Key Points
Macrophage-derived microvesicles induced cellular differentiation in naive monocytes.
Macrophage-derived microvesicles shuttle of miRNAs to target cells.
Abstract
Microvesicles are small membrane-bound particles comprised of exosomes and various-sized extracellular vesicles. These are released by several cell types. Microvesicles have a variety of cellular functions from communication to mediating growth and differentiation. Microvesicles contain proteins and nucleic acids. Previously, we showed that plasma microvesicles contain microRNAs (miRNAs). Based on our previous report, the majority of peripheral blood microvesicles are derived from platelets, while mononuclear phagocytes, including macrophages, are the second most abundant population. Here, we characterized macrophage-derived microvesicles and explored their role in the differentiation of naive monocytes. We also identified the miRNA content of the macrophage-derived microvesicles. We found that RNA molecules contained in the macrophage-derived microvesicles were transported to target cells, including mono cytes, endothelial cells, epithelial cells, and fibroblasts. Furthermore, we found that miR-223 was transported to target cells and was functionally active. Based on our observations, we hypothesize that microvesicles bind to and activate target cells. Furthermore, we find that microvesicles induce the differentiation of macrophages. Thus, defining key components of this response may identify novel targets to regulate host defense and inflammation.
Introduction
Microvesicles are small (0.05-1 μm) membrane-bound particles released by healthy and diseased cells.1-9 Microvesicles are also known as exosomes, microparticles, or shedding vesicles. While relatively little is known about their function, microvesicles are enriched in bioactive molecules that play roles in cellular growth, differentiation, and transformation.1,3-13
Microvesicle production is energy-dependent and enhanced by environmental stimuli including oxidative stress, hypoxia, and injury.7,9,14-17 Microvesicles contain proteins, RNA molecules, lipids or even organelles from the cells of origin to directly activate target cells.6-8,13,18-21 We and others found that microRNAs (miRNAs) are encapsulated in peripheral blood microvesicles.6,19-21 While early reports suggested that microvesicle content is nonspecifically derived, recent evidence suggests specificity in their production and shows that cellular activation impacts their content.3,12,13,17,22
One of the first reports shows in vitro evidence that microvesicles can exchange genetic information with target cells.8 For instance, microvesicles improve the chemotactic responsiveness of hematopoietic stem/progenitor cells.10 In addition, microvesicles direct embryonic stem cells to hematopoietic progenitor cell lineage by transferring mRNA and proteins.10,11,13,23,24
Microvesicles are normal constituents of blood plasma and many originate from activated platelets.25 In the peripheral blood, two-thirds of microvesicles are derived from platelets.6,25 Platelet-derived microvesicles play a role in angiogenesis and in metastatic spread of cancer.26,27 Treating lung cancer cell lines with platelet-derived microvesicles activate the MAPK signaling pathway and induce matrix metalloproteinase expression.26
Microvesicles released in vivo from cancer cells are also detected in the tumor microenvironment and promote tumor growth and metastases.7,14,26-29 Interestingly, the molecular profile of tumor-derived microvesicles is distinct from normal microvesicles; tumor-derived microvesicles express markers associated with inflammation, metastasis, and thrombosis, while normal microvesicles do not.3,19,20,28-30 Hence, microvesicles are likely important mediators of intercellular cross-talk.
Previously, we characterized miRNA expression in microvesicles of peripheral blood from normal healthy individuals.6 In addition to platelet-derived microvesicles, we found that myeloid-derived microvesicles constitute a large component of the peripheral blood microvesicles. Microvesicle and miRNAs pattern may reflect early disease diagnosis and progression.19-21,25,31 For example, elevated circulating expression of miR-141 or miR-21 predicts prostate cancer20 and neuroblastoma,19 respectively. These data suggest that microvesicle miRNA content may play a role in intercellular communication and serve as biomarkers of health and disease.
While data are emerging related to the content, function, and characterization of microvesicles, there are important knowledge gaps remaining. Therefore, we hypothesize that microvesicles provide novel mechanisms by which infiltrating mononuclear phagocytes in areas of inflammation amplify cellular activation, survival, and immune function. Interrogating the content of these microvesicles may allow an early window in disease prediction and intervention. Thus, the goal of our study was to characterize the function of macrophage-derived microvesicles and their ability to mediate cellular differentiation. Here, we report that macrophage-derived microvesicles induced cellular differentiation when added to naive monocytes, suggesting a potential amplification loop to enhance immune function. These data expand our knowledge of an important role for macrophage-derived microvesicles in cellular homeostasis.
Methods
Reagents
Reagents for antagomiR transfection, fluorescently labeling microvesicles, cell culture, RNA extraction, and miRNA expression were obtained from Life Technologies unless otherwise indicated. Housekeeping PCR primers except CAP1 were obtained from Real Time Primers LLC. Primers to measure gene expression were purchased from QIAGEN. Flow cytometry reagents including antibodies, annexin V, and instrumentation were from BD Biosciences. Transmission electron microscopes were from FEI. Chemicals were obtained from Sigma-Aldrich.
Human sample processing and cell culture
Peripheral human blood monocytes were isolated from American Red Cross buffy coats or healthy volunteers after informed consent per the Declaration of Helsinki (IRB no. 2011H0007) as previously described.6,32 Briefly, diluted blood was layered over Lymphocytes Separation Medium (d = 1.077; Cellgro). CD14+ monocytes were isolated using the Monocyte Isolation Kit from Miltenyi Biotec. Monocytes (1-10 × 106 cells/mL) in X-VIVO 15 serum-free media (Lonza) were supplemented with 10 μg/mL polymyxin B in the absence or presence of 50 ng/mL rhGM-CSF (Berlex Laboratories Inc). The monocytes were washed after 4 hours and cultured in fresh media without added rhGM-CSF. Primary cells were subjected to morphologic assessment to confirm response to GM-CSF before subsequent use and collection of microvesicles.
Human monocytic THP-1 cells, lung fibroblasts CCL-204 and lung epithelial A549 cells were obtained from ATCC. THP-1 cells were grown in RPMI 1640 supplemented with 10% FBS (Hyclone Laboratories) and 1% penicillin, streptomycin, and amphotericin B (PSA). To induce differentiation, THP-1 cells (1 × 106 cells/mL) were grown in X-VIVO 15 media and treated with 65nM phorbol 12-myristate 13-acetate (PMA). Cells treated with 0.001% dimethyl sulfoxide (DMSO) served as a vehicle control. Lung fibroblasts and epithelial cells were cultured in DMEM medium containing 10% FBS and 1% PSA. Human umbilical vein endothelial cells (HUVEC) and endothelial cell medium were purchased from ScienCell Research Laboratories. The cells were maintained in cell-culture flask coated with 15 μg/mL fibronectin.
Microvesicle isolation
Cell-free supernatants were obtained by a 1600g centrifugation at 4°C. To isolate RNA for miRNA profiling from the microvesicles, the microvesicle-containing supernatants (6-18 mL) were centrifuged at 160 000g in Nalgene Oak Ridge conical tubes (Thermo Fisher Scientific).6 For microvesicle transfer experiments, the supernatants (1.0 mL) were centrifuged at 16 000g for 30 minutes at 4°C. The microvesicle pellets were collected in sterile PBS.
AntagomiR transfection
THP-1 cells or monocytes were transfected with the miR-223 antagomiR (100nM; Life Technologies and SwitchGear Genomics) using the siPORT NeoFX transfection reagent according to manufacturer's instructions. The anti-miR and miRNA Inhibitor Negative Control 1 (100nM) served as a negative control. Cells were then cultured for 6-24 hours before analysis.
Transmission electron microscopy
Suspension THP-1 cells or freshly isolated monocytes were centrifuged at 1600g for 5 minutes then fixed in 2.5% glutaraldehyde in 0.1M phosphate buffer, pH 7.4, containing 0.1M sucrose for 30 minutes. Cells were rinsed, resuspended in 2% agarose, and dehydrated in graded ethanol concentrations for 10 minutes each. Sections were embedded in Spurr resin and polymerized overnight at 60°C. Treated adherent cells were grown in dual chamber slides then fixed and dehydrated in the same manner but embedded in silicon mold and resin filled Beam capsules. Transmission electron microscopy (TEM) sections were cut at 70 nm using EM UC6 ultra-microtome, and collected on 300-mesh grids. Sections were stained in 2% uranyl acetate and Reynolds lead citrate and imaged with Technai G2 Spirit TEM at 80 kV.
Cryo-TEM images of microvesicles were obtained from samples centrifuged at 16 000g and 160 000g. To prepare vitrified cryo-TEM specimens, 4-μL suspensions of microvesicles were applied to glow discharged lacey carbon coated copper 400-mesh grids (Pacific Grid-Tech) and flash-frozen in liquid ethane at 22°C and 95% relative humidity using an automated Vitrobot Mark IV (FEI). The vitrified samples were visualized with Tecnai G2 F20 ST TEM at 200 kV using low-dose mode. All images were imported to Adobe Illustrator CS2 to generate composite images.
Confocal microscopy studies
THP-1 cells or freshly isolated monocytes were suspended in X-VIVO 15 media and treated with either the phospholipid membrane dye, lipophilic carbocyanine DilC163 (D384, 1.25μM), or 0.625μM SytoRNA Select that specifically binds RNA molecules.33 After 10 minutes of incubation at 37°C, the cells were washed, resuspended in fresh media, and treated with PMA or GM-CSF for 4 hours. Cells were then washed and cultured for an additional 48 hours. Fluorescent microvesicles were collected and added to recipient cells. Both donor and recipient cells were fixed then mounted on slides using fluorescent mounting solution (DAKO) then imaged using the Zeiss LSM 510 multiphoton confocal microscope equipped with a C- Apochromat 63×/1.2 W Corr objective. To detect green and red fluorescence, an argon laser with a 488 nm excitation and 500-555 nm band pass (bp) filter and helium-neon laser with a 543 nm excitation and long pass (LP) 560 nm filter were used, respectively. A helium-neon laser with a 633 nm excitation was used for black and white imaging and designated as differential interference contrast (DIC). The Zeiss LCM Image Browser software was used for image acquisitions then the images were imported to Adobe Illustrator CS2 to generate composite images. Background fluorescence was subtracted using unstained cells.
