Abstract
Fifty percent of Diamond-Blackfan anemia (DBA) patients possess mutations in genes coding for ribosomal proteins (RPs). To identify new mutations, we investigated large deletions in the RP genes RPL5, RPL11, RPL35A, RPS7, RPS10, RPS17, RPS19, RPS24, and RPS26. We developed an easy method based on quantitative-PCR in which the threshold cycle correlates to gene copy number. Using this approach, we were able to diagnose 7 of 27 Japanese patients (25.9%) possessing mutations that were not detected by sequencing. Among these large deletions, similar results were obtained with 6 of 7 patients screened with a single nucleotide polymorphism array. We found an extensive intragenic deletion in RPS19, including exons 1-3. We also found 1 proband with an RPL5 deletion, 1 patient with an RPL35A deletion, 3 with RPS17 deletions, and 1 with an RPS19 deletion. In particular, the large deletions in the RPL5 and RPS17 alleles are novel. All patients with a large deletion had a growth retardation phenotype. Our data suggest that large deletions in RP genes comprise a sizable fraction of DBA patients in Japan. In addition, our novel approach may become a useful tool for screening gene copy numbers of known DBA genes.
Introduction
Diamond-Blackfan anemia (DBA; MIN# 105650) is a rare congenital anemia that belongs to the inherited BM failure syndromes, generally presenting in the first year of life. Patients typically present with a decreased number of erythroid progenitors in their BM.1 A main feature of the disease is red cell aplasia, but approximately half of patients show growth retardation and congenital malformations in the craniofacial, upper limb, cardiac, and urinary systems. Predisposition to cancer, in particular acute myeloid leukemia and osteogenic sarcoma, is also characteristic of the disease.2
Mutations in the RPS19 gene were first reported in 25% of DBA patients by Draptchinskaia et al in 1999.3 Since that initial finding, many genes that encode large (RPL) or small (RPS) ribosomal subunit proteins were found to be mutated in DBA patients, including RPL5 (approximately 21%), RPL11 (approximately 9.3%), RPL35A (3.5%), RPS7 (1%), RPS10 (6.4%), RPS17 (1%), RPS24 (2%), and RPS26 (2.6%).4-7 To date, approximately half of the DBA patients analyzed have had a mutation in one of these genes. Konno et al screened 49 Japanese patients and found that 30% (12 of 49) carried mutations.8 In addition, our data showed that 22 of 68 DBA patients (32.4%) harbored a mutation in ribosomal protein (RP) genes (T.T., K.T., R.W., and E.I., unpublished observation, April 16, 2011). These abnormalities of RP genes cause defects in ribosomal RNA processing, formation of either the large or small ribosome subunit, and decreased levels of polysome formation,4-6,9-12 which is thought to be one of the mechanisms for impairment of erythroid lineage differentiation.
Although sequence analyses of genes responsible for DBA are well established and have been used to identify new mutations, it is estimated that approximately half of the mutations remain to be determined. Because of the difficulty of investigating whole allele deletions, there have been few reports regarding allelic loss in DBA, and they have only been reported for RPS19 and RPL35A.3,6,13 However, a certain percentage of DBA patients are thought to have a large deletion in RP genes. Therefore, a detailed analysis of allelic loss mutations should be conducted to determine other RP genes that might be responsible for DBA.
In the present study, we investigated large deletions using our novel approach for gene copy number variation analysis based on quantitative-PCR and a single nucleotide polymorphism (SNP) array. We screened Japanese DBA patients and found 7 patients with a large deletion in an allele in RPL5, RPL35A, RPS17, or RPS19. Interestingly, all of these patients with a large deletion had a phenotype of growth retardation, including short stature and small-for-gestational age (SGA), which suggests that this is a characteristic of DBA patients with a large gene deletion in Japan.
Methods
Patient samples
Genomic DNA was extracted using the GenElute Blood Genomic DNA Kit (Sigma-Aldrich) according to the manufacturer's protocol. Clinical manifestation of patients from a Japanese DBA genomic library are listed elsewhere or are as reported by Konno et al.8 The study was approved by the institutional review board at the National Institute of Infectious Diseases and Hirosaki University.
