Abstract
Deregulated expression of CRLF2 (CRLF2-d) arises via its juxtaposition to the IGH@ enhancer or P2RY8 promoter. Among 865 BCP-ALL children treated on MRC ALL97, 52 (6%) had CRLF2-d, but it was more prevalent among Down syndrome patients (54%). P2RY8-CRLF2 (n = 43) was more frequent than IGH@-CRLF2 (n = 9). CRLF2-d was not associated with age, sex, or white cell count, but IGH@-CRLF2 patients were older than P2RY8-CRLF2 patients (median 8 vs 4 years, P = .0017). Patients with CRLF2-d were more likely to present with enlarged livers and spleens (38% vs 18%, P < .001). CRLF2-d was not seen in conjunction with established chromosomal translocations but 6 (12%) cases had high hyperdiploidy, and 5 (10%) had iAMP21. Univariate analysis suggested that CRLF2-d was associated with an inferior outcome: (event-free survival [EFS] hazard ratio 2.27 [95% confidence interval 1.48-3.47], P < .001; OS 3.69 [2.34-5.84], P < .001). However, multivariate analysis indicated that its effect was mediated by other risk factors such as cytogenetics and DS status (EFS 1.45 [0.88-2.39], P = .140; OS 1.90 [1.08-3.36], P = .027). Although the outcome of IGH@-CRLF2 patients appeared inferior compared with P2RY8-CRLF2 patients, the result was not significant (EFS 2.69 [1.15-6.31], P = .023; OS 2.86 [1.15-6.79], P = .021). Therefore, we concluded that patients with CRLF2-d should be classified into the intermediate cytogenetic risk group.
Introduction
Deregulated expression of the cytokine receptor gene, CRLF2, can arise via either a cryptic chromosomal translocation [t(X;14)(p22;q32)/t(Y;14)(p11;q32)] or a cryptic interstitial deletion within the pseudoautosomal region, that is, del(X)(p22.33p22.33)/del(Y)(p11.32p11.32).1 Overexpression of CRLF2 is driven by its juxtaposition to either the IGH@ enhancer (IGH@-CRLF2) or the P2RY8 promoter (P2RY8-CRLF2).1,2 Rare activating mutations have also been reported, for instance, CRLF2-F232C.3,4 Functional experiments have confirmed the leukemogenic potential of CRLF2.1,2,4,5 Overall, CRLF2 deregulation (CRLF2-d) occurs in 5%-7% of childhood acute lymphoblastic leukemia (ALL) but is more frequent among Down syndrome (DS) patients, where it occurs in 50% of cases.1,2,4 To assess the incidence and clinical and prognostic relevance of CRLF2-d in pediatric ALL, we screened 865 B-cell precursor ALL (BCP-ALL) patients treated in the trial Medical Research Council (MRC) ALL976-9 for the presence of this abnormality and evaluated its effect on outcome within the context of other risk factors.
Methods
Study population
Between April 1997 and June 2002, 1725 children with BCP-ALL ages 1-18 years were treated on the MRC ALL97 trial. Patients were selected for this study on the basis of the availability of material for testing. Fixed-cell suspension from the diagnostic bone marrow was available for 865 (50%) patients. There was no difference between the tested and nontested cohorts with respect to age, sex, and outcome, but tested patients had a slightly greater white cell count (WCC; supplemental Table 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Full details of the treatment protocol and cytogenetic analyses have been published.6-9 All participating centers obtained local ethical committee approval and written informed consent from patients or parents in accordance with the Declaration of Helsinki. Both phases, ALL97 and ALL99, included a steroid (prednisolone/dexamethasone) and purine (mercaptopurine/thioguanine) randomization in induction and continuing therapy. In ALL97, patients received a 3-drug induction followed by 2 or 3 intensification blocks, central nervous system–directed treatment and continuing therapy for a total of 2 years. High-risk patients were identified by the Oxford Hazard Score (on the basis of age, sex, and WCC) or by the presence of high-risk cytogenetics, that is, t(9;22)(q34;q11.2)/BCR-ABL1, near haploidy (< 30 chromosomes), low hypodiploidy (30-39 chromosomes), or MLL/11q23 translocations (< 2 years old), and were transferred to HR1.6,9 In ALL99, children were stratified on the basis of age and WCC to regimen A (< 10 years and < 50 × 109/L) or regimen B (all others). Patients received a 3/4 (regimen A/B) drug induction and were classified as a slow early responder if the day 15/8 (regimen A/B) marrow contained 25% or more blasts. Patients who failed to remit, were slow early responders, or who had high-risk cytogenetics were transferred to regimen C. After induction, patients received consolidation, 2 interim maintenance blocks, 2 delayed intensification blocks, and continuing therapy for up to 2 years (girls) or 3 years (boys). All patients treated on HR1 were eligible for sibling allogeneic transplantation, but only failure to achieve remission at day 29 and presence of t(9;22) were indications in ALL99.