Dynamic light scattering
Microvesicle size distributions were measured using a Nano Zetasizer Zen3600 (Malvern Instruments Ltd). Samples were diluted to 50-300 kilocounts per second and equilibrated at 25°C. All measurements were done in triplicates.
Crystal violet assay
Cellular adherence (6 × 103 cells/well) was measured using crystal violet uptake in a 12-well plate.34 After 48 hours of treatment, cells were washed, fixed in 4% paraformaldehyde, then stained with 0.5% crystal violet in 2% ethanol for 10 minutes at room temperature. The cells were washed 4 times with PBS. The incorporated stain was eluted with 0.1M sodium citrate (pH 4.2) in 50% ethanol. Absorbance was measured at 550 nm using an assay ELISA plate reader (SLT Lab Instruments GmbH).
Flow cytometry
FITC-congujated beads 0.2-1 μm (Bangs Laboratory Inc) and nonfluoroscent bead standards 0.2-2.0 μm (Spherotech Inc) were used to gate on microvesicles < 1 μm using the LSRII flow cytometer. Microvesicle concentration was calculated as described.35 Background from sheath fluid and media only samples was subtracted.
To determine DNA, RNA, and annexin V expression, microvesicles were incubated with 0.625μM SytoRNA Green Select or 2.5 μL of annexin V–FITC and/or 4.86μM Hoechst 33 342 for 30 minutes on ice then analyzed on the LSRII flow cytometer.
Surface expression of macrophage lineage markers was quantified by flow cytometry using the Aria flow cytometer (BD Biosciences).32 Cells were washed twice with cold buffer (HBSS containing 0.1% NaN3 and 1% bovine serum albumin [BSA] then resuspended in the buffer and blocked with 100 μg/mL human IgG solution; Jackson ImmunoResearch Laboratories). Antibodies recognizing CD16 PE-Cy5, CD206 PE-Cy5, and CCR5-PE-Cy7 were used according to the manufacturer's recommendation. Isotype antibody controls were purchased from Cedarlane.
GM-CSF ELISA
Microvesicle-containing supernatants and protein lysates from concentrated microvesicles were analyzed using the Quantikine human GM-CSF ELISA (R&D Systems). The signal was read at 450 nm on a PerkinElmer Victor X3 Multilabel Plate Reader.
RNA extraction and quantitative real-time PCR
Total RNA was isolated using Trizol.6,36 An average of 500 ng of RNA was obtained from microvesicles from cells cultured in a 6-well plate (5 × 106 cells/well). Profiling for all known and individual mature miRNAs was performed and analyzed as previously described using TaqMan MicroRNA assays.6,36
RNA (0.5-1 μg) was subjected to cDNA synthesis and quantitative real-time (qRT)–PCR using SYBR Green Master Mix.32,36 The average cycle threshold (Ct) value ± SEM (15.432 ± 1.72) of GAPDH and CAP-1 (forward primer 5′-ATTCCCTGGATTGTGAAATAGTC-3′ and reverse primer 5′-ATTAAAGTCACCGCCTTCTGTAG-3′) were used for normalization. Small nuclear (sn) RNA U6 was used to normalize miRNA expression and data is presented as ΔCt. For microvesicle treated samples, gene and miRNA is expression reported as the relative fold-increase 2(−ΔΔCt) over nonstimulated freshly isolated samples. For antagomir studies, the data were presented as relative copy number (RCN) using 2(−ΔCt).
Phagocytosis assay
Phagocytosis assay was described previously.37 Briefly, the fluorescent antibody coated SRBC were incubated with freshly isolated monocytes or macrophages (1 × 106/μL coated sheep red blood cells [SRBCs]) at 37°C for 30 minutes then washed with PBS and fixed in 1% paraformaldehyde. One hundred cells were counted per slide using a fluorescent microscope to enumerate phagocytes and calculate phagocytic index.
Caspase-3 analysis
Adherent and nonadherent THP-1 cells and monocytes were collected 18 hours and 6 hours, respectively, posttransfection with miR-223 antagomiR for caspase-3 activity.32 Briefly, protein lysates were incubated with the fluorogenic substrate Ac-DEVD-AMC [N-acetyl- (Asp-Glu-Val-Asp)-(7-amino-4-methylcoumarin); EMD Millipore] and measured with the Cytofluor 4000 fluorometer (Perseptive). Caspase-3 activity is presented as the relative fold-increase change in fluorescence over untreated samples per total protein and is expressed as the mean ± SEM.
Microarray analysis
Changes in gene expression were analyzed in cytokine and microvesicle-treated cells using Human Genome U133 Plus 2.0 Affymetrix GeneChips. The mRNA (30 ng) was processed using the Ovation RNA Amplification System and the FL-Ovation cDNA Biotin Module V2 (both from NuGEN Technologies, Inc). At all steps, the quantity and quality of the preparations were controlled using Nanodrop (NanoDrop Technologies) and Experion Automated Electrophoresis System (Bio-Rad Laboratories). The arrays were scanned by the GeneChip Scanner 3000 (Affymetrix) using the GeneChip Operating Software (GCOS) Version 1.4.0.036. Partek Genomic Suite was used for data analysis on .cel files imported using robust multichip average with GC content adjustment (GC-RMA) for background correction and normalized by quantile normalization. Genes that were P (present) or M (marginal) in at least 2 experimental samples of the triplicate per experimental conditions were analyzed and used to perform principal component analysis (PCA) and ANOVA. Genes were statistically corrected by Bonferroni adjustment were further analyzed and used to generate the pairwise Venn diagrams. All microarray data were deposited to GEO with the accession no. GSE41889.
Pathway analysis and prediction
Genes identified to be commonly expressed between cells treated with either the positive control stimulus or microvesicles were subjected to pathway exploration using the Ingenuity Pathway Analysis software (Ingenuity Systems) as previously described.6 Top-ranked pathways and their associated biologic functions were determined.
Luciferase reporter assay
A549 cells were transfected with 100 ng of 3′-UTR luciferase reporter vector containing miR-223 target seed sequence or an empty vector (SwitchGear Genomics) using FuGene HD transfection reagent (Roche). As a positive control, as set of cells were cotransfected with miR-223 precursor (150nM; Life Technologies). After 18-20 hours, cells were washed and cultured in X-VIVO media alone or microvesicle-containing media generated from GM-CSF–stimulated monocytes for 24 hours before measuring luciferase activity with LightSwitch Assay Reagent (SwitchGear Genomics) and Victor X3 Microplate Reader.
Statistical analysis
For all comparisons, data are expressed as the mean ± SEM derived from at least 3 independent studies. SPSS17 software was used to generate an independent sample t test, and P < .05 was considered significant. Nonparametric t test was used for statistical analysis for the analysis of qRT-PCR and caspase-3 assay using greater than 3 independent experiments.
Results
Microvesicle production during macrophage differentiation
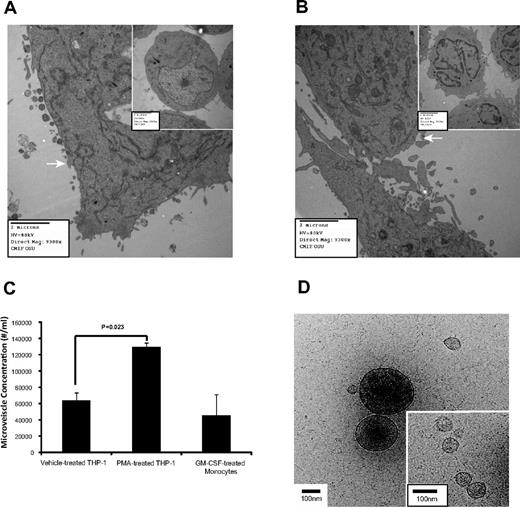
Microvesicle production is induced on cell activation and growth.3,9-15,38 Using electron microscopy, increased microvesicle production was apparent in PMA-treated THP-1 cells (Figure 1A) compared with cells treated with the vehicle (inset). Similarly, GM-CSF–treated monocytes had more microvesicles than untreated cells (Figure 1B). The vesicles ranging between 0.2 and 1 μm were RNA positive (supplemental Figure 1A-B, available on the Blood Web site; see the Supplemental Materials link at the top of the article). The majority of the gated microvesicles were annexin V negative (71.58% ± 2.70%), while only 6.87% ± 1.52% were positive for DNA (supplemental Figure 1C right panel). Interestingly, microvesicle concentration from the 0.2-1 μm gate increased during macrophage differentiation (Figure 1C).
Microvesicle production during macrophage differentiation. (A) THP-1 cells were treated with vehicle (inset) or alternatively with PMA to induce differentiation and observed by electron microscopy, n = 3. (B) Freshly isolated peripheral blood monocytes were untreated for 30 minutes (inset) or GM-CSF for 4 hours then cultured overnight, n = 6. Images were captured with an AMT camera (Advanced Microscopy Techniques). Representative images are shown in panels A and B, and microvesicles are indicated by the white arrows in both panels. (C) THP-1 cells and peripheral blood monocytes were treated with PMA and GM-CSF, respectively. Microvesicle concentration containing both annexin V positive and negative events from the MV-gated region (supplemental Figure 1A) was quantified by flow cytometry. Shown is the average concentration ± SEM (n = 6). (D) The vitrified microvesicle samples were transferred to a Gatan Cryo holder and visualized with Tecnai G2 F20 ST TEM at 200 kV. Images were captured using a 4k × 4k Gatan Ultrascan CCD camera at a magnification of ×38 000. Representative cryo-TEM images of microvescicles collected using a 16 000g centrifugation from the culture supernatant of PMA-treated THP-1 cells are shown. Large (> 200 nm diameter) and small microvesicles (30-100 nm diameter; inset) are present. Scale bar = 100 nm in the figure and inset.
