Abstract
The genetics of classical Hodgkin lymphoma (cHL) is poorly understood. The finding of a JAK2-involving t(4;9)(q21;p24) in 1 case of cHL prompted us to characterize this translocation on a molecular level and to determine the prevalence of JAK2 rearrangements in cHL. We showed that the t(4;9)(q21;p24) leads to a novel SEC31A-JAK2 fusion. Screening of 131 cHL cases identified 1 additional case with SEC31A-JAK2 and 2 additional cases with rearrangements involving JAK2. We demonstrated that SEC31A-JAK2 is oncogenic in vitro and acts as a constitutively activated tyrosine kinase that is sensitive to JAK inhibitors. In vivo, SEC31A-JAK2 was found to induce a T-lymphoblastic lymphoma or myeloid phenotype in a murine bone marrow transplantation model. Altogether, we identified SEC31A-JAK2 as a chromosomal aberration characteristic for cHL and provide evidence that JAK2 rearrangements occur in a minority of cHL cases. Given the proven oncogenic potential of this novel fusion, our studies provide new insights into the pathogenesis of cHL and indicate that in at least some cases, constitutive activation of the JAK/STAT pathway is caused by JAK2 rearrangements. The finding that SEC31A-JAK2 responds to JAK inhibitors indicates that patients with cHL and JAK2 rearrangements may benefit from targeted therapies.
Introduction
The nonreceptor protein tyrosine kinase (PTK) JAK2 is an important mediator of cytokine signaling. Activation of JAK2 by binding of a ligand to its receptor predominantly activates STATs, which regulate the expression of target genes related to cell survival, proliferation and differentiation, including SOCS, BCL-XL, CDKN1A (p21), CCND1, and MYC.1
The JAK2 gene is often implicated in gain-of-function mutations and chromosomal translocations in hematologic malignancies, the latter leading to the formation of chimeric oncoproteins. To date, 6 variant JAK2 fusions affecting TEL(ETV6),2,3 PCM1,4 BCR,5 SSBP2,6 , STRN3,7 and PAX58 have been described in myeloproliferative neoplasms (MPNs) and acute leukemias. All these fusions, except for PAX5-JAK2, lead to constitutive phosphorylation of the JAK2 tyrosine kinase by partner-mediated oligomerization and constitutive activation of downstream pathways. The PAX5-JAK2 fusion, occurring in B-cell acute lymphoblastic leukemia (B-ALL), possibly acts as a constitutive repressor of transcriptional activity of PAX5.9,10 Because aberrant activation of PTKs is important for cancer cell survival, these kinases form primary targets for rational cancer therapies based on the use of small molecule inhibitors. Indeed, the discovery of JAK2 mutations in MPNs prompted efforts toward the development of JAK2 inhibitors, resulting in numerous JAK2 antagonists currently being tested in phase 1-3 clinical trials.11
Classical Hodgkin lymphoma (cHL) is one of the most common types of malignant lymphoma in the Western world that mainly affects young adults. It is characterized by the presence of rare neoplastic Reed-Sternberg (RS) cells that typically comprise less than 1% of the tumor mass, which is predominantly composed of reactive cells. In most cases, RS cells originate from preapoptotic germinal center (GC) or post-GC B cells12 that have lost the B-cell phenotype.13 Because of the rarity, high variability, and low proliferation index of RS cells, the genetics and molecular biology of this lymphoma are poorly understood. So far, cHL-associated genetic aberrations have not been identified. Molecular studies, mainly performed on HL-derived cell lines, showed that multiple signaling pathways and transcription factor networks—for example, the NFκB, PI3K/AKT, MAPK/ERK, AP-1, NOTCH1, and JAK/STAT pathways—are deregulated in RS cells, providing these cells with additional proliferation signals and protection against apoptosis.14 The finding of genetic aberrations affecting the NFκB-related genes REL, BCL3, NFKBIA, NFKBIE, and TNFAIP314 , and mutations of the SOCS1 gene, a negative regulator of JAK/STAT signaling, in cHL15 indicate that aberrant activation of these pathways may be caused by genetic defects of the involved players. Whether gain and amplification of the 9p24/JAK2 region, detected in more than 30% of cHL cases, activate the JAK/STAT pathway, remains unclear.16-18 Of note, the common JAK2V617F mutation seems to be absent in cHL.19 We report here the results of the molecular characterization of a novel JAK2-involving t(4;9)(q21;p24) identified in 1 case of cHL. We further attempted to determine the prevalence of JAK2 rearrangements in cHL.
Methods
Case selection
One hundred one cases of cHL, admitted between 2003 and 2010 and subjected to cytogenetic analysis, were selected from the archive of the Department of Pathology and the Center for Human Genetics (Katholieke Universiteit Leuven, Leuven, Belgium). This series was extended by 23 cHL cases with a high number of RS cells from the archives of the same department collected before 2003 and with 8 cases from the Department of Pathology (AZ St Jan, Bruges, Belgium). In all cases, cytogenetic pellets, fresh frozen tumor tissue and/or formalin- or B5-fixed paraffin-embedded blocks were available. Cultured fixed cells were used for FISH, frozen material for RT-PCR, and paraffin-embedded material for immunohistochemistry. Ready-to-use antisera against ALK1, CD3, CD4, CD8, CD15, CD30, PAX5 (B cell–specific activator protein 5), and EBV-LMP were purchased from DAKO. The goat anti–TIA1 antiserum (sc-1751) was purchased from Santa Cruz Biotechnology. Cytogenetic pellets and frozen samples have been stored at −20°C and −80°C, respectively. All cases were histologically reviewed and diagnosed as cHL according to the recent criteria of the 2008 WHO classification of lymphoid neoplasms.20 Eighty-seven (65.9%) cases were diagnosed as nodular sclerosis cHL, 16 (12.1%) as mixed cellularity cHL, 7 (5.3%) as lymphocyte-rich cHL, 4 (3%) as lymphocyte-depleted cHL, and 18 (13.6%) as cHL not otherwise specified. In all cases, CD15 and CD30 was expressed by RS cells. In the majority of cases, the RS cells also expressed PAX5. Of interest, in 14 cases (10.6%), not distinguishable from the others on H&E-stained sections, RS cells lacked PAX5 expression. The latter cases were further investigated for the presence of several T-cell markers. In 7 of 14 cases (including case 1 and case 2), TIA1 expression, associated with or without the expression of perforin and granzyme B, was found in these RS cells, while other T-cell markers (for example, CD2, CD3, CD4 and CD8) were absent. Evidence for clonal TCR rearrangement was not found. Although a diagnosis of unusual peripheral T-cell lymphoma is not completely excluded in these cases, most histologic characteristics were in favor of HL with RS cells probably of T-cell type.