DBA gene copy number assay by s-q-PCR
For s-q-PCR, primers were designed using Primer Express Version 3.0 software (Applied Biosystems). Primers are listed in Tables 1 and 2. Genomic DNA in water was denatured at 95°C for 5 minutes and immediately cooled on ice. The composition of the s-q-PCR mixture was as follows: 5 ng of denatured genomic DNA, 0.4mM forward and reverse primers, 1× SYBR Premix Ex Taq II (Takara), and 1× ROX reference dye II (Takara) in a total volume of 20 μL (all experiments were performed in duplicate). Thermal cycling was performed using the Applied Biosystems 7500 fast real-time PCR system. Briefly, the PCR mixture was denatured at 95°C for 30 seconds, followed by 35 cycles of 95°C for 5 seconds, 60°C for 34 seconds, and then dissociation curve measurement. Threshold cycle (Ct) scores were determined as the average of duplicate samples. The technical errors of Ct scores in the triplicate analysis were within 0.2 cycles (supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). The sensitivity and specificity of this method was evaluated with 15 healthy samples. Any false positive was not observed in all primer sets in all healthy samples (supplemental Figure 2). We performed direct sequencing of the s-q-PCR products. The results of the sequence analysis were searched for using BLAST to confirm uniqueness. Sequence data were obtained from GenBank (http://www.ncbi.nlm.nih.gov/gene/) and Ensemble Genome Browser (http://uswest.ensembl.org).
Genomic PCR
Genomic PCR was performed using KOD FX (Toyobo) according to the manufacturer's step-down PCR protocol. Briefly, the PCR mixture contained 20 ng of genomic DNA, 0.4mM forward and reverse primers, 1mM dNTP, 1× KOD FX buffer, and 0.5 U KOD FX in a total volume of 25 μL in duplicate. Primers are given in supplemental Figure 3 and Table 2. PCR mixtures were denatured at 94°C for 2 minutes, followed by 4 cycles of 98°C for 10 seconds, 74°C for 12 minutes, followed by 4 cycles of 98°C for 10 seconds, 72°C for 12 minutes followed by 4 cycles of 98°C for 10 seconds, 70°C for 12 minutes, followed by 23 cycles of 98°C for 10 seconds and 68°C for 12 minutes. PCR products were loaded on 0.8% agarose gels and detected by LAS-3000 (Fujifilm).
DNA sequencing analysis
The genomic PCR product was purified by the GenElute PCR clean-up kit (Sigma-Aldrich) according to the manufacturer's instructions. Direct sequencing was performed using the BigDye Version 3 sequencing kit. Sequences were read and analyzed using a 3120x genetic analyzer (Applied Biosystems).
SNP array-based copy number analysis
SNP array experiments were performed according to the standard protocol of GeneChip Human Mapping 250K Nsp arrays (Affymetrix). Microarray data were analyzed for determination of the allelic-specific copy number using the CNAG program, as described previously.14 All microarray data are available at the EGA database (www.ebi.ac.uk/ega) under accession number EGAS00000000105.
Results
Construction of a convenient method for RP gene copy number analysis based on s-q-PCR
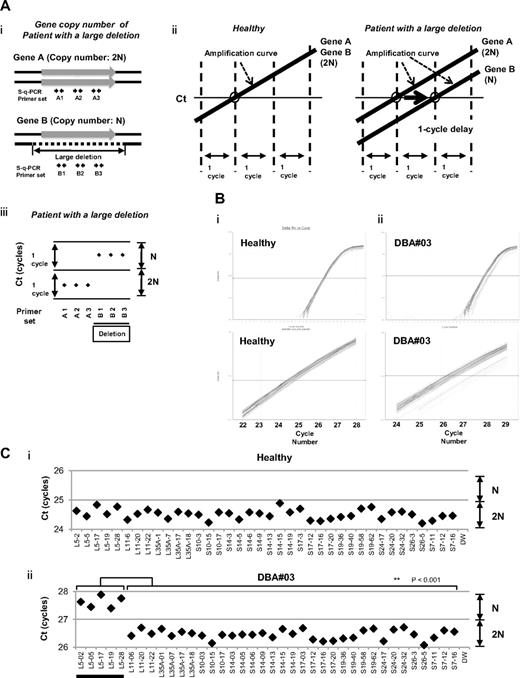
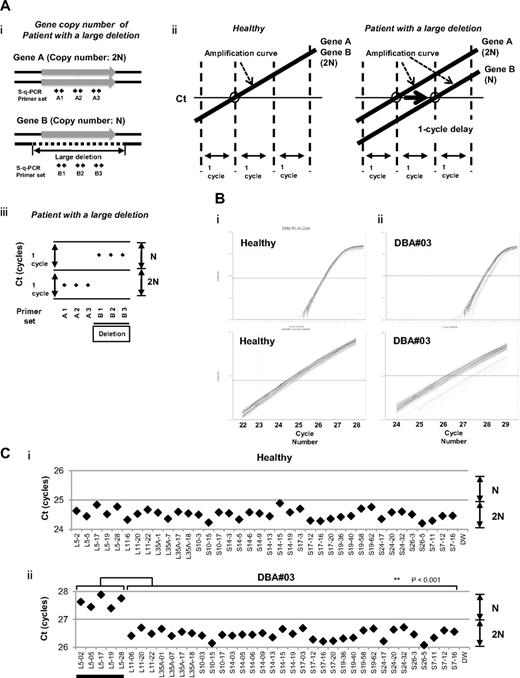