Cytogenetics and fluorescence in situ hybridization
Cytogenetic and fluorescence in situ hybridization (FISH) analyses were performed on pretreatment samples by the use of standard methodologies and were reported with established nomenclature and definitions.9,10 The presence of IGH@-CRLF2 and P2RY8-CRLF2 was determined by interphase FISH by use of the following validated1 in-house and commercial probes: (1) break-apart CRLF2 probe (XX-87136C11/XX-82904A1 in spectrum red and RP4-674K6/RP13-309C18 in spectrum green); (2) LSI IGH Dual Color Break-Apart Rearrangement Probe (Vysis/Abbott Diagnostics); (3) break-apart P2RY8 probe (RP11-261P04/G248P81970A3/G248P82018G10/G248P86203A11 in spectrum red and G248P81114G2/G248P800157A9/G248P8154G2/G248P87990D1 in spectrum green). The normal signal pattern for these break-apart probes is 0 red, 0 green, and 2 fusion signals (0R0G2F). Indicative signal patterns for the presence of IGH@-CRLF2 were CRLF2-1R1G1F and IGH@-1R1G1F and for P2RY8-CRLF2 were CRLF2-1R0G1F and P2RY8-0R1G1F. The actual signal patterns observed varied according to the number of normal and derived X and Y chromosomes present in each case, as previously described.1 The presence of IKZF1 deletions was determined by the following in-house probe at 7p12 (G248P800745C8/WI2-3001F15 in spectrum green) hybridized together with the commercial centromeric probe for chromosome 7 (Vysis) in spectrum red. The normal signal pattern for this probe combination is 2R2G0F, whereas a 2R1G0F signal pattern indicated a deletion of IKZF1. All probes were grown, labeled, scored, and interpreted as previously described.1
Multiplex ligation-dependent probe amplification analysis
DNA extracted from pretreatment samples was analyzed by use of the SALSA Multiplex ligation-dependent probe amplification (MLPA) kit P335-A1 (MRC Holland) according to the manufacturer's instructions and as previously described.11 Data were analyzed by the use of GeneMarker V1.85 analysis software (SoftGenetics). This kit includes probes for IKZF1 (8 probes), CDKN2A/B (3 probes), PAX5 (6 probes), EBF1 (4 probes), ETV6 (6 probes), BTG1 (4 probes), and RB1 (5 probes). The relative copy number, obtained after normalization against controls, was used to determine genomic copy number of each one as follows: 0 (< 0.25, ie, bialleclic loss), 1 (< 0.75, ie, monoallelic loss), 2 (0.75-1.30, ie, normal copy number) and 3 or more (> 1.3, ie, gain).
JAK2 mutational analysis
Genomic DNA was extracted from fixed cell suspensions or bone marrow samples.12 Primers were used to amplify exon 14 of the JAK2 gene as described previously.12 Polymerase chain reaction products were purified directly by magnetic bead separation (Ampure; Agencourt) and mutations detected by direct sequencing with BigDye Terminator sequencing chemistry (Applied Biosystems) or denaturing high-performance liquid chromatography with Transgenomic patented separation DNASep cartridges (TransgenomicWave system). Normal DNA samples were used as negative controls.