Microvesicle production during macrophage differentiation. (A) THP-1 cells were treated with vehicle (inset) or alternatively with PMA to induce differentiation and observed by electron microscopy, n = 3. (B) Freshly isolated peripheral blood monocytes were untreated for 30 minutes (inset) or GM-CSF for 4 hours then cultured overnight, n = 6. Images were captured with an AMT camera (Advanced Microscopy Techniques). Representative images are shown in panels A and B, and microvesicles are indicated by the white arrows in both panels. (C) THP-1 cells and peripheral blood monocytes were treated with PMA and GM-CSF, respectively. Microvesicle concentration containing both annexin V positive and negative events from the MV-gated region (supplemental Figure 1A) was quantified by flow cytometry. Shown is the average concentration ± SEM (n = 6). (D) The vitrified microvesicle samples were transferred to a Gatan Cryo holder and visualized with Tecnai G2 F20 ST TEM at 200 kV. Images were captured using a 4k × 4k Gatan Ultrascan CCD camera at a magnification of ×38 000. Representative cryo-TEM images of microvescicles collected using a 16 000g centrifugation from the culture supernatant of PMA-treated THP-1 cells are shown. Large (> 200 nm diameter) and small microvesicles (30-100 nm diameter; inset) are present. Scale bar = 100 nm in the figure and inset.
Previously, we used an ultracentrifugation spin to isolate peripheral blood microvesicles6 ; however, to ensure that microvesicles of various sizes were intact and recovered, we reduced the centrifugal force as reported.39 Cryo-TEM images confirmed that secreted microvesicles from PMA-treated THP-1 cells centrifuged at 16 000g (Figure 1D) and at 160 000g (supplemental Figure 2A) had similar physical properties. As previously reported,40,41 both samples contained small (< 150 nm diameter) and large (> 200 nm diameter) spherical microvesicles (supplemental Figure 2B) with nonuniform internal electron densities and similar distributions. Protrusions on the microvesicle surfaces suggested the presence of membrane proteins.40 The cryo-TEM images also revealed that 3% to 5% of the > 200 nm microvesicles were ruptured in both centrifugation samples, and a greater fraction ruptured in the 160 000g sample (data not shown).
Microvesicles uptake and transfer of RNA molecules in monocytes
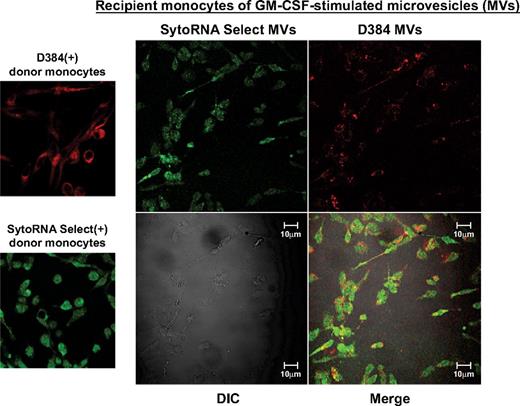
Because microvesicles contain both mRNA and miRNA molecules,6,8,11,12,19-21 we were interested in determining whether the microvesicles transferred RNA molecules to recipient cells using similar concentrations of microvesicles reported for human plasma.39 Fluorescent-labeled microvesicles were incubated with freshly isolated unstained monocytes. Confocal imaging revealed delivery of labeled microvesicles as indicated by the presence of the fluorescent membrane and RNA dyes in unlabeled recipient monocytes (Figure 2). We observed a similar transfer of membrane and RNA dyes from THP-1 cell-derived microvesicles to naive, unlabeled THP-1 cells (supplemental Figure 3A).
Microvesicle uptake by peripheral blood monocytes. Shown are confocal images from GM-CSF–treated donor monocytes stained with D384, a phospholipid membrane dye (red stain; top left panel) and SytoRNA Select, an RNA-select stain (green stain; bottom left panel). After 48 hours, microvesicles from GM-CSF–stimulated monocytes were collected. Based on previous studies,39 100 000 microvesicles/mL were incubated with unstained naive recipient cells. Cells were imaged using the Zeiss LSM 510 multiphoton confocal microscope. Background fluorescence was subtracted using unstained cells. Transfer of fluorescent microvesicles from the GM-CSF–stimulated cells was apparent in the recipient cell membranes and cytoplasm as indicated by the appearance of the red dye as well as transferred RNA as indicated by the green stain (right panel). The differential interference contrast (DIC) was used to visual the morphology of the cells without fluorescence. Data are representative of 3 independent monocyte donors and 3 monocyte recipients.
Microvesicle uptake by peripheral blood monocytes. Shown are confocal images from GM-CSF–treated donor monocytes stained with D384, a phospholipid membrane dye (red stain; top left panel) and SytoRNA Select, an RNA-select stain (green stain; bottom left panel). After 48 hours, microvesicles from GM-CSF–stimulated monocytes were collected. Based on previous studies,39 100 000 microvesicles/mL were incubated with unstained naive recipient cells. Cells were imaged using the Zeiss LSM 510 multiphoton confocal microscope. Background fluorescence was subtracted using unstained cells. Transfer of fluorescent microvesicles from the GM-CSF–stimulated cells was apparent in the recipient cell membranes and cytoplasm as indicated by the appearance of the red dye as well as transferred RNA as indicated by the green stain (right panel). The differential interference contrast (DIC) was used to visual the morphology of the cells without fluorescence. Data are representative of 3 independent monocyte donors and 3 monocyte recipients.
Microvesicle treatment induces differentiation in recipient cells
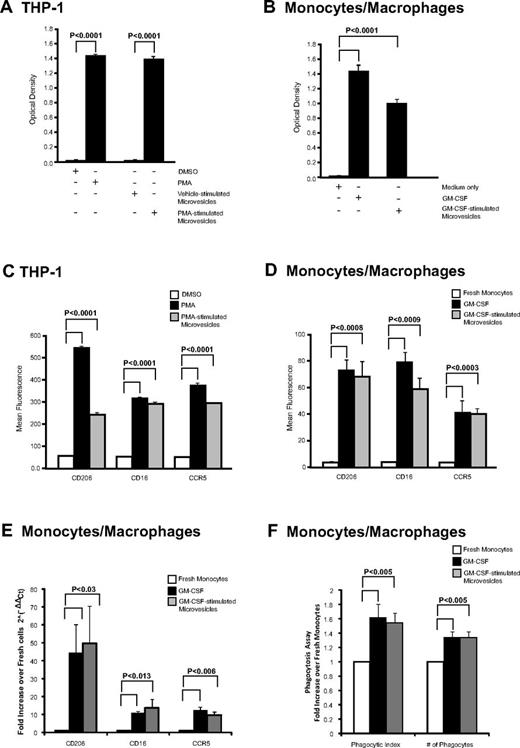
We next examined whether macrophage-derived microvesicles induced activation in recipient cells. Both PMA and microvesicles from PMA-stimulated THP-1 cells induced naive THP-1 cell adherence as measured by crystal violet staining, while DMSO (vehicle) treatment or microvesicles from vehicle-stimulated THP-1 cells did not (Figure 3A). Similarly, microvesicles from GM-CSF–stimulated cells added to naive monocytes significantly increased their adherence similar to GM-CSF–stimulated cells (Figure 3B). These observations suggested that the cells were differentiating in the presence of microvesicles.
Cellular differentiation induced by microvesicles. (A) THP-1 cells and (B) monocytes were treated with microvesicles from PMA- or GM-CSF–stimulated cells, respectively. After 48 hours, adherent cells possessing macrophage-like phenotype were stained with crystal violet and dye uptake was measured at 550 nm. As controls, THP-1 cells were treated with vehicle (DMSO), PMA, or microvesicles from untreated cells, while monocytes were left untreated (media only) or treated with GM-CSF. The absorbance was measured and shown as the average optical density ± SEM (n = 3). (C-D) CD206, CD16, and CCR5 surface expression by flow cytometry. Quantification of surface marker expression is presented as average fluorescence ± SEM (n = 3). Significance was determined comparing to vehicle or freshly isolated monocytes as indicated. (E) Monocytes cells were treated with GM-CSF or microvesicles from GM-CSF–stimulated cells for 48 hours. qRT-PCR was performed using primers for CD16, CD206, or CCR5. Data are normalized over the average Ct value of housekeeping genes GAPDH and CAP-1 and expressed as relative fold increase of in gene expression compared with freshly isolated monocytes. Data represents the average fold change in expression ± SEM for 3 independent experiments. (F) Freshly isolated monocytes were treated with GM-CSF or microvesicles from GM-CSF–stimulated cells for 48 hours. Phagocytic SRBC were counted from 100 cells under fluorescent microscope. Total number of phagocytes and total number of SRBC within 100 phagocytes is presented as phagocytic index. Data shown are the average fold change ± SEM from independent recipient blood donors treated with microvesicles generated from 3 different donors.