We focused on the heterozygous large deletions in DBA-responsible gene. The difference in copy number of genes between a mutated DBA allele and the intact allele was 2-fold (N and 2N; Figure 1Ai). If each PCR can synchronously amplify DNA fragments when the template genomic DNA used is of normal karyotype, it is possible to conveniently detect a gene deletion with a 1-cycle delay in s-q-PCR analysis (Figure 1Aii-iii).
s-q-PCR can determine a large gene deletion in DBA. (A) Concept of the DBA s-q-PCR assay. The difference in gene copy number between a healthy sample and that with a large deletion is 2-fold (i). When all genomic s-q-PCR for genes of interest synchronously amplify DNA fragments, a 2-fold difference in the gene copy number is detected by a 1-cycle difference of the Ct scores of the s-q-PCR amplification curves (ii). Also shown is a dot plot of the Ct scores (iii). (B) Results of the amplification curves of s-q-PCR performed with a healthy person (i) and a DBA patient (patient 3; ii). The top panel shows the results of PCR cycles; the bottom panel is an extended graph of the PCR cycles at logarithmic amplification. (C) Graph showing Ct scores of s-q-PCR. If all specific primer sets for DBA genes show a 1-cycle delay relative to each other, this indicates a large deletion in the gene. Gene primer sets with a large deletion are underlined in the graph. **P < .001.
s-q-PCR can determine a large gene deletion in DBA. (A) Concept of the DBA s-q-PCR assay. The difference in gene copy number between a healthy sample and that with a large deletion is 2-fold (i). When all genomic s-q-PCR for genes of interest synchronously amplify DNA fragments, a 2-fold difference in the gene copy number is detected by a 1-cycle difference of the Ct scores of the s-q-PCR amplification curves (ii). Also shown is a dot plot of the Ct scores (iii). (B) Results of the amplification curves of s-q-PCR performed with a healthy person (i) and a DBA patient (patient 3; ii). The top panel shows the results of PCR cycles; the bottom panel is an extended graph of the PCR cycles at logarithmic amplification. (C) Graph showing Ct scores of s-q-PCR. If all specific primer sets for DBA genes show a 1-cycle delay relative to each other, this indicates a large deletion in the gene. Gene primer sets with a large deletion are underlined in the graph. **P < .001.
To apply this strategy for allelic analysis of DBA, we prepared primers for 16 target genes, RPL5, RPL11, RPL35A, RPS10, RPS19, RPS26, RPS7, RPS17, RPS24, RPL9, RPL19, RPL26, RPL36, RPS14, RPS15, and RPS27A, under conditions in which the Ct of s-q-PCR would occur within 1 cycle of that of the other primer sets (Tables 1 and 2). At the same time, we defined the criteria of a large deletion in our assay as follows. If multiple primer sets for one gene showed a 1-cycle delay from the other gene-specific primer set at the Ct score, we assumed that this represented a large deletion. As shown in Figure 1Bii and 1Cii, the specific primer sets for RPL5 (L5-02, L5-05, L5-17, L5-19, and L5-28) detected a 1-cycle delay with respect to the mutated allele of patient 3. This assessment could be verified by simply confirming the difference of the cycles with the s-q-PCR amplification curves.