Statistics and end points
Event-free survival (EFS) and overall survival (OS) were defined as the time from the start of treatment to relapse/death and death, respectively. Relapse-free survival (RFS) was calculated only for patients who achieved a complete remission and was defined as the time from the date of complete remission until relapse, with deaths in first remission being censored. Patients without an event of interest were censored at the date of last contact or date of second neoplasm, whichever was earlier. The median follow-up time for RFS, EFS, and OS was 7.75, 7.90, and 7.95 years, respectively. RFS, EFS, and OS estimates were calculated by use of the Kaplan-Meier method. Hazard ratios between subgroups were calculated with univariate Cox models. The proportional hazard assumption was assessed by visual inspection of the log-log plot and the Grambsch and Therneau proportional hazards test.13 Multivariate analysis was performed by use of a Cox regression model, which was adjusted for other risk factors: steroid randomization, phase of trial, age, a log-transformed WCC factor, sex, and cytogenetic risk group. Heterogeneity between subgroups was determined by investigating the appropriate interaction term in Cox proportional hazards modeling. The difference in −2*Log likelihood was used to assess whether there was any additional benefit to the addition of an interaction term in a forward selection modeling process. Other comparisons were performed using the χ2, Fisher exact test, or Wilcoxon rank-sum test, as appropriate. Because of the investigative nature of this analysis, we did not apply a stringent multiple comparisons adjustment. However, all tests were conducted at the 1% significance level. All analyses were performed by the use of Intercooled Stata 11.0 for Windows (Stata Corporation).
Results
Fifty-two patients with CRLF2-d were identified either with P2RY8-CRLF2 (n = 43) or IGH@-CRLF2 (n = 9). The overall incidence was 6% (52/865), but it was greater among DS patients at 54% (14/26; P < .001). Patients with CRLF2-d had few distinguishing demographic or clinical features and there was no evidence for a difference by DS status (Table 1). However, CRLF2-d patients were more likely to present with grossly enlarged livers and spleens. There was no association between CRLF2-d and age; however, IGH@-CRLF2 patients were significantly older than those with P2RY8-CRLF2 (median 8 vs 4 years, P = .0017).
CRLF2-d was not observed in patients with any of the major chromosomal translocations but it was seen in a small number of high hyperdiploid karyotypes, all were P2RY8-CRLF2 (Table 2). Aside from the observation that all 6 high hyperdiploid cases with CRLF2-d had gained an extra copy of a normal chromosome X, they did not have any distinguishing karyotypic features. The gain of chromosome X is a recurring theme in CRLF2-d cases,1 even outside the context of high hyperdiploidy. Overall, it was observed in 22/44 (50%) cases with successful cytogenetics. Among 20 patients with iAMP21, a total of 5 (25%) had CRLF2-d; all were P2RY8-CRLF2. Following our previous observations that CRLF2-d was associated with IKZF1 deletions and JAK2 mutations,1 we screened 43 CRLF2-d patients by MLPA for deletions of IKZF1, CDKN2A/B, PAX5, EBF1, ETV6, BTG1, and RB1. In addition, we screened 43 patients for IZKF1 deletions by FISH and 18 CRLF2-d patients for JAK2 mutations. Deletions of IKZF1, CDKN2A/B and PAX5 were common, occurring in approximately 40% of CRLF2-d patients with little difference between non-DS and DS (Table 2). IKZF1 deletions were seen in 6/9 (67%) IGH@-CRLF2 and 13/40 (33%) P2RY8-CRLF2 cases (P = .07).