Cellular differentiation induced by microvesicles. (A) THP-1 cells and (B) monocytes were treated with microvesicles from PMA- or GM-CSF–stimulated cells, respectively. After 48 hours, adherent cells possessing macrophage-like phenotype were stained with crystal violet and dye uptake was measured at 550 nm. As controls, THP-1 cells were treated with vehicle (DMSO), PMA, or microvesicles from untreated cells, while monocytes were left untreated (media only) or treated with GM-CSF. The absorbance was measured and shown as the average optical density ± SEM (n = 3). (C-D) CD206, CD16, and CCR5 surface expression by flow cytometry. Quantification of surface marker expression is presented as average fluorescence ± SEM (n = 3). Significance was determined comparing to vehicle or freshly isolated monocytes as indicated. (E) Monocytes cells were treated with GM-CSF or microvesicles from GM-CSF–stimulated cells for 48 hours. qRT-PCR was performed using primers for CD16, CD206, or CCR5. Data are normalized over the average Ct value of housekeeping genes GAPDH and CAP-1 and expressed as relative fold increase of in gene expression compared with freshly isolated monocytes. Data represents the average fold change in expression ± SEM for 3 independent experiments. (F) Freshly isolated monocytes were treated with GM-CSF or microvesicles from GM-CSF–stimulated cells for 48 hours. Phagocytic SRBC were counted from 100 cells under fluorescent microscope. Total number of phagocytes and total number of SRBC within 100 phagocytes is presented as phagocytic index. Data shown are the average fold change ± SEM from independent recipient blood donors treated with microvesicles generated from 3 different donors.
To confirm that adherence signified cellular differentiation, recipient cells were stained for macrophage-lineage markers: CD16, CD206, and CCR5 that are highly expressed on macrophages compared with monocytes.32,42-44 Treating naive THP-1 cells with PMA or microvesicles from PMA-stimulated THP-1 cells significantly increased the surface expression of macrophage differentiation markers compared with vehicle-treated samples as quantified by flow cytometry (Figure 3C). For THP1 cells, > 97% of the PMA and microvesicle-treated cells were positive for the surface antigens. The mRNA was also increased for these surface antigens (data not shown). Similarly, monocytes treated with microvesicles from GM-CSF–stimulated cells also exhibited enhanced surface and mRNA expression for CD16, CD206, and CCR5 by flow cytometry and qRT-PCR, respectively (Figure 3D-E, respectively). Notably, GM-CSF or microvesicles treated monocytes were 50% to 55% positive for CCR5 expression, while the percent-positive cells for CD16 were 11.06% ± 3.17% and 24.04% ± 7.23% for GM-CSF and microvesicle-treated cells, respectively. However, CD206 percent-positive cells were greater in GM-CSF–treated cells (69.42% ± 15.37%) compared with microvesicle-treated cells (17.56% ± 1.64%). Furthermore, these genes were absent in the microvesicles as confirmed by qRT-PCR (data not shown).
We further investigated whether the macrophages derived from microvesicle-treated cells were functionally active. We found a significant increase in phagocytic activity in macrophages derived from either GM-CSF or GM-CSF–derived microvesicles compared with freshly isolated monocytes (Figure 3F).
To verify that this effect was directly attributed to the microvesicles, we confirmed that there was no carryover of either GM-CSF or PMA in the microvesicles inducing recipient cell activity. To address whether GM-CSF was retained in the microvesicles preparation, we used a human GM-CSF ELISA. The GM-CSF concentration was < 30 pg/mL in both protein lysates from the microvesicles as well as the GM-CSF–stimulated microvesicle preparation used to treat monocytes and less than the minimal doses of GM-CSF required to activate monocytes.45 Furthermore, we did not detect the presence of GM-CSF mRNA or other genes that cluster with GM-CSF including IL-4, -5, and -13 in the microvesicles (data not shown).
To determine whether PMA carry over from THP-1 cell stimulation was responsible for the recipient cell adherence and differentiation, we found that while microvesicles from PMA-stimulated THP-1 cells induced adherence of naive primary human monocytes, adding PMA directly to these primary monocytes did not induce their differentiation (supplemental Figure 3B). Thus, our observations suggested that the phenotypic and differentiation changes in recipient cells treated with microvesicles from GM-CSF–stimulated monocytes or PMA-stimulated THP-1 cells were from the microvesicle content. Because THP-1 cells are growth factor independent, collectively these observations further suggested that something other than a growth factor was shuttled by the microvesicle to stimulate the differentiation process.
Gene changes in microvesicle-treated cells detected by microarray analysis
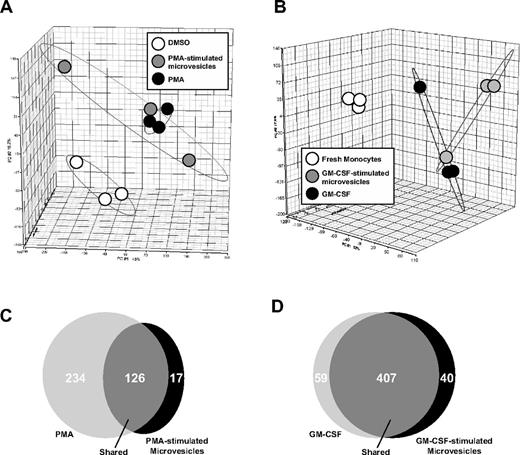
Next, we determined whether microvesicles from GM-CSF–stimulated monocytes or PMA-treated THP-1 cells induced similar gene expression profiles in primary human monocytes or THP-1 cells treated directly with either GM-CSF or PMA, respectively. Principle component analysis (PCA) of the cDNA expression data were performed to determine the percentage of variance among the samples. The gene expression profile of THP-1 cells directly stimulated with PMA compared with cells treated with PMA-stimulated microvesicles with THP-1 cells were highly similar and resided in a relative similar dimension (Figure 4A).
Gene expression profiling after microvesicle treatment. Microarray was performed using Human Genome U133 Chips from RNA isolated from THP-1 cells (n = 3) and peripheral blood monocytes from 3 individual monocyte donors to 3 different monocyte recipients. Each treatment was performed in 3 independent experiments. (A) Principle component analysis (PCA) mapping between THP-1 cells treated with vehicle (DMSO), PMA, or microvesicles from PMA-stimulated cells. (B) Freshly isolated monocytes treated with GM-CSF or microvesicles from GM-CSF–stimulated monocytes were compared with freshly isolated monocytes by PCA. THP-1 cells treated with PMA or microvesicles from PMA-stimulated THP-1 cells expressed similar genes and were different from vehicle-treated cells. Gene expression was similar in monocytes incubated with GM-CSF or microvesicles from GM-CSF–stimulated cells but different compared with freshly isolated monocytes. (C) Venn diagrams generated using pairwise comparisons for significantly expressed genes in cells treated as indicated showed 126 significantly coexpressed genes between THP-1 cells treated with PMA or microvesicles from PMA-stimulated THP-1 cells. (D) 407 significantly coexpressed genes were found between the monocytes incubated with either GM-CSF or microvesicles from GM-CSF–stimulated monocytes.
Gene expression profiling after microvesicle treatment. Microarray was performed using Human Genome U133 Chips from RNA isolated from THP-1 cells (n = 3) and peripheral blood monocytes from 3 individual monocyte donors to 3 different monocyte recipients. Each treatment was performed in 3 independent experiments. (A) Principle component analysis (PCA) mapping between THP-1 cells treated with vehicle (DMSO), PMA, or microvesicles from PMA-stimulated cells. (B) Freshly isolated monocytes treated with GM-CSF or microvesicles from GM-CSF–stimulated monocytes were compared with freshly isolated monocytes by PCA. THP-1 cells treated with PMA or microvesicles from PMA-stimulated THP-1 cells expressed similar genes and were different from vehicle-treated cells. Gene expression was similar in monocytes incubated with GM-CSF or microvesicles from GM-CSF–stimulated cells but different compared with freshly isolated monocytes. (C) Venn diagrams generated using pairwise comparisons for significantly expressed genes in cells treated as indicated showed 126 significantly coexpressed genes between THP-1 cells treated with PMA or microvesicles from PMA-stimulated THP-1 cells. (D) 407 significantly coexpressed genes were found between the monocytes incubated with either GM-CSF or microvesicles from GM-CSF–stimulated monocytes.
Similarly, monocytes treated with GM-CSF or GM-CSF–stimulated microvesicles were similar using PCA mapping of the microarray data (Figure 4B). Using pairwise comparisons, we observed 126 coexpressed genes between cells treated directly with PMA or with microvesicles from PMA-stimulated THP-1 cells (Figure 4C). Notably, 234 genes were exclusively expressed in the THP-1 cells treated with PMA, while 17 genes were unique in cells treated microvesicles from PMA-stimulated THP-1 cells (Figure 4C). On the other hand, treating monocytes with GM-CSF or microvesicles from GM-CSF–stimulated monocytes induced the coexpression of 407 genes (Figure 4D). In contrast, 59 genes and 40 genes were exclusively expressed in monocytes treated with GM-CSF or microvesicles from GM-CSF–treated cells, respectively (Figure 4D).
Using the coexpressed genes in each group from the Venn diagrams in Figure 4C-D, the predicted top associated network functions, molecular and cellular functions, physiologic and systems development functions were analyzed using Ingenuity Pathway Analysis (IPA) software (Tables 1, 2, and 3). It is noteworthy to mention that the IPA software identified hematologic system development as a top-ranked pathway in THP-1 cells treated with PMA-stimulated microvesicles and in monocytes treated with GM-CSF–stimulated microvesicles.