Study of large gene deletions in a Japanese DBA genomic DNA library
Sixty-eight Japanese DBA patients were registered and blood genomic DNA was collected at Hirosaki University. All samples were first screened for mutations in RPL5, L11, L35A, S10, S14, S17, S19, and S26 by sequencing. Among these patients, 32.4% (22 of 68) had specific DBA mutations (Table 3 and data not shown). We then screened for large gene deletions in 27 patients from the remaining 46 patients who did not possess mutations as determined by sequencing (Table 4).
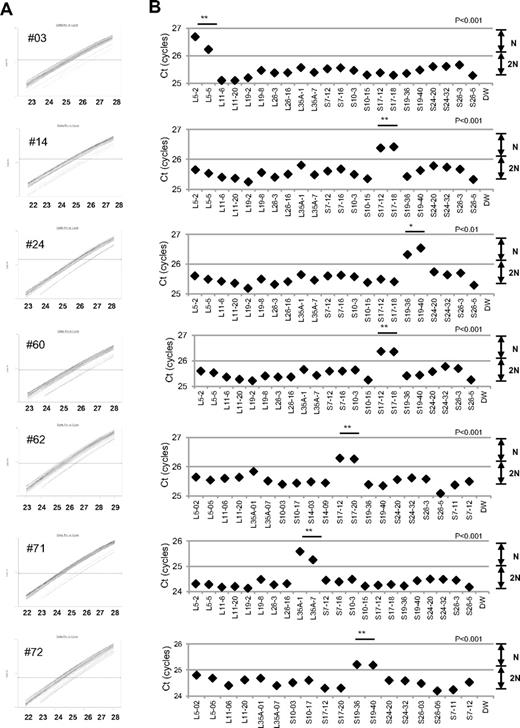
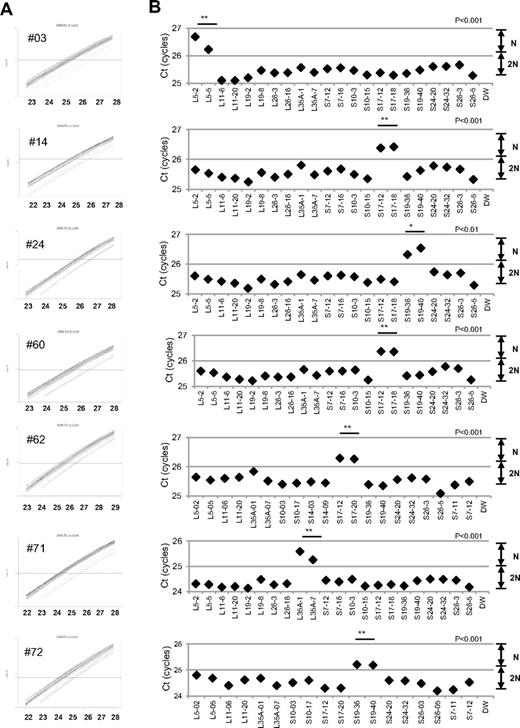
When we performed the s-q-PCR DBA gene copy number assay, 7 of 27 samples displayed a 1-cycle delay of Ct scores: 1 patient had RPL5 (patient 14), 1 had RPL35A (patient 71), 3 had RPS17 (patients 3, 60, 62), and 2 had RPS19 (patients 24 and 72; Figure 2 and Table 4). Among these patients, the large deletions in the RPL5 and RPS17 genes are the first reported cases of allelic deletions in DBA. From these results, we estimate that a sizable number of Japanese DBA patients have a large deletion.