This study was approved by the institutional ethics commission of the Katholieke Universiteit Leuven. Informed consent was provided according to the Declaration of Helsinki principles.
Cytogenetics
Conventional G-banding chromosomal analysis of lymph node biopsy samples followed routine protocols. The karyotype was described according to the International System for Human Cytogenetic Nomenclature (ISCN 2005).21 Results of cytogenetic analysis performed in 3 of 4 reported cases are shown in Table 1.
FISH followed standard protocols. Experiments were performed on fixed cells from remaining cytogenetic harvests. Multicolor FISH (M-FISH) was performed according to the manufacturer's protocols (MetaSystems). FISH analysis was mainly performed on interphase cell level because the number of metaphases was limited. Only huge atypical nuclei (3-10 per experiment), likely representing RS cells, were evaluated. A list of the applied probes can be found in supplemental Table 1 (available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Bacterial artificial chromosome (BAC) and fosmid clones were selected from the Ensembl22 and UCSC23 Genome Browsers, respectively. These probes were directly labeled with SpectrumOrange- and SpectrumGreen-dUTP (Abbott Molecular) by random prime labeling. FISH images were acquired with a 63×/1.40 oil-immersion objective in an Axioplan 2 fluorescence microscope equipped with an Axiophot 2 camera (Carl Zeiss Microscopy) and a MetaSystems Isis imaging system (Meta Systems).
Molecular analysis
RT-PCR.
The presence of the SEC31A-JAK2 fusion transcript was confirmed by nested RT-PCR followed by sequencing as previously described.24 The first round of PCR was carried out with the primers SEC31A-F1 and JAK2-R1. In the second round of PCR, SEC31A-F2 and JAK2-R2 were used (supplemental Table 2). In addition, this strategy was used to screen 14 NScHL cases rich in RS cells for the presence of the SEC31A-JAK2 fusion.
Functional analysis
Constructs.
The generation of the SEC31A fragment of the construct has previously been described.24 The open reading frame of exon 17 to 25 of JAK2 was generated by PCR amplification with the Phusion High Fidelity PCR kit (Finnzymes) from a previously described plasmid containing the full-length JAK2 gene.25
Several deletion constructs were generated that fuse fragments of SEC31A present in the fusion to the JAK2 portion of the fusion. All SEC31A fragments were obtained by PCR from tissue cDNA. The schematic representation of their protein structure is given (see Figure 2B); primer sequences can be found in the supplemental Table 2. The generated SEC31A and JAK2 fragments were ligated into the retroviral pMSCV-puro vector (Clontech). The constructs were verified by sequencing.
Cell culture and retroviral transduction.
HEK293T and Ba/F3 cells were cultured, transfected and transduced as previously described.26 Transduced Ba/F3 cells were selected with puromycin (2.5 μg/mL). Ba/F3 cells expressing the SEC31A-ALK fusion or the JAK2V617F mutation have already been described.24,25
For growth curves, Ba/F3 cells were deprived of IL3 and seeded at 1 × 105 cells/mL of medium. Viable cells were counted on 5 consecutive days with a Vi-CELL XR cell viability analyzer (Beckman Coulter). For dose-response curves, 2 × 105 cells were seeded in 1 mL of medium and incubated in the presence of the JAK inhibitors CP690550 (Pfizer) and JAK inhibitor I (Calbiochem/Merck4Biosciences) for 24 (SEC31A-JAK2 and SEC31A-ALK) or 48 (JAK2V617F) hours. Viable cell numbers were determined by the Celltiter AQueousOne Solution (Promega). For Western blotting, 4 × 106 cells were incubated with CP690550 and JAK inhibitor I for 90 minutes and lysed in cold lysis buffer containing 1% Triton X-100 and protease and phosphatase inhibitors.
Western blotting.
Standard Western blotting procedures were used to analyze total cell lysates with the antibodies anti–phospho-JAK2 (Tyr1007/1008), anti-JAK2 (D2E12), and anti–phospho-STAT5 (Tyr694; C11C5; Cell Signaling Technology), and with previously described antibodies for SEC31A, (phospho)–MAPK (ERK1/2), (phospho)–AKT, (phospho)–STAT3, and STAT5.24
Murine bone marrow transplantations
Murine bone marrow transplantation assays were carried out as previously described using Balb/c mice.27 For secondary transplantation experiments, 1 × 106 viable spleen cells isolated from a diseased SEC31A-JAK2– transplanted mouse were injected into the tail vein of sublethally irradiated (4 Gy) female syngeneic recipient mice.
Analysis of diseased animals
A blood sample was taken by submandibular cheek pouch punction from animals showing signs of disease (decreased activity, ruffled fur, swollen appearance, and/or shortness of breath). White blood cell counts and differentials were measured with a hematology analyzer (Scil VET Abs; Scil Animal Care Company BV). Animals with abnormal blood cell counts were killed and subjected to macroscopic analysis of the organs. One femur and a part of the spleen, thymus, lymph nodes, and of gross anomalies in other organs (for example, liver) were embedded in paraffin and histopathologically analyzed. Single-cell suspensions were prepared from the same material; bone marrow cells were isolated from femur and tibia. The cells were subsequently subjected to erythrocyte lysis and used for flow cytometric analysis, and for RNA, DNA, and protein extraction.