Associated network functions of coexpressed genes
| THP-1 cells . | Score . | Monocytes . | Score . |
|---|---|---|---|
| Gene expression, carbohydrate metabolism, digestive system development and function | 44 | Cell morphology, lipid metabolism, small molecule biochemistry | 40 |
| Protein synthesis, antigen presentation, cell-to-cell signaling and interaction | 42 | Cell-to-cell signaling and interaction, tissue development, cellular movement | 36 |
| Cellular development, organismal injury and abnormalities, respiratory disease | 34 | Gene expression, hematologic system development and function, immune cell trafficking | 33 |
| Cell-to-cell signaling and interaction, cellular function and maintenance, hematologic system development and function | 32 | Free radical scavenging, molecular transport, energy production | 29 |
| Drug metabolism, lipid metabolism, small molecule biochemistry | 24 | Tissue development, nucleic acid metabolism, small molecule biochemistry | 21 |
| THP-1 cells . | Score . | Monocytes . | Score . |
|---|---|---|---|
| Gene expression, carbohydrate metabolism, digestive system development and function | 44 | Cell morphology, lipid metabolism, small molecule biochemistry | 40 |
| Protein synthesis, antigen presentation, cell-to-cell signaling and interaction | 42 | Cell-to-cell signaling and interaction, tissue development, cellular movement | 36 |
| Cellular development, organismal injury and abnormalities, respiratory disease | 34 | Gene expression, hematologic system development and function, immune cell trafficking | 33 |
| Cell-to-cell signaling and interaction, cellular function and maintenance, hematologic system development and function | 32 | Free radical scavenging, molecular transport, energy production | 29 |
| Drug metabolism, lipid metabolism, small molecule biochemistry | 24 | Tissue development, nucleic acid metabolism, small molecule biochemistry | 21 |
Molecular and cellular functions of coexpressed genes
| THP-1 cells . | P . | Monocytes . | P . |
|---|---|---|---|
| Cell death | .0000432 | Lipid metabolism | .000832 |
| Cellular function and maintenance | .0000432 | Molecular transport | .000832 |
| Lipid metabolism | .000272 | Small molecule biochemistry | .000832 |
| Small molecule biochemistry | .000272 | RNA posttranscriptional modification | .00147 |
| Cellular development | .0016 | Carbohydrate metabolism | .00378 |
| THP-1 cells . | P . | Monocytes . | P . |
|---|---|---|---|
| Cell death | .0000432 | Lipid metabolism | .000832 |
| Cellular function and maintenance | .0000432 | Molecular transport | .000832 |
| Lipid metabolism | .000272 | Small molecule biochemistry | .000832 |
| Small molecule biochemistry | .000272 | RNA posttranscriptional modification | .00147 |
| Cellular development | .0016 | Carbohydrate metabolism | .00378 |
Associated systems and functions of coexpressed genes
| THP-1 cells . | P . | Monocytes . | P . |
|---|---|---|---|
| Hematologic system development and function | .0012 | Hematologic system development and function | .0192 |
| Reproductive system development and function | .0016 | Cardiovascular system development and function | .0199 |
| Immune cell trafficking | .00251 | Cell-mediated immune response | .0199 |
| Tissue development | .00308 | Digestive system development and function | .0199 |
| Nervous system development and function | .00353 | Embryonic development | .0199 |
| THP-1 cells . | P . | Monocytes . | P . |
|---|---|---|---|
| Hematologic system development and function | .0012 | Hematologic system development and function | .0192 |
| Reproductive system development and function | .0016 | Cardiovascular system development and function | .0199 |
| Immune cell trafficking | .00251 | Cell-mediated immune response | .0199 |
| Tissue development | .00308 | Digestive system development and function | .0199 |
| Nervous system development and function | .00353 | Embryonic development | .0199 |
Nonmyeloid uptake of GM-CSF–stimulated microvesicles
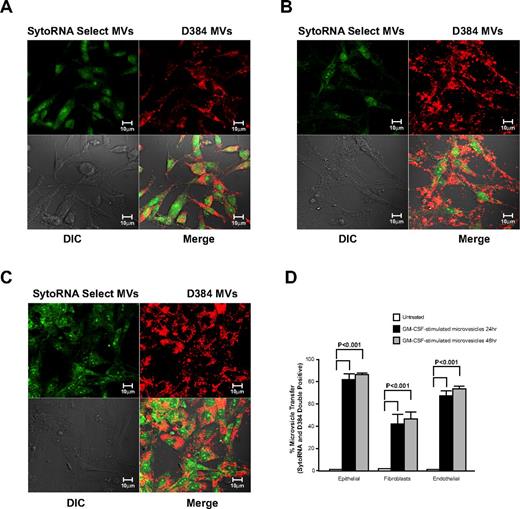
We next wanted to determine whether GM-CSF–stimulated microvesicles bind to cells other than myeloid lineages and transferred RNA molecules. Therefore, fluorescently labeled GM-CSF–stimulated microvesicles were incubated with epithelial cells (A549), fibroblasts (CCL-204) or endothelial cells (HUVECs). Confocal imaging revealed that membrane- and RNA-labeled microvesicles from GM-CSF–stimulated cells were readily taken up by all 3 cell types (Figure 5A-C). As observed by flow cytometry, significant increase in fluorescence of double-positive cells for the membrane and RNA dye was apparent after 24 hours of incubation. No further increase in uptake was observed at 48 hours in the microvesicle-treated cells (Figure 5D).
Cellular uptake of microvesicles from GM-CSF–stimulated monocytes. Donor monocytes cells were stained with D384 and SytoRNA Select then treated with GM-CSF. Microvesicles from GM-CSF–stimulated monocytes were collected and added to different cells lines as indicated. Representative confocal images using the Zeiss LSM 510 multiphoton confocal microscope are shown. Microvesicles obtained from 3 individual monocyte donors revealed the uptake of the microvesicles by (A) A549 lung epithelial cells, (B) CCL-204 lung fibroblasts, and (C) human umbilical vein endothelial cells (HUVECs). (D) Quantification of microvesicle transfer from 3 independent experiments is shown (average ± SEM). Cellular morphology was visualized without fluorescence using differential interference contrast (DIC).
Cellular uptake of microvesicles from GM-CSF–stimulated monocytes. Donor monocytes cells were stained with D384 and SytoRNA Select then treated with GM-CSF. Microvesicles from GM-CSF–stimulated monocytes were collected and added to different cells lines as indicated. Representative confocal images using the Zeiss LSM 510 multiphoton confocal microscope are shown. Microvesicles obtained from 3 individual monocyte donors revealed the uptake of the microvesicles by (A) A549 lung epithelial cells, (B) CCL-204 lung fibroblasts, and (C) human umbilical vein endothelial cells (HUVECs). (D) Quantification of microvesicle transfer from 3 independent experiments is shown (average ± SEM). Cellular morphology was visualized without fluorescence using differential interference contrast (DIC).
GM-CSF–stimulated microvesicles shuttle functionally active miR-223 to target cells
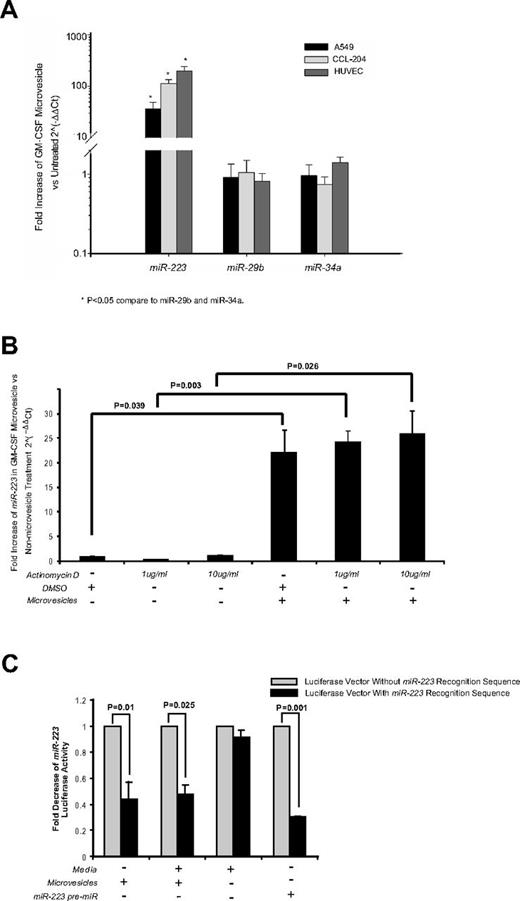
To determine whether the microvesicles could transfer miRNAs to target cells, we first characterized the miRNA expression in the microvesicles. As shown in Table 4, we detected 186 miRNAs expressed in the GM-CSF–stimulated microvesicles. Previously, we identified that miR-223 was among the highest expressed miRNA in peripheral blood mononuclear cells and microvesicles, while miR-29b was expressed at low levels in these microvesicles.6 Notably, GM-CSF–stimulated microvesicles had a similarly high expression of miR-223 (mean normalized expression (ΔCt) 53 018.46 ± 33 473.39 SEM, n = 5, Table 4), while miRs-29b and -34a were undetectable.