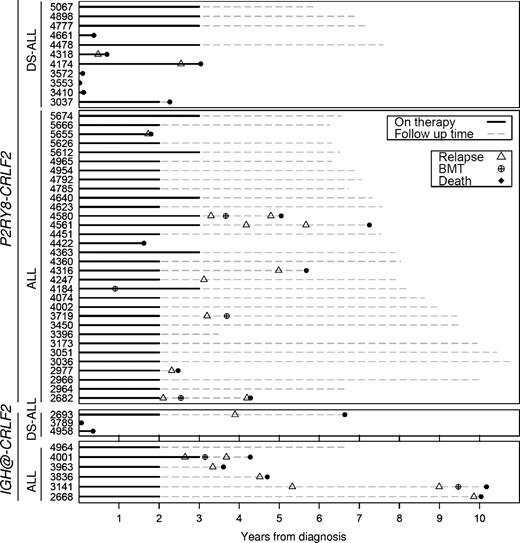
Table 3 describes the outcome of BCP-ALL patients and provides survival estimates stratified by CRLF2-d and DS status. Univariate analysis of the whole cohort revealed that CRLF2-d was associated with an inferior EFS (hazard ratio [HR] 2.27. 95% confidence interval [95% CI] 1.48-3.47, P < .001) and OS (3.69, 95% CI 2.34-5.84, P < .001) but not RFS (1.83. 95% CI 1.10-3.06, P = .021). CRLF2-d is strongly associated with DS-ALL (Table 1), and it is well known that DS patients respond differently to therapy.14 Therefore, we compared the effect of CRLF2-d among DS and non-DS patients. Although the prognostic effect of CRLF2-d on outcome appeared stronger among DS patients (Table 3) the test for heterogeneity was not statistically significant (data not shown). Overall patients treated on the ALL99 phase of the trial had a better outcome compared with those treated on the ALL97 phase.6 Thus, we compared the effect of CRLF2-d in the 2 phases but the test for heterogeneity was not significant for any of the 3 outcomes measures (data not shown). Hence, there was no evidence for a differential effect of CRLF2-d in the 2 phases of the trial. Multivariate analysis of the whole cohort adjusting for treatment, age, sex, WCC, cytogenetic risk group, and DS demonstrated that CRLF2-d was not an independent risk factor for EFS, RFS, or OS (Table 4). It is noteworthy that 14 of 16 (88%) CRLF2-d who relapsed have subsequently died, 5 after a second relapse (Figure 1).
Temporal distribution of major adverse events and transplants among children with acute lymphoblastic leukemia and CRLF2 deregulation.
Temporal distribution of major adverse events and transplants among children with acute lymphoblastic leukemia and CRLF2 deregulation.
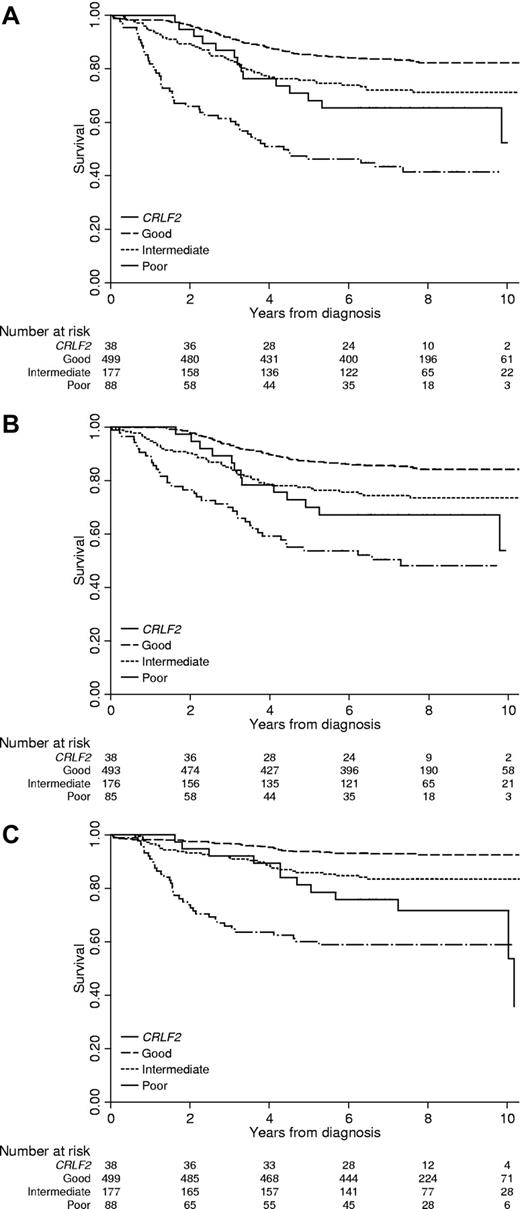
As a result of the strong association between DS-ALL and CRLF2-d, 27% of our CRLF2 cohort had DS; even though DS-ALL accounts for only 2% BCP-ALL overall (supplemental Table 1). Therefore, we excluded DS patients when plotting the outcome of CRLF2-d patients against the 3 cytogenetic risk groups (Figure 2). The majority of non-DS CRLF2-d patients (25/38, 66%) would have been classified in the intermediate cytogenetic risk group (Table 2). Only 1 of 6 high hyperdiploid patients with CRLF2-d relapsed whereas the remaining 5 remain in first remission after more than 6 years. Seven patients would have been classified in the poor-risk cytogenetic group: iAMP21 (n = 5), near haploidy (n = 1), and abnormal 17p (n = 1). Three of the iAMP21 patients and the patient with abnormal 17p have relapsed, whereas the remaining cases remain in first remission after more than 6 years. It should be noted that patients with iAMP21 or abnormal 17p were not included in the risk stratification algorithm for this trial, whereas those with near haploidy were transferred to the high-risk arm.