Flow cytometry
Isolated murine bone marrow, spleen, thymus, and lymph node cells were stained with 7-amino-actinomycin D (7-AAD), mouse T-lymphocyte subset antibody cocktail (PE-Cy7 CD3e, PE CD4, and APC or FITC CD8a), PE-Cy7 Mac1, PE Gr1, and APC B220 (BD Biosciences). Cells were detected on a FACSCanto flow cytometer and analyzed with the FACSDiva software (BD Biosciences). Dead cells identified by 7-AAD staining were discarded during data analysis.
Results
SEC31A-JAK2 is a novel fusion associated with the t(4;9)(q21;p24)
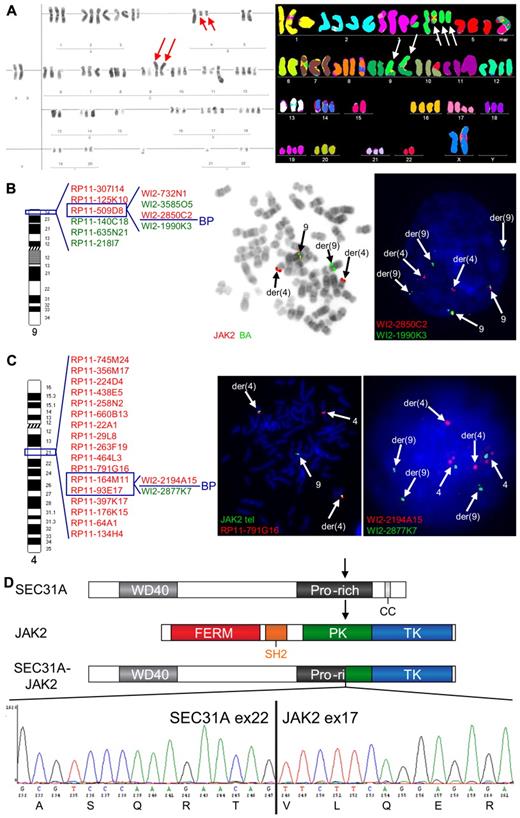
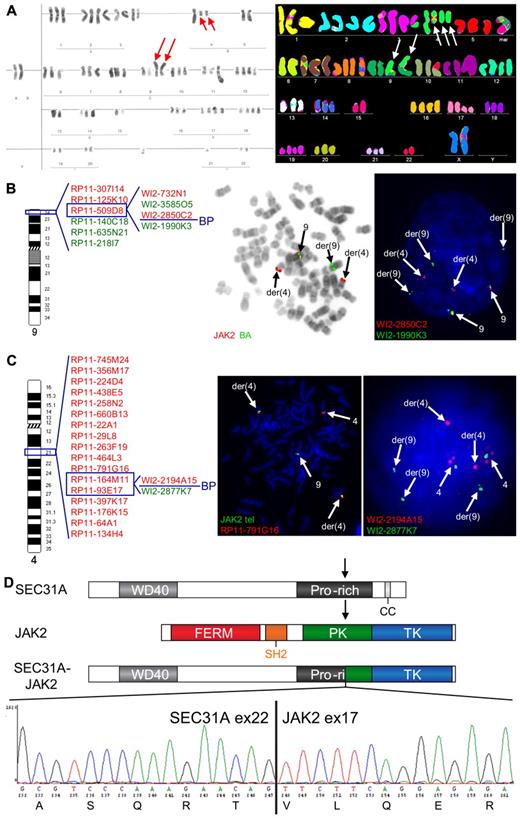
Cytogenetic analysis of 1 cHL case (case 1) revealed a complex karyotype, including a t(4;9)(q21;p24) (Table 1; Figure 1A). Given the involvement of 9p24, the region harboring JAK2, this translocation was further extensively investigated by FISH, which demonstrated a rearrangement of JAK2 and mapped the 9p24 breakpoint between exon 9 and exon 18 of the gene (Figure 1B). The partner 4q21 breakpoint was investigated with a BAC/fosmid walking interphase FISH strategy that eventually narrowed down the breakpoint to the region of SEC31A (Figure 1C). Rearrangement of this gene was further confirmed by a break-apart (BA) FISH assay for SEC31A (Figure 1C). Further RT-PCR analysis followed by sequencing identified an in-frame fusion of exon 22 of SEC31A to exon 17 of JAK2 (Figure 1D).
Cytogenetic and molecular analysis of the t(4;9)(q21;p24) found in case 1. (A) Conventional (left) and M-FISH karyotype (right). Note 2 copies of the t(4;9)(q21;p24) in both karyotypes (red and white arrows) and 1 additional copy of the der(4) in the M-FISH karyotype (white arrows). (B) An ideogram of chromosome 9 with probes used for the FISH characterization of the t(4;9) (left) and examples of FISH experiments (right). BAC probes included in the dual color JAK2 break-apart (JAK2 BA) assay are shown in the middle figure, fosmid probes covering JAK2 are shown in the right figure. Split of JAK2 BA signals on der(4) and der(9) (metaphase cell) illustrates the t(4;9)-associated rearrangement of JAK2. Further analysis with fosmid probes covering JAK2 mapped the breakpoint in the region flanked by WI2-2850C2 and WI2-1990K3 (split of red and green fosmid signal in an interphase cell). (C) An ideogram of chromosome 4 with probes used for the FISH characterization of the 4q21 breakpoint in the t(4;9) (left) and examples of FISH experiments (right). The 4q21 breakpoint was mapped with a BAC/fosmid walking interphase FISH strategy. The telomeric (5′) JAK2 probes (labeled with SpectrumGreen-dUTP) marking the der(4) were combined with particular 4q21 probes (labeled with SpectrumOrange-dUTP). The colocalized red and green signals (metaphase cell) indicate that the analyzed 4q21 probe hybridized with the der(4), while separated signals indicate that the probe is translocated to the der(9). Using this strategy, we narrowed down the 4q21 breakpoint to the region flanked by RP11-164M11 and RP11-93E17 that harbors SEC31A. Fosmid probes flanking the SEC31A gene (WI2-2194A15 and WI2-2877K7) were used to prove rearrangement of this gene (interphase cell; separated red and green signals). (D) Schematic representation of the SEC31A, JAK2 and SEC31A-JAK2 protein structures (top panel). Sequencing of the fragment amplified by SEC31A-JAK2 nested RT-PCR identified an in-frame fusion between exon 22 of SEC31A and exon 17 of JAK2 as shown in the electropherogram (bottom panel). FISH probes were directly labeled with SpectrumOrange- and SpectrumGreen-dUTP (Abbott Molecular). FISH images were acquired with a 63×/1.40 oil-immersion objective in an Axioplan 2 fluorescence microscope equipped with an Axiophot 2 camera (Carl Zeiss Microscopy) and a MetaSystems Isis imaging system (MetaSystems).