Normalized miRNA expression in microvesicles from GM-CSF–treated monocytes (n = 3)
| miRNA . | Mean (ΔCt) ± SD . |
|---|---|
| hsa-miR-223 | 53 018.46 ± 33 473.39 |
| hsa-miR-191 | 1012.46 ± 655.37 |
| hsa-miR-193a | 732.27 ± 855.55 |
| hsa-miR-484 | 569.21 ± 128.13 |
| hsa-miR-324-5p | 565.25 ± 675.85 |
| hsa-miR-182* | 466.01 ± 591.37 |
| hsa-miR-222 | 393.56 ± 264.48 |
| hsa-miR-016 | 281.18 ± 197.40 |
| hsa-miR-432 | 280.28 ± 366.88 |
| hsa-miR-026a | 258.65 ± 174.98 |
| hsa-miR-146a | 255.12 ± 251.93 |
| hsa-miR-024 | 209.18 ± 160.62 |
| hsa-miR-150 | 190.38 ± 331.16 |
| hsa-miR-574 | 186.52 ± 113.85 |
| hsa-miR-019b | 185.97 ± 156.39 |
| hsa-miR-328 | 183.57 ± 215.33 |
| hsa-miR-585 | 166.96 ± 206.43 |
| hsa-miR-197 | 115.92 ± 59.58 |
| hsa-miR-146b | 115.88 ± 113.56 |
| hsa-miR-614 | 113.68 ± 140.24 |
| hsa-miR-572 | 96.70 ± 123.69 |
| hsa-miR-092 | 96.64 ± 38.47 |
| hsa-miR-155 | 90.73 ± 70.86 |
| hsa-miR-020a | 87.20 ± 70.18 |
| hsa-miR-093 | 81.17 ± 74.65 |
| hsa-miR-030b | 78.82 ± 37.54 |
| hsa-miR-140 | 77.48 ± 48.37 |
| hsa-miR-030c | 64.98 ± 31.53 |
| hsa-miR-149 | 53.86 ± 67.93 |
| hsa-miR-021 | 48.26 ± 48.10 |
| hsa-miR-142-3p | 47.38 ± 38.98 |
| hsa-miR-132 | 45.97 ± 46.86 |
| hsa-let-7b | 44.44 ± 23.13 |
| hsa-miR-331 | 42.97 ± 14.95 |
| hsa-miR-030a-5p | 33.47 ± 24.29 |
| hsa-miR-629 | 31.47 ± 40.41 |
| hsa-miR-142-5p | 30.04 ± 18.93 |
| hsa-miR-342 | 29.81 ± 19.52 |
| hsa-miR-027a | 27.62 ± 18.24 |
| hsa-miR-025 | 21.99 ± 12.24 |
| hsa-miR-186 | 20.80 ± 15.72 |
| hsa-miR-015b | 20.45 ± 14.06 |
| hsa-miR-423 | 17.05 ± 9.01 |
| hsa-miR-133b | 16.86 ± 20.11 |
| hsa-miR-026b | 16.75 ± 15.05 |
| hsa-miR-195 | 15.02 ± 12.17 |
| hsa-miR-422a | 13.51 ± 11.12 |
| hsa-miR-029a | 12.33 ± 13.22 |
| hsa-miR-532 | 12.02 ± 6.82 |
| hsa-miR-345 | 11.81 ± 7.65 |
| hsa-miR-099b | 11.62 ± 13.22 |
| hsa-miR-613 | 10.59 ± 13.66 |
| hsa-miR-029c | 10.55 ± 5.55 |
| hsa-miR-181d | 9.78 ± 4.52 |
| hsa-miR-019a | 9.68 ± 9.49 |
| hsa-miR-125a | 9.39 ± 9.36 |
| hsa-miR-320 | 8.13 ± 3.67 |
| hsa-miR-425-5p | 7.51 ± 5.87 |
| hsa-miR-425-3p | 7.04 ± 2.54 |
| hsa-miR-106b | 6.76 ± 4.11 |
| hsa-miR-015a | 6.68 ± 6.01 |
| hsa-miR-374 | 6.62 ± 4.96 |
| hsa-miR-030d | 6.54 ± 4.49 |
| hsa-miR-618 | 5.99 ± 6.92 |
| hsa-let-7a | 5.44 ± 5.05 |
| hsa-miR-487a | 5.15 ± 6.26 |
| hsa-let-7g | 4.28 ± 4.64 |
| hsa-let-7c | 4.25 ± 2.79 |
| hsa-miR-486 | 4.03 ± 2.52 |
| hsa-miR-376a | 3.86 ± 5.20 |
| hsa-miR-126 | 3.47 ± 2.36 |
| hsa-miR-103 | 3.31 ± 3.60 |
| hsa-miR-020b | 3.20 ± 2.90 |
| hsa-miR-550 | 3.16 ± 2.49 |
| hsa-miR-657 | 3.06 ± 3.88 |
| hsa-miR-660 | 2.91 ± 2.80 |
| hsa-miR-597 | 2.72 ± 3.19 |
| hsa-miR-362 | 2.68 ± 1.91 |
| hsa-miR-579 | 2.67 ± 3.94 |
| hsa-let-7d | 2.64 ± 2.23 |
| hsa-miR-554 | 2.50 ± 3.11 |
| hsa-miR-571 | 2.42 ± 3.09 |
| hsa-miR-378 | 2.35 ± 1.09 |
| hsa-miR-628 | 2.34 ± 2.79 |
| hsa-miR-151 | 2.24 ± 1.78 |
| hsa-miR-017-5p | 2.22 ± 2.18 |
| hsa-miR-339 | 2.19 ± 0.49 |
| hsa-miR-565 | 2.05 ± 1.04 |
| hsa-miR-361 | 2.03 ± 2.03 |
| hsa-miR-632 | 1.98 ± 2.14 |
| hsa-miR-096 | 1.91 ± 3.51 |
| hsa-miR-200c | 1.89 ± 1.55 |
| hsa-miR-340 | 1.87 ± 1.82 |
| hsa-miR-604 | 1.72 ± 1.65 |
| hsa-miR-193b | 1.66 ± 1.99 |
| hsa-miR-371 | 1.62 ± 3.16 |
| hsa-miR-586 | 1.61 ± 2.23 |
| hsa-miR-636 | 1.60 ± 2.87 |
| hsa-miR-610 | 1.59 ± 2.33 |
| hsa-miR-028 | 1.58 ± 1.40 |
| hsa-miR-365 | 1.52 ± 1.40 |
| hsa-miR-302d | 1.50 ± 2.51 |
| miRNA . | Mean (ΔCt) ± SD . |
|---|---|
| hsa-miR-223 | 53 018.46 ± 33 473.39 |
| hsa-miR-191 | 1012.46 ± 655.37 |
| hsa-miR-193a | 732.27 ± 855.55 |
| hsa-miR-484 | 569.21 ± 128.13 |
| hsa-miR-324-5p | 565.25 ± 675.85 |
| hsa-miR-182* | 466.01 ± 591.37 |
| hsa-miR-222 | 393.56 ± 264.48 |
| hsa-miR-016 | 281.18 ± 197.40 |
| hsa-miR-432 | 280.28 ± 366.88 |
| hsa-miR-026a | 258.65 ± 174.98 |
| hsa-miR-146a | 255.12 ± 251.93 |
| hsa-miR-024 | 209.18 ± 160.62 |
| hsa-miR-150 | 190.38 ± 331.16 |
| hsa-miR-574 | 186.52 ± 113.85 |
| hsa-miR-019b | 185.97 ± 156.39 |
| hsa-miR-328 | 183.57 ± 215.33 |
| hsa-miR-585 | 166.96 ± 206.43 |
| hsa-miR-197 | 115.92 ± 59.58 |
| hsa-miR-146b | 115.88 ± 113.56 |
| hsa-miR-614 | 113.68 ± 140.24 |
| hsa-miR-572 | 96.70 ± 123.69 |
| hsa-miR-092 | 96.64 ± 38.47 |
| hsa-miR-155 | 90.73 ± 70.86 |
| hsa-miR-020a | 87.20 ± 70.18 |
| hsa-miR-093 | 81.17 ± 74.65 |
| hsa-miR-030b | 78.82 ± 37.54 |
| hsa-miR-140 | 77.48 ± 48.37 |
| hsa-miR-030c | 64.98 ± 31.53 |
| hsa-miR-149 | 53.86 ± 67.93 |
| hsa-miR-021 | 48.26 ± 48.10 |
| hsa-miR-142-3p | 47.38 ± 38.98 |
| hsa-miR-132 | 45.97 ± 46.86 |
| hsa-let-7b | 44.44 ± 23.13 |
| hsa-miR-331 | 42.97 ± 14.95 |
| hsa-miR-030a-5p | 33.47 ± 24.29 |
| hsa-miR-629 | 31.47 ± 40.41 |
| hsa-miR-142-5p | 30.04 ± 18.93 |
| hsa-miR-342 | 29.81 ± 19.52 |
| hsa-miR-027a | 27.62 ± 18.24 |
| hsa-miR-025 | 21.99 ± 12.24 |
| hsa-miR-186 | 20.80 ± 15.72 |
| hsa-miR-015b | 20.45 ± 14.06 |
| hsa-miR-423 | 17.05 ± 9.01 |
| hsa-miR-133b | 16.86 ± 20.11 |
| hsa-miR-026b | 16.75 ± 15.05 |
| hsa-miR-195 | 15.02 ± 12.17 |
| hsa-miR-422a | 13.51 ± 11.12 |
| hsa-miR-029a | 12.33 ± 13.22 |
| hsa-miR-532 | 12.02 ± 6.82 |
| hsa-miR-345 | 11.81 ± 7.65 |
| hsa-miR-099b | 11.62 ± 13.22 |
| hsa-miR-613 | 10.59 ± 13.66 |
| hsa-miR-029c | 10.55 ± 5.55 |
| hsa-miR-181d | 9.78 ± 4.52 |
| hsa-miR-019a | 9.68 ± 9.49 |
| hsa-miR-125a | 9.39 ± 9.36 |
| hsa-miR-320 | 8.13 ± 3.67 |
| hsa-miR-425-5p | 7.51 ± 5.87 |
| hsa-miR-425-3p | 7.04 ± 2.54 |
| hsa-miR-106b | 6.76 ± 4.11 |
| hsa-miR-015a | 6.68 ± 6.01 |
| hsa-miR-374 | 6.62 ± 4.96 |
| hsa-miR-030d | 6.54 ± 4.49 |
| hsa-miR-618 | 5.99 ± 6.92 |
| hsa-let-7a | 5.44 ± 5.05 |
| hsa-miR-487a | 5.15 ± 6.26 |
| hsa-let-7g | 4.28 ± 4.64 |
| hsa-let-7c | 4.25 ± 2.79 |
| hsa-miR-486 | 4.03 ± 2.52 |
| hsa-miR-376a | 3.86 ± 5.20 |
| hsa-miR-126 | 3.47 ± 2.36 |
| hsa-miR-103 | 3.31 ± 3.60 |
| hsa-miR-020b | 3.20 ± 2.90 |
| hsa-miR-550 | 3.16 ± 2.49 |
| hsa-miR-657 | 3.06 ± 3.88 |
| hsa-miR-660 | 2.91 ± 2.80 |
| hsa-miR-597 | 2.72 ± 3.19 |
| hsa-miR-362 | 2.68 ± 1.91 |
| hsa-miR-579 | 2.67 ± 3.94 |
| hsa-let-7d | 2.64 ± 2.23 |
| hsa-miR-554 | 2.50 ± 3.11 |
| hsa-miR-571 | 2.42 ± 3.09 |
| hsa-miR-378 | 2.35 ± 1.09 |
| hsa-miR-628 | 2.34 ± 2.79 |
| hsa-miR-151 | 2.24 ± 1.78 |
| hsa-miR-017-5p | 2.22 ± 2.18 |
| hsa-miR-339 | 2.19 ± 0.49 |
| hsa-miR-565 | 2.05 ± 1.04 |
| hsa-miR-361 | 2.03 ± 2.03 |
| hsa-miR-632 | 1.98 ± 2.14 |
| hsa-miR-096 | 1.91 ± 3.51 |
| hsa-miR-200c | 1.89 ± 1.55 |
| hsa-miR-340 | 1.87 ± 1.82 |
| hsa-miR-604 | 1.72 ± 1.65 |
| hsa-miR-193b | 1.66 ± 1.99 |
| hsa-miR-371 | 1.62 ± 3.16 |
| hsa-miR-586 | 1.61 ± 2.23 |
| hsa-miR-636 | 1.60 ± 2.87 |
| hsa-miR-610 | 1.59 ± 2.33 |
| hsa-miR-028 | 1.58 ± 1.40 |
| hsa-miR-365 | 1.52 ± 1.40 |
| hsa-miR-302d | 1.50 ± 2.51 |
Because epithelial cells, fibroblasts, and endothelial cells have low basal expression of miRs-223, -29b, and -34a, they are an amendable model to measure the shuttling of miR-223. On transfer and uptake of GM-CSF–stimulated microvesicles, these target cells expressed significantly higher levels of miR-223 but levels of miRs-34a and -29b remained unchanged (Figure 6A). These data are consistent with the direct transfer of miR-223 from microvesicles to target cells. To ensure that the target cells did not up-regulate the expression of miR-223 on microvesicle treatment, A549 cells were treated with actinomycin D before microvesicle treatment. As shown in Figure 6B, similar miR-223 was detected in the microvesicle-treated cells regardless of the presence of actinomycin D.