EFS, RFS, and OS of children with non-DS ALL and CRLF2 deregulation in comparison with other children, stratified by cytogenetic risk group. (A) EFS. (B) RFS. (C) OS.
EFS, RFS, and OS of children with non-DS ALL and CRLF2 deregulation in comparison with other children, stratified by cytogenetic risk group. (A) EFS. (B) RFS. (C) OS.
Among CRLF2-d patients, there was no evidence for a prognostic effect associated with deletions of IKZF1, PAX5 or CDKN2A/B, or JAK2 mutations (Table 5). It should be noted that most patients with JAK2 mutations (6/10) have DS-ALL who are more likely to die in remission (Table 3).
Although the results were not statistically significant, there was some evidence to suggest that IGH@-CRLF2 patients have an inferior outcome compared with P2RY8-CLRF2 patients (Figure 1): EFS, 33% (95% CI 8%-62%) versus 63% (46%-75%), HR 2.69 (95% CI 1.15-6.31), P = .023; RFS, 43% (10%-73%) versus 73% (56%-85%), HR 3.32 (1.24-9.5), P = .018; OS, 44% (14%-72%) versus 74% (59%-85%), HR 2.86 (1.17-6.95), P = .021. Among 9 IGH@-CRLF2 patients, 6 relapsed, all of whom subsequently died; 2 died after early infections within 5 months of diagnosis; and 1 remains in first remission after more than 6 years (Figure 1).
Discussion
We present the incidence and clinical relevance of CRLF2-d in the largest cohort reported to date. Overall we observed CRLF2-d in 5% of non-DS ALL, an incidence that agrees well with previous estimates.1,2,15 In this unselected and representative cohort, P2RY8-CRLF2 was approximately 5 times more prevalent than IGH@-CRLF2. The Berlin-Frankfurt-Muenster (BFM)–2000 study15 also showed a predominance of P2RY8-CRLF2, but the Children's Oncology Group (COG) P9906 study16 found IGH@-CRLF2 to be twice as prevalent as P2RY8-CRLF2. Our observation that IGH-CRLF2 patients are older than P2RY8-CRLF2 patients is the likely explanation for the interstudy variability. The BFM study showed a similar age profile to this one, whereas the COG study comprised a significantly older population of “high-risk” children, with a median age of 12.8 years. Furthermore, the COG study comprised 25% Hispanic patients and reported that these patients were 5 times more likely to have CRLF2-d compared with non-Hispanic patients. They did not report whether this association was stronger for IGH@-CRLF2 or P2RY8-CRLF2.