Cytogenetic and molecular analysis of the t(4;9)(q21;p24) found in case 1. (A) Conventional (left) and M-FISH karyotype (right). Note 2 copies of the t(4;9)(q21;p24) in both karyotypes (red and white arrows) and 1 additional copy of the der(4) in the M-FISH karyotype (white arrows). (B) An ideogram of chromosome 9 with probes used for the FISH characterization of the t(4;9) (left) and examples of FISH experiments (right). BAC probes included in the dual color JAK2 break-apart (JAK2 BA) assay are shown in the middle figure, fosmid probes covering JAK2 are shown in the right figure. Split of JAK2 BA signals on der(4) and der(9) (metaphase cell) illustrates the t(4;9)-associated rearrangement of JAK2. Further analysis with fosmid probes covering JAK2 mapped the breakpoint in the region flanked by WI2-2850C2 and WI2-1990K3 (split of red and green fosmid signal in an interphase cell). (C) An ideogram of chromosome 4 with probes used for the FISH characterization of the 4q21 breakpoint in the t(4;9) (left) and examples of FISH experiments (right). The 4q21 breakpoint was mapped with a BAC/fosmid walking interphase FISH strategy. The telomeric (5′) JAK2 probes (labeled with SpectrumGreen-dUTP) marking the der(4) were combined with particular 4q21 probes (labeled with SpectrumOrange-dUTP). The colocalized red and green signals (metaphase cell) indicate that the analyzed 4q21 probe hybridized with the der(4), while separated signals indicate that the probe is translocated to the der(9). Using this strategy, we narrowed down the 4q21 breakpoint to the region flanked by RP11-164M11 and RP11-93E17 that harbors SEC31A. Fosmid probes flanking the SEC31A gene (WI2-2194A15 and WI2-2877K7) were used to prove rearrangement of this gene (interphase cell; separated red and green signals). (D) Schematic representation of the SEC31A, JAK2 and SEC31A-JAK2 protein structures (top panel). Sequencing of the fragment amplified by SEC31A-JAK2 nested RT-PCR identified an in-frame fusion between exon 22 of SEC31A and exon 17 of JAK2 as shown in the electropherogram (bottom panel). FISH probes were directly labeled with SpectrumOrange- and SpectrumGreen-dUTP (Abbott Molecular). FISH images were acquired with a 63×/1.40 oil-immersion objective in an Axioplan 2 fluorescence microscope equipped with an Axiophot 2 camera (Carl Zeiss Microscopy) and a MetaSystems Isis imaging system (MetaSystems).
JAK2 rearrangements are recurrent in cHL
To identify additional cHL cases with the SEC31A-JAK2 fusion, we have preliminarily analyzed 14 cases showing an increased number of RS cells, like the index case, by nested RT-PCR for the SEC31A-JAK2 transcript. One case was positive and showed 4q21 and 9p24 breakpoints similar to the breakpoints observed in case 1 (fusion between exon 22 of SEC31A and exon 17 of JAK2; Table 1; data not shown). In the next step, we screened 131 unselected cHL cases (including the cases analyzed by RT-PCR) by interphase FISH with the JAK2 BA assay and found JAK2 rearrangements in 2 cases (Table 1). Involvement of SEC31A or any of the remaining 5 known JAK2 partners in these 2 cases was excluded by FISH with differentially labeled probes for the 5′ end of the partner genes and the 3′ end of JAK2 (Table 1; data not shown).
In total, 4 cases with JAK2 rearrangements were identified from a set of 132 cHL cases (3%), with the SEC31A-JAK2 fusion present in 2 cases and novel unknown JAK2 fusions present in the 2 other cases. Lack of residual, appropriate material excluded further detailed molecular analysis of the 2 latter cases.
SEC31A-JAK2 is a constitutively activated tyrosine kinase that is sensitive to treatment with JAK inhibitors
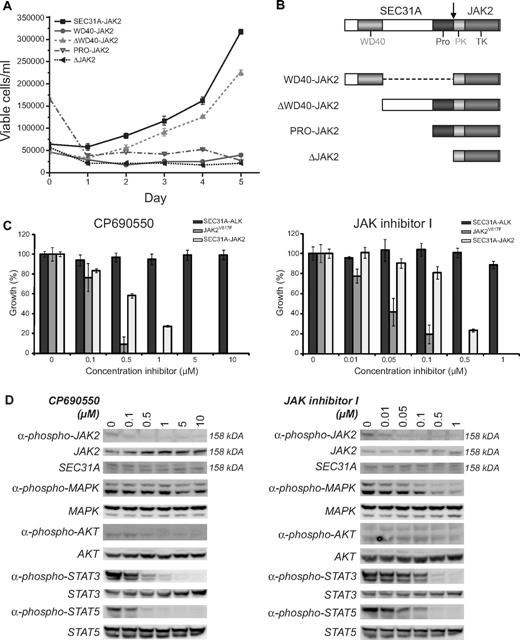
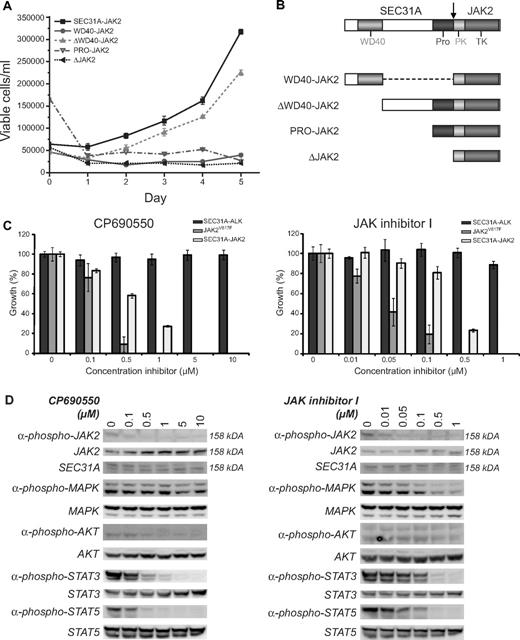
To study the functional consequences of the SEC31A-JAK2 fusion transcript, an expression construct was designed and introduced into the murine hematopoietic IL3-dependent Ba/F3 cell line. SEC31A-JAK2 was able to transform Ba/F3 cells to cytokine independent growth, demonstrating its implication in oncogenic transformation (Figure 2A).
