+ cells from CUX1 shRNA A, CUX1 shRNA B, or control shRNA transduced xenografts were stained for the lineage markers CD13/CD33 (myeloid cells) and CD3 (T cells).
Knockdown of CUX1 leads to increased engraftment of human hematopoietic cells in immunodeficient mice. (A) CUX1 is highly expressed in normal human hematopoietic stem cells. The CUX1 transcript expression profile was extracted from published microarray data.33 Hematopoietic stem cells (HSC, lineage−, CD133+, CD34dim, n = 10), common myeloid progenitors (CMP, lin−, CD34+, CD38+, IL-3Rlo, CD45RA−, n = 4), granulocytes (FSChi, SSChi, CD16+ CD11b+, n = 4), monocytes (FSChi, SSClo, CD14+, CD45dim, n = 5), megakaryocytes (CD34−, CD41+, CD61+, CD45−, n = 6), and erythrocytes (CD34−, CD71−, GlyA+, n = 6). Data were normalized such that the mean of each gene, across all 38 hematopoietic cell types tested, is zero. (B) shRNA constructs targeting CUX1 lead to haploinsufficient CUX1 protein levels in K562 cells. Western blot was performed on lysates of K562 cells stably expressing CUX1 shRNA A, shRNA B, or control shRNA. Data represent the mean ± SD from 3 experiments. (C) Residual CUX1 transcript levels in human hematopoietic stem cells transduced with lentivirus expressing shRNA targeting CUX1. Lineage-depleted human cord blood cells were transduced with 2 different shRNA constructs targeting CUX1 (shRNA A or shRNA B), or a nonspecific control shRNA. RT-PCR was performed on sorted GFP+ cells on the day of transplantation. Each sample was performed in triplicate, and 1 of 2 experiments is shown. (D) Transduction efficiency was similar across the 3 groups of transplanted cord blood stem cells. Transduction efficiency was determined by flow cytometry for GFP+ cells before transplantation. (E) Knockdown of CUX1 leads to increased engraftment of human hematopoietic cells in NSG mice. Peripheral blood from mice transplanted with human xenografts was collected on week 10 after transplantation and tested for human CD45 expression (left bars) and coexpression of human dCD45 and GFP (right bars). The numbers of mice per group are indicated. Data are mean ± SEM. (F) Peripheral blood human CD45+/GFP+ cells from CUX1 shRNA A, CUX1 shRNA B, or control shRNA transduced xenografts were stained for the lineage markers CD13/CD33 (myeloid cells) and CD3 (T cells).
We used 2 different CUX1-targeting shRNAs, shRNA A and shRNA B. K562 cells expressing shRNA A or shRNA B retain 24.1% ± 3.7% or 55.3% ± 10.9% residual CUX1 protein, respectively (Figure 6B). These shRNAs were transduced in a GFP-expressing lentiviral vector into lineage-depleted human cord blood progenitors and transplanted into NOD scid γ (NSG) mice. CUX1 shRNA-transduced progenitors retained 47.0%-66.9% residual CUX1 transcripts at the time of transplantation (Figure 6C). Transduction efficiency was similar in all groups (Figure 6D). Total engraftment of all human cells was not significantly different in CUX1-targeted recipient mice (Figure 6E). However, there was a significant expansion of the GFP+ population in the CUX1-knockdown xenografts (Figure 6E). Compared with nonspecific shRNA-transduced cells, there is a 40% increase in the percentage of CUX1-shRNA–transduced cells. All lineages appeared to be equally expanded in the peripheral blood (Figure 6F), suggesting that CUX1 TSG activity is not restricted to myeloid cells.
Discussion
Loss of part or all of chromosome 7 is a frequent event in myeloid malignancies and is associated with a poor prognosis, yet it is unknown how these deletions contribute to leukemogenesis. We used high-density SNP arrays and transcriptome sequencing to identify a transcription factor gene, CUX1, as a haploinsufficient TSG within a CDS of chromosome 7. We also characterized a t-MNs sample with an inactivated CUX1 gene resulting from a translocation (Figure 4). CUX1 is normally highly expressed in multipotent hematopoietic progenitors and exhibits dynamic dosage changes during the course of differentiation (Figure 6A). Deletion of a single allele of CUX1 is associated with haploinsufficient transcript and protein levels (Figure 3). To test the TSG activity of CUX1, we first used Drosophila to demonstrate that haploinsufficient levels of the highly conserved Drosophila homolog, ct, leads to hemocyte overproliferation and melanotic tumor formation in developing larvae (Figure 5). In a second in vivo model, partial knockdown of CUX1 in human hematopoietic progenitors leads to increased hematopoietic engraftment on transplantation into NSG mice (Figure 6). These findings indicate that CUX1/cut is a conserved, haploinsufficient TSG that is essential for the regulation of normal hematopoietic cell growth. A proliferative gene signature was identified within CUX1-associated genes, a finding that was replicated in an independent dataset of normal hematopoietic cells (data not shown). A significant number of these genes are direct targets of CUX1 binding.34 This suggests that CUX1 may act as a TSG in myeloid cells by regulating cell cycle components at the transcriptional level.
Deletions of chromosome 7 are typically large with heterogeneity in the breakpoints in myeloid diseases, making it difficult to map the CDS. It is probable that multiple TSGs on chromosome 7 cooperate in leukemogenesis. There may be at least 3 different CDS on chromosome 7, including an interval at 7q36 containing the EZH2 gene.4,16 A precedent for this is seen in 5q deletions, which are also heterogeneous.4
We have not excluded the possibility that other differentially expressed genes within the CDS of 7q22.1 may also play a role in disease pathogenesis. However, CUX1, which was implicated as early as 1996,40 was identified in a narrower CDS in 2 recent studies of AML arising from myeloproliferative neoplasms.7,41 Four cryptic deletions were identified: 3 samples had deletions that spanned just the CUX1 and SH2B2 genes,41 and 1 sample had a deletion containing only CUX1.7 The CDS defined in the current study is larger, most probably because of smaller sample size.
We note that biallelic inactivation of CUX1 appears to be infrequent (Figure 1).7,41 The remaining CUX1 allele is unlikely to be silenced by epigenetic modifications, as samples with −7/del(7q) express CUX1 at 46.3% ± 20.2% of the level of samples with both copies of CUX1 (Figure 3), consistent with a gene dosage effect and expression from the remaining allele. However, rare instances of homozygous deletions of the CUX1 locus and a CUX1 homeodomain mutation in a region of acquired uniparental disomy have been reported in AML arising from myeloproliferative neoplasms.7,30 Mutations in CUX1 were not demonstrated by Sanger sequencing of CUX1 exons in 144 AML/t-MNs samples, 39 with −7/del(7q) (data not shown). A previous study also failed to find CUX1 mutations in pediatric myeloid neoplasias with −7.42 CUX1 is therefore most often inactivated by chromosome loss or deletion but can infrequently be inactivated by mutation30 or translocation (Figure 4). As biallelic inactivation of CUX1 occurs only rarely in myeloid diseases, CUX1 appears to be a haploinsufficient TSG.
Studies in murine models support the hypothesis that CUX1 is a myeloid TSG in mammals. Cux1ΔHD mice, which have a Cux1 truncation, have increased myeloid cell numbers and myeloid colony-forming units.43 Transgenic expression of the p75 isoform of Cux1, containing 1 Cut repeat and the homeodomain, leads to a myeloproliferative disease in 33% of mice.44 It is not clear whether the myeloid expansion is the result of increased Cux1 activity or a dominant negative effect of the transgene.
CUX1/cut was first described in Drosophila, wherein ct is essential for lineage specification in multiple tissues.31 Consistent with a developmental role in several cell types, altered Cux1 levels led to abnormal growth of multiple cell types in murine models.31 In humans, CUX1 mutations have been reported in 6.7% of lung squamous cell carcinoma45 and 2.8% of colorectal carcinoma46 ; however, the functional significance of these mutations remains to be tested. The TSG activity of CUX1 may have several mechanisms, including regulation of differentiation, cell cycle progression, genomic stability, and apoptosis, and the mechanism may be cell type specific.31 In one report, ct deletion in a Drosophila cancer model promoted invasive tumorigenesis because of both a block in differentiation and increased proliferation.47
In conclusion, this is the first biologic confirmation of a haploinsufficient myeloid TSG on chromosome band 7q22. The concentrations of hematopoietic transcription factors play a critical role in hematopoietic cell fate, regulating lineage decisions, proliferative signals, and leading to leukemia when concentrations are above or below crucial thresholds. Examples of the detrimental consequences of abnormal gene product dosage can be found in many master regulators of hematopoiesis, including SPI1, RUNX1, and CBFB.48 Intriguingly, these genes are typically not completely inactivated when mutated in leukemia but instead retain some level of activity.48 CUX1 levels during normal human hematopoiesis appear to change significantly, depending on the developmental stage and lineage (Figure 6A). A gradient of Ct has been shown to be important in neural development in Drosophila: high, medium, and low levels of Ct, measured at the single-cell level, lead to distinct classes of neural cells.49 Thus, changes in CUX1 levels may have important phenotypes during hematopoiesis and deleterious consequences when altered.
There is an Inside Blood commentary on this article in this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Kevin Shannon for critical reading of the manuscript, Ronald Hause for assistance with statistical analysis, and the TRiP at Harvard Medical School for transgenic RNAi fly stocks. Next-generation sequencing was performed at the University of Chicago High-throughput Genome Analysis Core. Illumina SNP array services were performed at the Northwestern University Genomics Core. Sanger sequencing was performed at the University of Chicago Comprehensive Cancer Center Genomics Core.
This work was supported by a Leukemia & Lymphoma Society Fellow award (M.E.M), the Cancer Research Foundation, National Institutes of Health (CA40046; M.M.L.), and the Chicago Cancer Genomes Project.
National Institutes of Health
Authorship
Contribution: M.E.M. designed research, performed experiments, analyzed and interpreted data, and wrote the manuscript; C.D.B. designed research, analyzed, and interpreted RNA-sequencing data, and edited the manuscript; X.W. performed Drosophila crosses and hemocyte counting; E.T.B. assisted in analyzing RNA-sequencing data; S.K. assisted in generating sequencing libraries; C.B. identified fusion genes; S.Y. assisted in analyzing SNP data and fusion validation; B.P.S., J.K., and J.M.C. designed and performed xenograft experiments; T.S. assisted in designing research; J.A. performed morphologic analysis and collected biospecimens; M.M.L. performed cytogenetic analysis of leukemia samples, designed research, collected biospecimens, interpreted data, and edited the manuscript; R.L.G. provided computational infrastructure and bioinformatics support; and K.P.W. designed research, interpreted data, and edited the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Megan E. McNerney, Institute for Genomics and Systems Biology, University of Chicago, 900 East 57th St, KCBD 10100A, Chicago, IL 60637; e-mail: megan.mcnerney@uchospitals.edu; and Kevin P. White, Institute for Genomics and Systems Biology, University of Chicago, 900 East 57th St, KCBD 10100A, Chicago, IL 60637; e-mail: kpwhite@uchicago.edu.