Detection of 7 mutations with a large deletion in DBA patients. Genomic DNA of 27 Japanese DBA patients with unknown mutations were subjected to the DBA gene copy number assay. (A) Amplification curve of s-q-PCR of a mutation with a large deletion. The deleted gene can be easily distinguished. (B) Ct score (cycles) of representative s-q-PCR with DBA genomic s-q-PCR primers. Results of the 2 gene-specific primer pairs indicated in the graph are representative of at least 2 sets for each gene-specific primer (carried out in the same run). **P < .001; *P < .01
Detection of 7 mutations with a large deletion in DBA patients. Genomic DNA of 27 Japanese DBA patients with unknown mutations were subjected to the DBA gene copy number assay. (A) Amplification curve of s-q-PCR of a mutation with a large deletion. The deleted gene can be easily distinguished. (B) Ct score (cycles) of representative s-q-PCR with DBA genomic s-q-PCR primers. Results of the 2 gene-specific primer pairs indicated in the graph are representative of at least 2 sets for each gene-specific primer (carried out in the same run). **P < .001; *P < .01
Based on our findings, the rate of large deletions was approximately 25.9% (7 of 27) in a category of unspecified gene mutations. Such mutations have typically gone undetected by conventional sequence analysis. We could not find any additional gene deletions in the analyzed samples.
Confirmation of the gene copy number for DBA genes by genome-wide SNP array
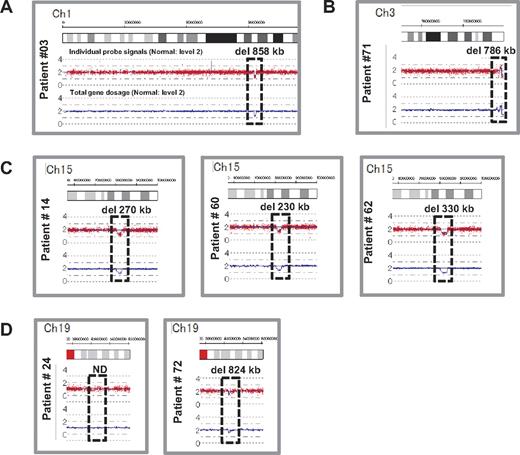
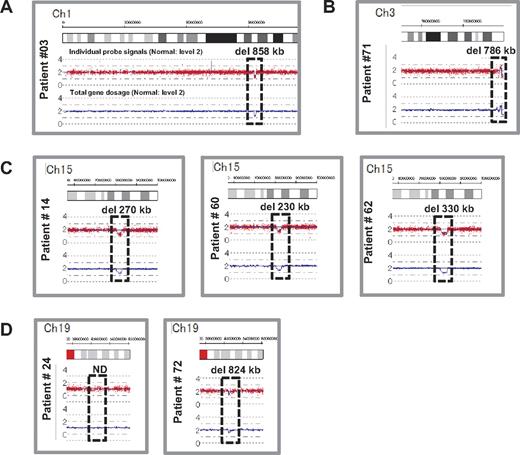
We performed genome-wide copy number analysis of the 27 DBA patients with a SNP array to confirm our s-q-PCR results. SNP array showed that patient 3 had a large deletion in chromosome 1 (ch1) spanning 858 kb (Figure 3A); patient 71 had a large deletion in ch3 spanning 786 kb (Figure 3B); patients 14, 60, and 62 had a large deletion in ch15 spanning 270 kb, 260 kb, and 330 kb, respectively (Figure 3C); and patient 72 had a large deletion in ch19 spanning 824 kb (Figure 3D). However, there were no deletions detected in ch19 in patient 24 (Figure 3D). Genes estimated to reside within a large deletion are listed in supplemental Table 1. Consistent with these s-q-PCR results, 6 of 7 large deletions were detected and confirmed as deleted regions, and these large deletions contained RPL5, RPL35A, RPS17, and RPS19 (Table 4 and supplemental Table 1). Other large deletions in RP genes were not detected by this analysis. From these results, we conclude that the synchronized multiple PCR amplification method has a detection sensitivity comparable to that of SNP arrays.
Results of SNP genomic microarray (SNP-chip) analysis. Genomic DNA of 27 Japanese DBA patients with unknown mutations was examined using a SNP array. Six patients had large deletions in their chromosome (ch), which included one DBA-responsible gene. Patient 3 has a large deletion in ch1 (A), patient 71 has a deletion in ch3 (B), patients 14, 60, and 62 have deletions in ch15 (C), and patient 72 has a deletion in ch19 (D).
Results of SNP genomic microarray (SNP-chip) analysis. Genomic DNA of 27 Japanese DBA patients with unknown mutations was examined using a SNP array. Six patients had large deletions in their chromosome (ch), which included one DBA-responsible gene. Patient 3 has a large deletion in ch1 (A), patient 71 has a deletion in ch3 (B), patients 14, 60, and 62 have deletions in ch15 (C), and patient 72 has a deletion in ch19 (D).