Functional analysis of SEC31A-JAK2. (A) IL3 deprivation of Ba/F3 cells transduced with SEC31A-JAK2 and with the deletion mutant ΔWD40-JAK2 resulted in transformation to growth factor–independent growth. When transduced with the deletion mutants WD40-JAK2, PRO-JAK2, and ΔJAK2, Ba/F3 cells were not able to proliferate in the absence of IL3. The mean growth ± SEM of 3 separate measurements over 5 consecutive days is shown. (B) Schematic representation of the protein structures of the SEC31A-JAK2 deletion mutants. (C) SEC31A-JAK2, SEC31A-ALK, and JAK2V617F–expressing Ba/F3 cells, cultured in the absence of IL3, were treated with the indicated concentrations of the JAK inhibitors CP690550 (left panel) and JAK inhibitor I (right panel), and cell survival was quantified after 24 (SEC31A-JAK2 and SEC31A-ALK) or 48 (JAK2V617F) hours. Ba/F3 cells expressing SEC31A-JAK2 and JAK2V617F, but not SEC31A-ALK, were inhibited in a dose-dependent manner by treatment with JAK inhibitors. IC50 values for CP690550 were observed between 500nM and 1μM for SEC31A-JAK2 and between 100nM and 500nM for JAK2V617F. Treatment with JAK inhibitor I resulted in IC50 values between 100nM and 500nM for SEC31A-JAK2 and between 10nM and 50nM for JAK2V617F. Cell survival in the absence of inhibitor was set at 100%. Mean ± SEM of 4 independent measurements is shown. (D) Western blot analysis showing the effect of CP690550 (left panel) and JAK inhibitor I (right panel) treatment on Ba/F3 cells transduced with SEC31A-JAK2, grown in the absence of IL3. In addition to SEC31A-JAK2 phosphorylation (158 kDa), phosphorylation of AKT, STAT3, and STAT5 decreased with increasing CP690550 concentration. In cells treated with JAK inhibitor I, phosphorylation of MAPK (ERK1 + ERK2), STAT3, and STAT5 reduced with increasing inhibitor concentrations. Expression of total SEC31A-JAK2 (158 kDa), MAPK (ERK1 + ERK2), AKT, STAT3, and STAT5 remained unaffected.
Functional analysis of SEC31A-JAK2. (A) IL3 deprivation of Ba/F3 cells transduced with SEC31A-JAK2 and with the deletion mutant ΔWD40-JAK2 resulted in transformation to growth factor–independent growth. When transduced with the deletion mutants WD40-JAK2, PRO-JAK2, and ΔJAK2, Ba/F3 cells were not able to proliferate in the absence of IL3. The mean growth ± SEM of 3 separate measurements over 5 consecutive days is shown. (B) Schematic representation of the protein structures of the SEC31A-JAK2 deletion mutants. (C) SEC31A-JAK2, SEC31A-ALK, and JAK2V617F–expressing Ba/F3 cells, cultured in the absence of IL3, were treated with the indicated concentrations of the JAK inhibitors CP690550 (left panel) and JAK inhibitor I (right panel), and cell survival was quantified after 24 (SEC31A-JAK2 and SEC31A-ALK) or 48 (JAK2V617F) hours. Ba/F3 cells expressing SEC31A-JAK2 and JAK2V617F, but not SEC31A-ALK, were inhibited in a dose-dependent manner by treatment with JAK inhibitors. IC50 values for CP690550 were observed between 500nM and 1μM for SEC31A-JAK2 and between 100nM and 500nM for JAK2V617F. Treatment with JAK inhibitor I resulted in IC50 values between 100nM and 500nM for SEC31A-JAK2 and between 10nM and 50nM for JAK2V617F. Cell survival in the absence of inhibitor was set at 100%. Mean ± SEM of 4 independent measurements is shown. (D) Western blot analysis showing the effect of CP690550 (left panel) and JAK inhibitor I (right panel) treatment on Ba/F3 cells transduced with SEC31A-JAK2, grown in the absence of IL3. In addition to SEC31A-JAK2 phosphorylation (158 kDa), phosphorylation of AKT, STAT3, and STAT5 decreased with increasing CP690550 concentration. In cells treated with JAK inhibitor I, phosphorylation of MAPK (ERK1 + ERK2), STAT3, and STAT5 reduced with increasing inhibitor concentrations. Expression of total SEC31A-JAK2 (158 kDa), MAPK (ERK1 + ERK2), AKT, STAT3, and STAT5 remained unaffected.
In addition, we tested the sensitivity of SEC31A-JAK2 to 2 JAK inhibitors. We used CP690550, a JAK3 inhibitor that also inhibits JAK2 and is known to suppress the growth of human polycythemia vera cells carrying the JAKV617F mutation,28 and the JAK inhibitor I, a potent inhibitor of all JAK family members, including JAK2.29 Treatment of Ba/F3 cells transformed by SEC31A-JAK2 with CP690550 and JAK inhibitor I resulted in a dose-dependent response with 50% inhibitor concentrations (IC50) between 500nM and 1μM for CP690550, and between 100nM and 500nM for JAK inhibitor I (Figure 2C). In contrast, Ba/F3 cells transformed by the SEC31A-ALK fusion did not show any response to the JAK inhibitors, indicating the specific effect of these inhibitors on the SEC31A-JAK2–transformed cells (Figure 2C). In addition, JAK2V617F-expressing Ba/F3 cells (positive control) also responded to JAK inhibitor treatment, but with slightly lower IC50 values (between 100nM and 500nM for CP690550 and between 10nM and 50nM for JAK inhibitor I). Western blot analysis for SEC31A-JAK2 confirmed a decrease in JAK2 phosphorylation with an increasing dose of both CP690550 and JAK inhibitor I while total JAK2 protein expression remained unaffected (Figure 2D). In addition, phosphorylation of STAT3, STAT5, and to a lesser extent AKT was reduced in SEC31A-JAK2–expressing Ba/F3 cells treated with CP690550 (Figure 2D). When treated with JAK inhibitor I, SEC31A-JAK2–expressing Ba/F3 cells displayed reduced MAPK (ERK1 + ERK2), STAT3, and STAT5 activity (Figure 2D).
The SEC31A WD40-like repeats are dispensable for constitutive JAK2 activation
Given that the known JAK2 fusion proteins function as constitutively activated tyrosine kinases, because of partner-mediated oligomerization of JAK2, we attempted to identify the SEC31A domain responsible for the constitutive JAK2 activation. Deletion of the entire SEC31A part of the SEC31A-JAK2 fusion completely abolished the transforming properties of the fusion protein, indicating that a part of SEC31A was indeed required for activation of the fusion kinase (Figure 2A). In the next step, several deletion mutants were designed (Figure 2B) and tested for their oncogenic properties. These experiments excluded the known WD40-like and proline-rich domains as being the driving force of constitutive JAK2 activation, and identify the region between these domains as a critical region for kinase activation of the SEC31A-JAK2 fusion (Figure 2A), indicating that other, still unknown, domain(s) of SEC31A are important for constitutive kinase activity, or that JAK2 activation is not mediated by SEC31A-induced oligomerization.
SEC31A-JAK2 induces a fatal T-lymphoblastic lymphoma and myeloid phenotype in a murine bone marrow transplantation model
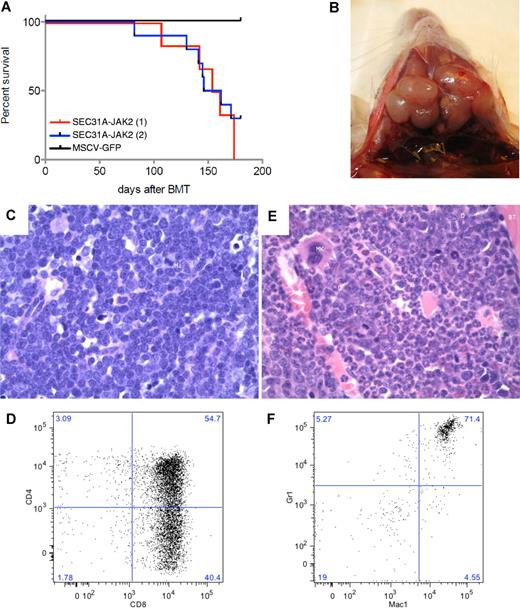
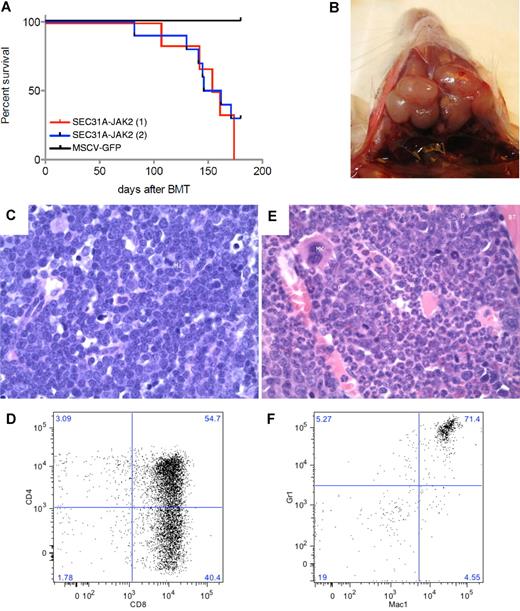
To assess the in vivo role of SEC31A-JAK2, a murine bone marrow transplantation model was established. The experiment was carried out on a group of 6 mice (M7-M12) and repeated independently on a group of 10 mice (N1-N10). From these 16 mice transplanted with SEC31A-JAK2–transduced bone marrow cells, 13 mice developed a fatal disease after 82-174 days (Figure 3A; supplemental Table 3), the other 3 mice were still alive at the end of the experiment (180 days posttransplantation). Furthermore, 3 mice were found dead in their cage and could not be analyzed anymore.
SEC31A-JAK2–derived murine bone marrow transplantation model. (A) Survival curves of 2 independent groups of mice transplanted with SEC31A-JAK2–expressing bone marrow cells show the development of a fatal disease (red and blue curve). Control mice transduced with the empty pMSCV-GFP vector did not show any signs of disease up to 180 days after transplantation. (B) Enlarged lymph nodes of the neck in a mouse presenting with T-lymphoblastic lymphoma. (C) The lymph nodes of mice diagnosed with SEC31A-JAK2–induced T-lymphoblastic lymphoma display effacement of the normal architecture by a monotonous medium-sized blastic proliferation with condensed nuclear chromatin, single nucleolus, and little basophilic cytoplasm (H&E, ×400). Mi indicates mitotic figure. (D) The spleen of mice diagnosed with SEC31A-JAK2–induced T-lymphoblastic lymphoma shows infiltration of mainly CD4−/CD8+ and CD4+/CD8+ cells by FACS analysis. (E) The bone marrow of mice presenting with the SEC31A-JAK2–associated myeloid phenotype displays an increased cellularity of the intertrabecular spaces, caused by a hyperplastic myelopoiesis (H&E, ×400). D indicates doughnut cells; Mk, megakaryocyte; G, granulocytes; and BT, bony trabeculae. (F) The bone marrow of mice presenting with the SEC31A-JAK2–associated myeloid phenotype shows an enrichment of gr1+/mac1+ myeloid cells by FACS analysis. Immunohistochemical images were captured with a Leica DM LB microscope (Leica) using a Leica PL FLUOTAR objective lens (40×/0.70) and a Leica DC200 camera. Images were imported directly into Photoshop CS (Adobe) using the Leica DC200 camera software (Version 2.51).
SEC31A-JAK2–derived murine bone marrow transplantation model. (A) Survival curves of 2 independent groups of mice transplanted with SEC31A-JAK2–expressing bone marrow cells show the development of a fatal disease (red and blue curve). Control mice transduced with the empty pMSCV-GFP vector did not show any signs of disease up to 180 days after transplantation. (B) Enlarged lymph nodes of the neck in a mouse presenting with T-lymphoblastic lymphoma. (C) The lymph nodes of mice diagnosed with SEC31A-JAK2–induced T-lymphoblastic lymphoma display effacement of the normal architecture by a monotonous medium-sized blastic proliferation with condensed nuclear chromatin, single nucleolus, and little basophilic cytoplasm (H&E, ×400). Mi indicates mitotic figure. (D) The spleen of mice diagnosed with SEC31A-JAK2–induced T-lymphoblastic lymphoma shows infiltration of mainly CD4−/CD8+ and CD4+/CD8+ cells by FACS analysis. (E) The bone marrow of mice presenting with the SEC31A-JAK2–associated myeloid phenotype displays an increased cellularity of the intertrabecular spaces, caused by a hyperplastic myelopoiesis (H&E, ×400). D indicates doughnut cells; Mk, megakaryocyte; G, granulocytes; and BT, bony trabeculae. (F) The bone marrow of mice presenting with the SEC31A-JAK2–associated myeloid phenotype shows an enrichment of gr1+/mac1+ myeloid cells by FACS analysis. Immunohistochemical images were captured with a Leica DM LB microscope (Leica) using a Leica PL FLUOTAR objective lens (40×/0.70) and a Leica DC200 camera. Images were imported directly into Photoshop CS (Adobe) using the Leica DC200 camera software (Version 2.51).
Of the remaining 10 informative animals, 6 (M7, M9, M12, N1, N2, and N4) developed a lymphoma and presented with enlarged spleen, thymus, lymph nodes, and liver (Figure 3B; supplemental Table 3). Examination of peripheral blood revealed elevated white blood cell counts with a marked increase of the lymphoid component and a decrease of cells from the myeloid lineage (neutrophils) in all 6 animals. A massive proliferation of lymphoblastic cells was found in the spleen, liver, bone marrow, and lymph nodes on histologic examination (Figure 3C). On FACS analysis, the lymphoblastic cells in the bone marrow and spleen were mainly CD4−/CD8+ in 2 mice, and CD4+/CD8+ and CD4−/CD8+ in 4 mice (Figure 3D; supplemental Table 3). TCR analysis demonstrated the presence of a mono- or oligoclonal T-cell population (supplemental Figure 1). Altogether, these data revealed the development of a mono- or oligoclonal T-lymphoblastic lymphoma (T-LL) in this subset of mice.
Four mice (M10, M11, N3, and N6) displayed a myeloid phenotype with a near-normal white blood cell count in the peripheral blood, but with an elevated level of neutrophils and a low percentage of lymphocytes, demonstrating an enrichment of the myeloid cell lineage (supplemental Table 3). Except for 1 mouse (N6), these mice did not have any organomegaly: the weight of the spleen, the thymus, and the lymph nodes was normal (supplemental Table 3). While the bone marrow histology showed prominent, hyperplastic myelopoiesis with full maturation up to granulocytes (Figure 3E), no abnormalities were found in the spleen. FACS analysis showed an increase in gr1+/mac1+ myeloid cells in spleen and bone marrow (Figure 3F; supplemental Table 3).
To investigate whether the disease phenotypes observed in the mice were transplantable, we performed secondary transplantation experiments. Spleen cells derived from the SEC31A-JAK2–associated T-LL (M12) and myeloid phenotype (M11) were transplanted to secondary recipients. Eighty-four days after transplantation, 4 of the 6 mice transplanted with cells from the mice with a lymphoblastic proliferation (M12.49, M12.50, M12.52, and M12.53) developed disease and were killed. These animals presented with high white blood cell counts, and enlarged spleen, thymus, and lymph nodes of the neck (supplemental Table 3). FACS analysis of the bone marrow and the spleen demonstrated the presence of a high number of CD4−/CD8+ T cells (supplemental Table 3). Clonality analysis showed the presence of a mono- or oligoclonal T-cell population (supplemental Figure 1). In addition, the pancreas was prominently enlarged because of massive involvement by lymphoblasts in all 4 diseased animals. Based on these findings, the disease in these mice was diagnosed as T-LL. In contrast, secondary recipient mice transplanted with cells from the SEC31A-JAK2–associated myeloid phenotype were killed 121 days after transplantation, but no abnormalities were found. In the group of mice developing a myeloid phenotype, a reactive hyperplasia can be considered, but an early stage of a myeloproliferation or myeloproliferative disease is not excluded.30
Discussion
The JAK2 gene is a known target of activating mutations and translocations identified in various hematologic malignancies.2-8,25,31-33 Our study demonstrates that JAK2 rearrangements also occur in cHL. We identified JAK2 aberrations in 4 cases of cHL, including 2 with the novel SEC31A-JAK2 fusion, created by a t(4;9)(q21;p24) in the index case. The SEC31A gene encodes a ubiquitously expressed protein that is localized in vesicular structures that are scattered throughout the cytoplasm, and that are involved in the vesicular transport from the endoplasmatic reticulum to the Golgi area.34 Of note, we have recently also identified SEC31A as a fusion partner of ALK in ALK-positive large B-cell lymphoma.24 The involvement of SEC31A in 2 different PTK-related fusions is not exceptional, as some genes, for example, TEL(ETV6), show a tendency to partner multiple PTKs.
Molecular studies showed that constitutive activation of JAK2 and other PTKs involved in chimeric oncoproteins is usually driven by an oligomerization motif provided by the partner protein. Although the affected N-terminal region of SEC31A contains WD40-like repeats and a part of the proline-rich region, both involved in protein-protein interactions,34-36 our molecular studies failed to prove a role of any of these motifs in the aberrant activation of the JAK2 kinase domain. Of interest, the same region of SEC31A (exons 1-22) was present in both the SEC31A-JAK2 fusion and in the SEC31A-ALK fusion.24 It is most likely that the same domain(s) are responsible for constitutive kinase activation in both fusions; however, for SEC31A-ALK, no attempts were made to identify the critical domain for constitutive ALK activity. These findings suggest that other, still unknown SEC31A domain(s) are important to confer constitutive JAK2 kinase activity to SEC31A-JAK2, or that JAK2 activation is not mediated by SEC31A-induced oligomerization. Similar findings were reported in the case of FIP1L1-PDGFRα and NUP214-ABL1 fusion kinases, where loss of the inhibitory juxtamembrane region or subcellular localization are important activating mechanisms.37,38
So far, mouse models derived from JAK2 fusions have only been established for the TEL-JAK2 fusion reported in patients with T-ALL,2 atypical CML and precursor B-ALL.3 Mice expressing TEL-JAK2 developed different pathologies, including a mixed myeloproliferative and T-cell proliferative disorder,26 T-cell leukemia,39 and T-cell or B-cell lymphoma40 depending on the model and the mouse strain used. In the light of these findings, development of a T-LL and a myeloid phenotype in SEC31A-JAK2 recipient mice is not unexpected. Of interest, RS cells in both SEC31A-JAK2–positive cHL cases lacked the expression of PAX5 (a B-cell marker commonly expressed in cHL41 ). They neither expressed any T-cell marker except for TIA1 in both cases, and granzyme B and perforin in case 1 (Table 1), which might suggest a T-cell origin of the RS cells in these cases.
Different phenotypes were found to develop in SEC31A-JAK2– expressing mice. It is difficult to predict what exactly drives each phenotype as many different factors, including acquired secondary aberrations, may influence the phenotype. Of interest, however, is the finding that GFP expression was generally lower for the mice with the myeloid phenotype than for the mice with a lymphoblastic lymphoma (supplemental Table 3).
Although the majority of cHL cases are expected to originate from B cells, a small fraction of cHL originate from T cells.42-48 Recent gene expression studies of B and T cell–derived HL cell lines support the existence of T-cHL and suggest that RS cells originating from B or T cells follow a similar gene expression program after malignant transformation.49 This concept is also supported by our finding of JAK2 rearrangements in cHL cases with and without PAX5 expression.
Multiple lines of evidence indicate that the development of cancer is a multi-step process. Because complex karyotypic aberrations accompanied the t(4;9) in the index case and given the relatively long latency before disease onset in the SEC31A-JAK2–recipient mice, it is likely that SEC31A-JAK2 is not the only driving force behind the development of cHL in humans and T-LL in mice. To get insight into secondary genomic aberrations associated with the SEC31A-JAK2 rearrangement, we performed array-comparative genome hybridization (aCGH) analysis of 4 primary proliferations (1 myeloid and 3 T-LLs) that developed in our murine SEC31A-JAK2 model (supplemental Results). Indeed, 2 of these cases showed a few acquired genetic imbalances that may contribute to the disease phenotype. Interestingly, the 14qA3 region duplicated in the myeloid M10 covers the adenosine kinase (Adk) gene found to be overexpressed in some human cancers,50 and the 12qF1 region deleted in 1 T-LL (and all 4 secondary transplants) harbors Bcl11b (supplemental Results). Bcl11b is a postulated tumor suppressor gene in murine and human T lymphomagenesis.51,52 FISH analysis of the index cHL case, however, failed to detect loss of the BCL11B region, as well as CDKN2A (p16) and TNFAIP3 (A20) in RS cells (data not shown). Although the 2 remaining T-LLs did not show tumor-related genomic imbalances (supplemental Results), other acquired genetic defects that escaped aCGH analysis (for example, balanced aberrations and mutations) cannot be excluded.
The finding that SEC31A-JAK2 is sensitive to JAK inhibitors is clinically important, and indicates that HL patients harboring JAK2 fusions might benefit from targeted treatment with JAK2 antagonists that are currently already under development for the treatment of other hematologic malignancies.11
In summary, we identified the SEC31A-JAK2 fusion as a recurrent chromosomal aberration in cHL, and we demonstrated that rearrangements of JAK2 are recurrent in cHL and may occur in 3% of cases. Oncogenic properties of this novel fusion were proved in vitro and in vivo, and, importantly, we demonstrated that SEC31A-JAK2 is sensitive to JAK inhibitors.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Ursula Pluys for her excellent technical assistance, Julio Finalet Ferreiro for his help with the murine aCGH studies, Dr Achiel Van Hoof for providing clinical data, and Rita Logist for her editorial assistance.
This work was supported by the concerted action grant from the KU Leuven no. 3M040406 (K.V.R., P.M., P.V., J.C., and I.W.), by a grant from the Stichting Emmanuel van der Schueren (K.V.R.), by IWT-Vlaanderen SBO grant no. 060848, and by FWO-Vlaanderen (G.0287.07). P.V. is a senior clinical investigator of the FWO-Vlaanderen.
Authorship
Contribution: K.V.R., L.C., J.C., and I.W. designed the research; K.V.R., T.T. and O.G. performed the research; K.V.R., L.C., T.T., P.V., C.D.W.-P., J.C., and I.W. analyzed the data; T.T., B.C., P.D.P., G.V., P.V., and C.D.W.-P. collected samples and provided clinical data; K.V.R., L.C., T.T., C.D.W.-P., J.C., and I.W. wrote the paper; and I.L. and P.M. advised in research design.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Iwona Wlodarska, Center for Human Genetics, Herestraat 49, B-3000 Leuven, Belgium; e-mail: iwona.wlodarska@uz.kuleuven.ac.be.
References
Author notes
K.V.R. and L.C. contributed equally to this study.