Cellular uptake of functional miR-223 contained in microvesicles from GM-CSF–stimulated monocytes does not require recipient cell transcriptional activity for expression. (A) A549, CCL-204, or HUVEC cells were incubated with microvesicles from GM-CSF–stimulated monocytes for 2 days. RNA was then isolated and expression for miRs-223, -29b, and -34a was measured by qRT-PCR. Shown is the average fold increase ± SEM (n = 4) in miR-223 levels compared with untreated cells. A significant increase after treatment with GM-CSF–stimulated microvesicles is observed for miR-223 (*P < .05), while miRs-29b and -34a levels remained unchanged. (B) A549 cells (5 × 106 cells/condition) were treated with vehicle (DMSO) or actinomycin D at the indicated concentrations for 6 hours. The cells were washed and culture media was replaced in the absence or presence of microvesicles. After 24 hours, RNA was isolated and miR-223 expression measured by qRT-PCR. Fold increase was determined by comparing the samples to the cells treated only with DMSO. Shown is the average ± SEM for cells treated with microvesicles from 4 independent donors. (C) A549 cells were transfected with the luciferase reporter vector containing miR-223 recognition sequence, a control luciferase vector lacking the sequence or cotransfected with miR-223 precursor. After 18-20 hours incubation, the cells were treated with or without microvesicles from GM-CSF–stimulated monocytes. The cells were cultured for another 24 hours then lysed and the luciferase activity was measured. The data are expressed as fold-decrease of luciferase activity of cells transfected with the luciferase reporter containing miR-223 recognition sequence over the vector lacking the recognition sequence for each culture condition. Shown is the average data ± SEM from A549 cells treated with microvesicles generated from 4 independent monocyte-derived microvesicle donors.
Cellular uptake of functional miR-223 contained in microvesicles from GM-CSF–stimulated monocytes does not require recipient cell transcriptional activity for expression. (A) A549, CCL-204, or HUVEC cells were incubated with microvesicles from GM-CSF–stimulated monocytes for 2 days. RNA was then isolated and expression for miRs-223, -29b, and -34a was measured by qRT-PCR. Shown is the average fold increase ± SEM (n = 4) in miR-223 levels compared with untreated cells. A significant increase after treatment with GM-CSF–stimulated microvesicles is observed for miR-223 (*P < .05), while miRs-29b and -34a levels remained unchanged. (B) A549 cells (5 × 106 cells/condition) were treated with vehicle (DMSO) or actinomycin D at the indicated concentrations for 6 hours. The cells were washed and culture media was replaced in the absence or presence of microvesicles. After 24 hours, RNA was isolated and miR-223 expression measured by qRT-PCR. Fold increase was determined by comparing the samples to the cells treated only with DMSO. Shown is the average ± SEM for cells treated with microvesicles from 4 independent donors. (C) A549 cells were transfected with the luciferase reporter vector containing miR-223 recognition sequence, a control luciferase vector lacking the sequence or cotransfected with miR-223 precursor. After 18-20 hours incubation, the cells were treated with or without microvesicles from GM-CSF–stimulated monocytes. The cells were cultured for another 24 hours then lysed and the luciferase activity was measured. The data are expressed as fold-decrease of luciferase activity of cells transfected with the luciferase reporter containing miR-223 recognition sequence over the vector lacking the recognition sequence for each culture condition. Shown is the average data ± SEM from A549 cells treated with microvesicles generated from 4 independent monocyte-derived microvesicle donors.
We next evaluated whether miR-223 transferred from the GM-CSF–stimulated microvesicles was functional in the recipient cells. To address this, we transfected A549 cells with a 3′-UTR luciferase reporter vector to which miR-223 binds. As shown in Figure 6C, transfected cells in media alone displayed robust luciferase production. However, when A549 cells were cultured with GM-CSF–stimulated microvesicles containing miR-223, luciferase activity was decreased significantly in cells transfected with the miR-223 3′-UTR luciferase reporter vector but not the control vector. Because these cells do not normally have miR-223, this indicated that miR-223 transferred from the microvesicles down-regulated luciferase production. As a positive control, we cotransfected the luciferase reporter vector and the miR-223 precursor. As expected, luciferase production was also decreased in these cells.
Inhibition of miR-223 reduces macrophage differentiation and survival
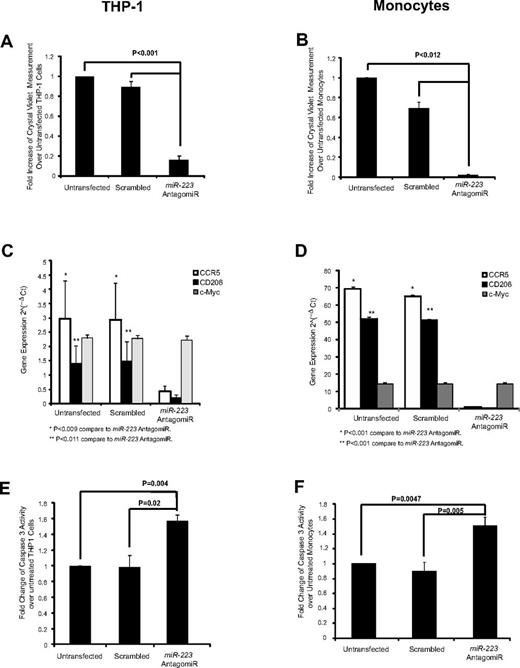
Because miR-223 is the most abundant miRNA in the microvesicles, we next examined if this miRNA was important in macrophage development. Using an antagomiR to miR-223, we specifically knocked down the expression of miR-223 in both THP-1 cells and monocytes (supplemental Figure 4A-B). Notably, miR-191 which is the second most abundant miRNA the microvesicles was unchanged in the treated cells. Phenotypically, miR-223 antagomiR-transfected THP-1 cells or monocytes did not become adherent in the presence of PMA or GM-CSF, respectively (Figure 7A-B). We further addressed whether the lack of cell adherence was associated with inhibition of cellular differentiation or survival. We found that the miR-223 antagomiR treatment prevented PMA or GM-CSF induction of macrophage markers CCR5 or CD206 (Figure 7C-D). As expected, c-Myc, which does not change during macrophage differentiation, was unaffected by the miR-223 antagomiR (Figure 7C-D). Moreover, we found that survival was compromised in the miR-233–transfected cells compared with untreated or scrambled-transfected cells as measured by an increase in caspase-3 activity (Figure 7E-F) and a concordant significant decrease in BCL-2 and BCL-xL expression (supplemental Figure 4C-D).