Detailed examination of a patient with intragenic deletion in the RPS19 allele (patient 24)
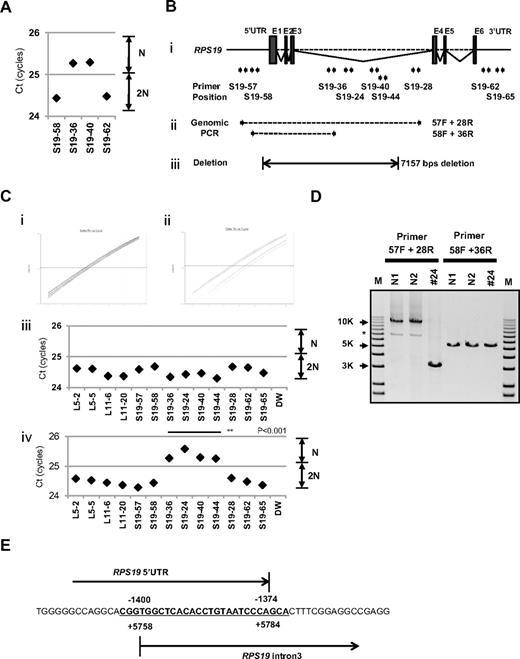
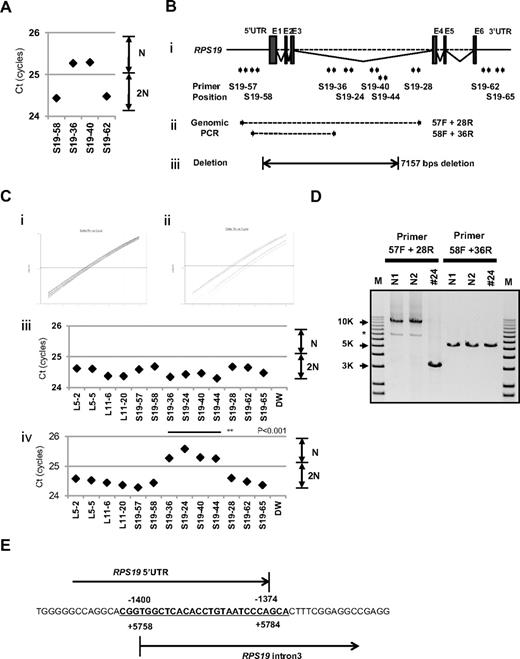
Interestingly, for patient 24, in whom we could not detect a large deletion by SNP array at s-q-PCR gene copy number analysis, 2 primer sets for RPS19 showed a 1-cycle delay (RPS19-36 and RPS19-40), but 2 other primer pairs (RPS19-58 and RPS19-62) did not show this delay (Figure 4A). We attempted to determine the deleted region in detail by testing more primer sets on RPS19. We tested a total of 9 primer sets for RPS19 (Figure 4B) and examined the gene copy numbers. Surprisingly, 4 primer sets (S19-24, S19-36, S19-40, and S19-44) for intron 3 of RPS19 indicated a 1-cycle delay, but the other primers for RPS19 located on the 5′untranslated region (5′UTR), intron 3, or 3′UTR did not show this delay (S19-57, S19-58, S19-28, S19-62, and S19-65; Figure 4B-C). These results suggest that the intragenic deletion occurred in the RPS19 allele. To confirm this deleted region precisely, we performed genomic PCR on RPS19, amplifying a region from the 5′UTR to intron 3 (Figure 4B). In patient 24, we observed an abnormally sized PCR product at a low molecular weight by agarose gel electrophoresis (Figure 4D). We did not detect a wild-type PCR product from the genomic PCR. This finding is probably because PCR tends to amplify smaller molecules more easily. However, we did detect a PCR fragment at the correct size using primers located in the supposedly deleted region. These bands were thought to be from the products of a wild-type allele. Sequencing of the mutant band revealed that intragenic recombination occurred at a homologous region of 27 nucleotides, from −1400 to −1374 in the 5′ region, to +5758 and +5784 in intron 3, which resulted in the loss of 7157 base pairs in the RPS19 gene (Figure 4E). The deleted region contains exons 1, 2, and 3, and therefore the correct RPS19 mRNA could not be transcribed.
Result of s-q-PCR gene copy number assay for patient 24. (A) Results of s-q-PCR gene copy number assay for RPS19 with 4 primer sets. (Bi) The RPS19 gene copy number was analyzed with 9 specific primer sets for RPS19 that span from the 5′UTR to the 3′UTR. (ii) Primer positions of genomic PCR for RPS19. (iii) Region determined to be an intragenic deletion in RPS19. (C) Results of gene copy number assay for RPS19 show a healthy person (i,iii) and a DBA patient (ii,iv), and Ct results are shown (iii-iv). Patient 24 showed a “1-cycle delay” with primers located in the intron 3 region, but other primer sets were normal. (D) Results of genomic PCR amplification visualized by agarose gel electrophoresis to determine the region of deletion. N1 and N2 are healthy samples. *Nonspecific band. (E) Results from the genomic sequence of the 3-kb DNA band from genomic PCR on patient 24 showing an intragenic recombination from −1400 to 5784 (7157 nt) in RPS19. **P < .001.
Result of s-q-PCR gene copy number assay for patient 24. (A) Results of s-q-PCR gene copy number assay for RPS19 with 4 primer sets. (Bi) The RPS19 gene copy number was analyzed with 9 specific primer sets for RPS19 that span from the 5′UTR to the 3′UTR. (ii) Primer positions of genomic PCR for RPS19. (iii) Region determined to be an intragenic deletion in RPS19. (C) Results of gene copy number assay for RPS19 show a healthy person (i,iii) and a DBA patient (ii,iv), and Ct results are shown (iii-iv). Patient 24 showed a “1-cycle delay” with primers located in the intron 3 region, but other primer sets were normal. (D) Results of genomic PCR amplification visualized by agarose gel electrophoresis to determine the region of deletion. N1 and N2 are healthy samples. *Nonspecific band. (E) Results from the genomic sequence of the 3-kb DNA band from genomic PCR on patient 24 showing an intragenic recombination from −1400 to 5784 (7157 nt) in RPS19. **P < .001.
Genotype-phenotype analysis and DBA mutations in Japan
Patients with a large deletion in DBA genes had common phenotypes (Table 4). Malformation with growth retardation (GR), including short stature or SGA, were observed in all 7 patients. In patients who had a mutation found by sequencing, half had GR (11 of 22; status data of DBA patients with mutations found by sequencing are not shown). GR may be a distinct phenotypic feature of large deletion mutations in Japanese DBA patients. Familial mutations were analyzed for parents for 5 DBA patients with a large deletion (patients 3, 24, 60, 62, and 72) by s-q-PCR. There are no large deletions in all 5 pairs of parents in DBA-responsible genes. Four of the 7 patients responded to steroid therapy. We have not observed significant phenotypic differences between patients with extensive deletions and other patients with regard to blood counts, responsiveness to treatment, or other malformations.
Discussion
Many studies have reported RP genes to be responsible for DBA. However, mutations have not been determined for approximately half of DBA patients analyzed. There are 2 possible reasons for this finding. One possibility is that patients have other genes responsible for DBA, and the other is that patients have a complicated set of mutations in RP genes that are difficult to detect. In the present study, we focused on the latter possibility because we have found fewer Japanese DBA patients with RP gene mutations (32.4%) compared with another cohort study of 117 DBA patients and 9 RP genes (approximately 52.9%).4 With our newly developed method, we identified 7 new mutations with a large deletion in RPL5, RPL35A, RPS17, and RPS19.
The frequency of a large deletion was approximately 25.9% (7 of 27) in our group of patients who were not found to have mutations by genomic sequencing. Therefore, total RP gene mutations were confirmed in 42.6% of these Japanese patients (Table 5). Interestingly, mutations in RPS17 have been observed at a high rate (5.9%) in Japan relative to that in other countries (1%).5,15,16 Although the percentage of DBA mutations differs among different ethnic groups,8,17-19 a certain portion of large deletions in DBA-responsible genes are likely to be determined in other countries by new strategies.
In the present study, we analyzed patient data to determine genotype-phenotype relations. To date, large deletions have been reported with RPS19 and RPL35A in DBA patients.3,6,13 RPS19 large deletions/translocations have been reported in 12 patients, and RPL35A large deletions have been reported in 2 patients.19 GR in patients with a large deletion has been observed previously with RPS19 translocations,3,19-21 but it was not found in 2 patients with RPL35A deletion.6 Interestingly, all of our patients with a large deletion had a phenotype of GR, including short stature and SGA, which suggests that this is a characteristic of DBA with a large gene deletion in Japan. Our study results suggest the possibility that GR is associated with extensive deletion in Japanese patients. Although further case studies will be needed to confirm this possibility, screening of DBA samples using our newly developed method will help to advance our understanding of the broader implications of the mutations and the correlation with the DBA genotype-phenotype.
Copy number variation analysis of DBA has been performed by linkage analysis, and the RPS19 gene was first identified as a DBA-susceptibility gene. Comparative genomic hybridization array technology has also been used to detect DBA mutations in RPL35A, and multiplex ligation-dependent probe amplification has been used for RPS19 gene deletion analysis.3,6,13,22 However, these analyzing systems have problems in mutation screening. Linkage analysis is not a convenient tool to screen for multiple genetic mutations, such as those in DBA, because it requires a high level of proficiency. Although comparative genomic hybridization technology is a powerful tool with which to analyze copy number comprehensively, this method requires highly specialized equipment and analyzing software, which limits accessibility for researchers. Whereas quantitative PCR–based methods for copy number variation analysis are commercially available (TaqMan), they require a standard curve for each primer set, which limits the number of genes that can be loaded on a PCR plate. To address this issue, a new method of analysis is needed. By stringent selection of PCR primers, the s-q-PCR method enables analysis of many DBA genes in 1 PCR plate and the ability to immediately distinguish a large deletion using the s-q-PCR amplification curve. In our study, 6 of 7 large deletions in the RP gene detected by s-q-PCR were confirmed by SNP arrays (Figure 3). Interestingly, we detected 1 large intragenic deletion in RPS19, which was not detected by the SNP array. This agreement between detection results suggests that the s-q-PCR copy number assay could be useful for detecting large RP gene deletions.
In the present study, 7 DBA patients carried a large deletion in the RP genes. This type of mutation could be underrepresented by sequencing analysis, although in the future, genome sequencing might provide a universal platform for mutation and deletion detection. We propose that gene copy number analysis for known DBA genes, in addition to direct sequencing, should be performed to search for a novel responsible gene for DBA. Although at present, it may be difficult to observe copy numbers on all 80 ribosomal protein genes in one s-q-PCR assay, our method allows execution of gene copy number assays for several target genes in 1 plate. Because our method is quick, easy, and low cost, it could become a conventional tool for detecting DBA mutations.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Momoka Tsuruhara, Kumiko Araki, and Keiko Furuhata for their expert assistance.
This work was partially supported by grants-in-aid for scientific research from the Ministry of Education, Culture, Sports, Science and Technology of Japan, and by Health and Labor Sciences research grants (Research on Intractable Diseases) from the Ministry of Health, Labor and Welfare of Japan.
Authorship
Contribution: M.K. designed and performed the research, analyzed the data, and wrote the manuscript; A.S-O. and S. Ogawa performed the SNP array analysis; T.M., M.T., and M.O designed the study; T.T, K. Terui, and R.W. analyzed the mutations and status data; H.K., S. Ohga, A.O., S.K., T.K., K.G., K.K., T.M., and N.M. analyzed the status data; A.M., H.M., K. Takizawa, T.M., and K.Y., performed the research and analyzed the data; E.I. and I.H. designed the study and analyzed the data; and all authors wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Isao Hamaguchi, MD, PhD, Department of Safety Research on Blood and Biological Products, National Institute of Infectious Diseases, 4-7-1, Gakuen, Musashimurayama, Tokyo 208-0011, Japan; e-mail: 130hama@nih.go.jp; or Etsuro Ito, MD, PhD, Department of Pediatrics, Hirosaki University Graduate School of Medicine, 5 Zaifucho, Hirosaki, Aomori 036-8562, Japan; e-mail: eturou@cc.hirosaki-u.ac.jp.