With the exception of iAMP21 and rare high hyperdiploid cases, CRLF2-d did not occur with other primary chromosomal abnormalities. Although the BFM study reported 2 BCR-ABL1 positive patients with “high” levels of CRLF2 expression, neither had IGH-CRLF2 or P2RY8-CRLF2.15 It should be noted that the BFM cohort is not analogous to ours or the COG study.2,16 The BFM study defined a “high” CRLF2 expression group by using an arbitrary cutoff value and thus their group comprises not only patients with IGH@-CRLF2 or P2RY8-CRLF2 but also those with no genomic alteration affecting CRLF2. Interestingly, several their “high” CRLF2 expression cases had more than 2 copies of CRLF2, which would be consistent with an extra copy of a sex chromosome. The COG study16 reported that more than 80% of CRLF2-d patients also had a deletion of IKZF1. In contrast, we found a much lower incidence of IKZF1 deletions (< 40%) among our patients. Our data suggest that IKZF1 deletions may be more prevalent among IGH@-CLRF2 than P2RY8-CRLF2 patients. If so, the high incidence of IGH@-CRLF2 in the COG study would explain this difference. The COG study also recorded a greater incidence of JAK2 mutations among CRLF2-d patients (75%) compared with this study (50%). This finding is somewhat surprising because our study had a greater proportion of DS patients (27% vs 7%) and JAK2 mutations are known to be strongly associated with DS-ALL.17
Although univariate analysis indicated that CRLF2-d patients had an inferior outcome, this premise was not supported by multivariate or stratified analyses. Thus, we concluded that CRLF2-d, per se, is not an independent risk factor in pediatric ALL. In the absence of another good or poor risk chromosomal abnormality, we suggest that these patients be included in the intermediate cytogenetic risk group.9 These findings are in stark contrast to recent reports by the COG16 and BFM15 groups, both of which found that CRLF2-d patients had a significantly inferior outcome. The COG study focused on a specific “high-risk” cohort of patients that was significantly older than patients in this study. In addition, it comprised a much greater proportion of Hispanic patients (25% vs < 1%). Hence, differences between these 2 studies are understandable, especially because in the COG study CRLF2-d was more prevalent in Hispanic patients, and there was a strong correlation between outcome and ethnicity. The COG study also found that IKZF1 deletions were a major prognostic factor, but there was no evidence for an effect in this study. An i-BFM group study reported that DS-ALL patients expressing CRLF2 tended to have a lower EFS; however, the difference was not statistically significant and patients were treated on several different protocols.4 Although our data also suggested an inferior outcome for CRLF2-d DS patients, the test for heterogeneity was not significant. Given the myriad of treatment complications experienced by DS-ALL patients, it will be difficult to tease out strong prognostic factors in this subgroup without large number of patients.
The BFM study is similar to this one, that is, a representative trial-based cohort of pediatric ALL. Although the initial BFM cohort was defined by expression rather than genomic alteration and included proportionally fewer DS patients, the directly comparable survival rates are very different. For example, the 6-year EFS for P2RY8-CRLF2 patients in BFM-2000 was 28% ± 15% (n = 21) compared with a 5-year EFS of 63% (46%-75%; n = 43) for P2RY8-CRLF2 patients in MRC-ALL97. Whereas our data suggest that IGH@-CRLF2 patients may have an inferior outcome, all IGH@-CRLF2 (n = 4) patients in BFM-2000 remain in first complete remission. The BFM study did not stratify by DS status but did state that their removal did not change the results. Although we cannot exclude a differential effect of the treatment protocol, this would seem unlikely as the drugs used are the same and many of the protocol elements are similar. In fact, the ALL99 phase of the trial adopted the risk-stratification strategy and intensification courses from the then U.S. Childhood Cancer Study Group and included more intensive consolidation blocks taken from the BFM protocols.7 In addition, there was no evidence for a differential effect of CRLF2-d between patients treated on the ALL97 and ALL99 phases of the trial. A more likely explanation is a combination of factors, including small numbers of cases, the strong association between CRLF2-d and DS, and the effect of other cytogenetic abnormalities. Although our cohort of CRLF2-d patients is the largest to date, none of the 3 studies are larger enough to address the issue of treatment heterogeneity. Further prospective studies and, potentially, meta-analyses will be required to further explore these findings. One of the major advantages of this study is that we were able to examine DS and non-DS patients separately and assess the prognostic effect of CRLF2-d in the context of cytogenetic risk groups. It will be important for future studies to incorporate these risk factors as well as minimal residual disease into the analysis.
One theory that may explain the divergent results from these 3 studies is that the poor outcome is associated with IGH@-CRLF2 rather than CRLF2-d per se. Although our results hint at a differential effect between IGH-CLRF2 and P2RY8-CRLF2 patients, the number of patients included in these analyses was small and larger independent studies are required. However, the idea that IGH@-CRLF2 patients have a poorer outcome compared with P2RY8-CRLF2 is consistent with the differences between this and the COG studies, especially the different frequencies of IGH@-CRLF2 and P2RY8-CLRF2. Moreover, there is a possible biologic explanation for this distinction, that CRLF2 expression is much greater in IGH@-CRLF2 patients compared with P2RY8-CRLF2 patients. The enhanced expression likely arises because the IGH@ enhancer is a more potent promoter of transcription than the P2RY8 promoter.1,16
In conclusion, we have presented a detailed analysis describing the incidence and clinical relevance of CRLF2-d in pediatric ALL. In agreement with previous studies,1,2,4,15,16 we have noted (1) an overall incidence of 6% that increases to more than 50% among DS patients; and (2) that CRLF2-d does not tend to coexist with established chromosomal abnormalities. We found that P2RY8-CRLF2 was substantially more prevalent than IGH@-CRLF2 in pediatric ALL, which confirms the results from BFM-2000 but not the COG study. However, in contrast to the findings of both BFM-2000 and COG, we found no evidence to suggest a clear and independent association between CRLF2-d and outcome. More extensive studies are urgently required to explore further the prognostic value of CRLF2-d in ALL and to investigate the potential differential prognostic effect of the 2 rearrangements giving rise to CRLF2 deregulation.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Leukemia & Lymphoma Research (formerly Leukemia Research) for financial support; member laboratories of the United Kingdom Cancer Cytogenetic Group (UKCCG) for providing cytogenetic data and material; Dr Elie Nacheva (University College London) for the IKZF1 FISH probe; Dr Julie Irving and Lynne Minto (Northern Institute for Cancer Research, Newcastle University) for help with the mutational analysis; and past and present members of the Leukemia Research Cytogenetics Group (LRCG) for their contribution to establishing this dataset. We thank all the members of the Childhood Leukemia Working Party during this period, especially the chairs: Professor Tim O. B. Eden (1991-2001) and Dr Brenda E. S. Gibson (2001-2010). Finally, we thank all the clinicians who entered patients into the trial and the children and families who agreed to take part.
The United Kingdom Medical Research Council funded the data management and statistical input into the trial and supported the Childhood Leukemia Working Party, which oversaw recruitment to the trial.
Authorship
Contribution: L.J.R., C.J.H., and A.V.M. performed study design and conception; C.S., L.J.R., H.M., D.M., and L.J. performed FISH and MLPA testing and analysis of cytogenetic data; L.J.R., H.M., and D.M. performed mutational analysis; H.M.E. and A.V.M. performed data analysis and interpretation; H.M.E. and A.V.M. wrote the manuscript; S.M.R., S.E.K., A.J.V., and C.D.M. performed trial coordination and provision of clinical and outcome data; and all authors performed critical appraisal and final approval of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Dr Anthony V. Moorman, Leukaemia Research Cytogenetics Group, Northern Institute for Cancer Research, Level 5, Sir James Spence Institute, Royal Victoria Infirmary, Newcastle-upon-Tyne, NE1 4LP, United Kingdom; e-mail: anthony.moorman@ncl.ac.uk.
References
Author notes
H.M.E., C.S., and L.J.R. contributed equally to this work.