AntagomiR to miR-223 reduces macrophage differentiation and cell survival. In 6-well plates, THP-1 cells (0.5 × 106) and monocytes (1.0 × 106) were plated in 2.4 mL of media then transfected with miR-223 antagomiR and incubated with PMA or GM-CSF, respectively, in X-VIVO 15 medium. Both nonadherent and adherent cells were collected and analyzed for cellular differentiation and survival. Cellular adherence was measured by crystal violet assay in the antagomiR-transfected (A) THP-1 cells and (B) monocytes. Shown is the average absorbance for crystal violet ± SEM (n = 3 in duplicate). AntagomiR-transfected (C) THP-1 cells and (D) monocytes were analyzed for gene expression as indicated. The average relative copy number is shown ± SEM from 3 independent experiments. Caspase-3 assay was performed for (E) THP-1 cells (n = 3) and (F) monocytes (n = 5) transfected with the miR-223 antagomiR then treated with PMA or GM-CSF, respectively. Shown is the average fold change ± SEM compared with untransfected cells.
AntagomiR to miR-223 reduces macrophage differentiation and cell survival. In 6-well plates, THP-1 cells (0.5 × 106) and monocytes (1.0 × 106) were plated in 2.4 mL of media then transfected with miR-223 antagomiR and incubated with PMA or GM-CSF, respectively, in X-VIVO 15 medium. Both nonadherent and adherent cells were collected and analyzed for cellular differentiation and survival. Cellular adherence was measured by crystal violet assay in the antagomiR-transfected (A) THP-1 cells and (B) monocytes. Shown is the average absorbance for crystal violet ± SEM (n = 3 in duplicate). AntagomiR-transfected (C) THP-1 cells and (D) monocytes were analyzed for gene expression as indicated. The average relative copy number is shown ± SEM from 3 independent experiments. Caspase-3 assay was performed for (E) THP-1 cells (n = 3) and (F) monocytes (n = 5) transfected with the miR-223 antagomiR then treated with PMA or GM-CSF, respectively. Shown is the average fold change ± SEM compared with untransfected cells.
Discussion
Microvesicles are important mediators of cell-cell communication. Released from the endosomal compartment or shed from the cell surface, microvesicles can directly stimulate target cells by receptor-mediated interactions or by transferring bioactive molecules including proteins, mRNAs, miRNAs, and organelles.1,3-5,7,8,10-13,23 Previously, we reported that circulating microvesicles in the plasma of normal human volunteers contain miRNAs and found that these microvesicles are most commonly derived from platelets and macrophages.6 The present data explores the function of macrophage-derived microvesicles in activating target cells and inducing their differentiation.
Notably, microvesicle production and release are signal and stimuli dependent. Factors like environmental stress and calcium concentration affect microvesicle release.2,7,9,15-17,30 In addition, cytokines like IL-1β induce microvesicle shedding from peripheral blood monocytes, demonstrating the impact of cytokines in microvesicle production.46 Accordingly, this report demonstrates that factors such as GM-CSF and PMA induces the production of microvesicles from monocytes and the myeloid leukemic cell line, THP-1, respectively. In accordance with others,4,7 treatment of tumor cells with PMA significantly alters calcium concentration and increases tumor-derived microvesicle release. Thus, we cannot rule out this mechanism of THP-1 cell microvesicle formation found in this study.
In the current study, microvesicle production increased as these stimuli induced cellular differentiation. Furthermore, these microvesicles drove the differentiation of THP-1 cells and naive monocytes. Previously, Valenti et al was the first to report that tumor-associated microvesicles impair monocytic differentiation to dendritic cells.14 In this study, myeloid cells generated by tumor microvesicles have an immunosuppressive phenotype and produce cytokines to promote tumor growth. In contrast, our study shows that normal macrophages produce microvesicles to maintain homeostasis and immune cell production. Thus, it appears that tumor cells have evolved to take advantage of this phenomenon.
Interestingly, dendritic cells use exosomes to communicate with each other.12 Many studies reveal cell-cell communication between derived microvesicles and cells of similar lineage.2,11,12,23,24 Our data suggest that macrophage-derived microvesicles communicate with a variety cell types, suggesting that the impact of these microvesicles may be widespread. We also found that microvesicle treatment of target cells induces genetic and phenotypic changes, appearing not to require the transcriptional machinery of the target cell.
Analysis of the gene profile from microvesicle-treated cells reveals several cellular processes including cellular metabolism, survival, signaling, membrane integrity, hematopoietic cell development, and immune cell homeostasis. We found that there was no carryover of GM-CSF in microvesicle preparations to contribute to these changes within the target cells. Furthermore, while microvesicles from PMA-stimulated THP-1 cells stimulated primary human monocyte differentiation, adding PMA directly to human monocytes did not induce these cells to differentiate. Because THP-1 cells are growth factor independent and microvesicles mediated their differentiation, our observations suggest that other critical factors were contained in macrophage-derived microvesicles. Therefore, we were interested in defining the molecules encapsulated in the microvesicles that contributed to cellular differentiation.
While microvesicles shed from apoptotic bodies can contain the morphogen, Hedgehog, to promote megakaryocyte differentiation,13 we find few apoptotic bodies in the GM-CSF– or PMA- stimulated microvesicles. Previously, Valadi et al reported that exosomes shuttle extracellular mRNAs or miRNAs to target cells.8 Because this concept of extracellular shuttle (es) RNAs was reported,8 numerous investigators report that exosomes and microvesicles from a variety of cells, including stem cells, contain both mRNAs and miRNAs.6,10-12,19,22 Of particular note, mesenchymal stem cell (MSC)–derived microvesicles appear to be selective in the miRNAs shuttled to target cells.11 In concordance with these studies, we confirm that the RNA molecules contained in the macrophage-derived microvesicles are shuttled not only to monocytic cells but other cell lineages.
The role of the esRNAs in target cells is just beginning to be elucidated. For example, monocyte-derived microvesicles contain miR-155 that mediate endothelial cell migration.22 Similar to our previous report of the miRNA content in peripheral blood microvesicles,6 we observed a similar pattern of miRNA expression in the macrophage-derived microvesicles with miR-223 being the most highly expressed. As we confirm here and as reported by others,47,48 miR-223 decreases as a monocyte differentiates to a macrophage (data not shown). Similarly miR-223 decreases during osteoclast and erythroid differentiation,49,50 while increased miR-223 is important in megakaryocyte production50 and progenitor cell proliferation.48 Furthermore, the absence of miR-223 is associated with incomplete granulopoiesis.48 Therefore, it is possible that during GM-CSF–induced macrophage differentiation, the monocytic cell selectively releases miR-223 to permit completion of the maturation process. On doing so, the miR-223 contained in the macrophage-derived microvesicles can be shuttled to other progenitor lineages to complete their terminal maturation such as granulocytes or megakaryocytes.
Because macrophage-derived microvesicles contain high levels of miR-223 and induced differentiation of recipient monocytes, we were interested in the impact of this miRNA on monocyte homeostasis. Several predicted targets of miR-223 include inositol phosphatases that are important in monocyte survival.45 As such, knocking down miR-223 in monocytes and THP-1 cells critically impacted cell survival. While cell viability was compromised in miR-223 antagomiR-transfected cells, we also observed that viable miR-223 antagomiR-transfected cells were unable to differentiate into macrophages in the presence of GM-CSF or PMA. Thus, current studies are underway to evaluate the precise mechanisms and time frame which miR-223 is required during macrophage differentiation. In addition, we are investigating whether the miR-223 antagomiR can reverse the differentiation process.
These data begin to uncover the function of microvesicles in target cells. In fact, continuous transfer of genetic information, including the shuttling of miRNAs between microvesicles and target cells, may be an important factor in this process. To our knowledge, the present article is the first to demonstrate that macrophage-derived microvesicles induced cellular differentiation in target cells. Because microvesicles are produced and released at high concentrations in inflammatory conditions,2,3,9,25,46 we speculate that macrophage-derived microvesicles are important mediators of the inflammatory response. We surmise that microvesicles produced from activated macrophages induce the differentiation of recruited monocytes, activate hematopoietic cell production in the marrow, and induce the release of more microvesicles. Thus, this feedback mechanism may be an innate response element that locally activates the immune system.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors appreciate the assistance Dr Baltazar Aguda provided in using the Ingenuity Pathway Analysis Software. The authors greatly appreciate the assistance provided to perform confocal and TEM microscopy through the Microscopy Core Laboratory (Davis Heart Lung Research Institute) and The Microscopy Shared Resource (The Ohio State University Comprehensive Cancer Center, James). The cryo-TEM data were obtained at the TEM facility at the Liquid Crystal Institute (Kent State University) supported by the Ohio Research Scholars Program Research Cluster on Surfaces in Advanced Materials. The authors thank Dr Min Gao for technical support provided for the TEM experiments. The authors also acknowledge the assistance of Mr Gary Philips (The Center for Biostatistics). The authors thank Ms Valerie Wright for assistance in generating graphs and Mr Kitch Kelly for help with performing the phagocytosis assay.
This work was supported by research funding from National Institutes of Health/National Heart, Lung, and Blood Institute (NHLBI) R01 HL067176 (C.B.M.).
National Institutes of Health
Authorship
Contribution: N.I., Y.W., D.D., L.M., K.B., P.S., J.W., S.T., and M.G.P. performed research; N.I., Y.W., L.M., T.D.E., S.T., M.G.P., and C.B.M. analyzed data; N.I., Y.W., M.G.P., and C.B.M. wrote the manuscript; K.A. and M.E.P. performed the DLS and cyro-TEM studies; and M.G.P. and C.B.M. conceived ideas and designed research.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for N.I. is Department of Internal Medicine-Nephrology, University of Michigan, 1150 W Medical Center Dr, 1574 MSRB II, Ann Arbor, MI 48109-5680.
Correspondence: Melissa Piper, PhD, and Clay Marsh, MD, 260 Meiling Hall, 370 West 9th Ave, Columbus, OH 43210; e-mail: melissa.piper@osumc.edu or clay.marsh@osumc.edu.
References
Author notes
N.I. and Y.W. contributed equally.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal