Abstract
We previously identified LDOC1 as one of the most significantly differentially expressed genes in untreated chronic lymphocytic leukemia (CLL) patients with respect to the somatic mutation status of the immunoglobulin heavy-chain variable region genes. However, little is known about the normal function of LDOC1, its contribution to the pathophysiology of CLL, or its prognostic significance. In this study, we have investigated LDOC1 mRNA expression in a large cohort of untreated CLL patients, as well as in normal peripheral blood B-cell (NBC) subsets and primary B-cell lymphoma samples. We have confirmed that LDOC1 is dramatically down-regulated in mutated CLL cases compared with unmutated cases, and have identified a new splice variant, LDOC1S. We show that LDOC1 is expressed in NBC subsets (naive > memory), suggesting that it may play a role in normal B-cell development. It is also expressed in primary B-cell lymphoma samples, in which its expression is associated with somatic mutation status. In CLL, we show that high levels of LDOC1 correlate with biomarkers of poor prognosis, including cytogenetic markers, unmutated somatic mutation status, and ZAP70 expression. Finally, we demonstrate that LDOC1 mRNA expression is an excellent predictor of overall survival in untreated CLL patients.
Introduction
In chronic lymphocytic leukemia (CLL), one of the best predictors of outcome is the somatic mutation status of the immunoglobulin heavy-chain variable region (IGHV) genes. Patients whose CLL cells have unmutated IGHV genes, approximately 40% of patients, have a median survival of 8 years; patients whose CLL cells have mutated IGHV genes, approximately 60% of patients, have a median survival of 25 years.1 To identify new prognostic biomarkers and molecular targets for therapy in untreated CLL patients, we reanalyzed the raw data from 4 published gene expression profiling microarray studies.2-5 Of 88 candidate biomarkers associated with somatic mutation status, we confirmed the expression of 37 using a highly sensitive quantitative RT-PCR assay performed on microfluidics cards (MF-QRT-PCR).6 Of these candidate biomarkers, the gene LDOC1 (leucine zipper down-regulated in cancer) was one of the most significantly differentially expressed genes that distinguished mutated from unmutated CLL cases.
The LDOC1 gene, on chromosome Xq27, encodes a 17-kDa protein about which little is known. A leucine zipper motif in the N-terminal region is followed by a short proline-rich region, which contains an SH3-binding consensus sequence, and then an acidic region in the C-terminus.7 Because leucine zipper and SH3-binding motifs mediate protein-protein interactions, LDOC1 protein may regulate transcription by homodimerization or by heterodimerization with other transcription factors through its leucine zipper domain. LDOC1 also may participate in cell signaling by providing a binding surface for signaling cascade proteins within its SH3 domain. Others have assessed LDOC1 mRNA expression in a wide range of normal tissues and in carcinoma cell lines.7 It is expressed ubiquitously in normal tissues, although at relatively low levels in leukocytes, liver, and placenta compared with other tissues. In tumor cell lines, LDOC1 is expressed in most breast cancer cell lines, but rarely in pancreatic or gastric carcinoma cell lines. Because LDOC1 is expressed in many normal tissues, but not in most pancreatic and gastric carcinoma cell lines, Nagasaki and colleagues8 have hypothesized that LDOC1 is a tumor suppressor gene. Others have reported that LDOC1 induces apoptosis in Jurkat lymphoma and K562 leukemia cell lines, but not in HeLa cervical carcinoma cells.9 Thus, it has been suggested that LDOC1 may have proapoptotic and antiproliferative effects. In CLL, it is possible that differences in LDOC1 expression may be related to the enhanced ability of unmutated cases to respond proliferative stimuli and resist apoptosis compared with mutated cases.10 However, the biologic functions of LDOC1 in normal B-cell development and the pathophysiology of CLL are unknown.
In the current study, we have investigated LDOC1 mRNA expression in a large cohort of untreated CLL patients, as well as in normal peripheral blood B-cell (NBC) subsets and primary small B-cell lymphoma samples. We have confirmed that LDOC1 is dramatically down-regulated in mutated CLL cases compared with unmutated cases, and have identified a new splice variant, LDOC1S. We show that LDOC1 is expressed in NBC subsets, suggesting that it may play a role in normal B-cell development. It is also expressed in primary small B-cell lymphoma samples, in which its expression is also associated with somatic mutation status. In CLL, we show that high levels of LDOC1 expression correlate with biomarkers of poor prognosis, including cytogenetic markers, unmutated somatic mutation status, and ZAP70 protein expression. Finally, we demonstrate that LDOC1 mRNA expression is an excellent predictor of overall survival in previously untreated CLL patients.
Methods
Patient and healthy donor sample collection
We collected peripheral blood samples from 131 previously untreated CLL patients and 6 healthy volunteers at The University of Texas M. D. Anderson Cancer Center after informed consent was obtained. The study was approved by the Institutional Review Board and conducted according to the principles expressed in the Declaration of Helsinki. CLL samples and CD19+ NBCs were purified by negative selection, and processed as described previously.11 We enriched NBCs from healthy donors for naive or memory B cells using a CD27 antibody column, according to the manufacturer's instructions (Miltenyi Biotec). In peripheral blood, approximately 60% of the B cells are antigen-naive (CD27−) and approximately 40% are antigen-experienced memory B cells (CD27+).12 The purity of all cell preparations was confirmed by flow cytometry.
Primary lymphoma samples were obtained from lymph node biopsy specimens from patients with follicular lymphoma (2 patients). Primary lymphoma samples were also obtained from the peripheral blood of patients in leukemic phase of follicular lymphoma (1 patient), mantle cell lymphoma (4 patients), splenic marginal zone lymphoma (2 patients), and marginal zone lymphoma (1 patient). CD19+ B cells were purified as described above. All samples contained ≥ 95% CD19+ B cells by flow cytometry.
Cell lines
The EBV-negative Burkitt lymphoma cell line, GA-10, and the T-cell lymphoblastic lymphoma cell line, Jurkat, were maintained in RPMI medium, supplemented with 10% FBS, 2mM l-glutamine, 100 IU/mL penicillin, and 100 μg/mL streptomycin; they were harvested during the exponential phase of growth. The breast carcinoma cell line, MCF-7, and the cervical carcinoma cell line, HeLa, were maintained in DMEM, supplemented as described, and harvested when they reached approximately 80% confluence.
Nucleic acid preparation
Total RNA was extracted using 2 rounds of guanidine isothiocyanate/phenol-chloroform extraction with TRIzol reagent (Life Technologies) according to the manufacturer's instructions, and its quality assessed by agarose gel electrophoresis. Total RNA was reverse transcribed into cDNA using random hexamers and a First-Strand cDNA synthesis kit (Amersham Pharmacia Biotech). DNA was extracted using a Qiagen DNeasy kit (Qiagen Sciences) according to the manufacturer's instructions.
Evaluation of the IGHV somatic mutation status
The somatic mutation status of the IGHV genes in CLL and primary B-cell lymphoma samples was assessed as described previously, with minor modifications.11 To determine the degree of somatic mutation, patient sequences were aligned to germline sequences in VBASE II.13 The somatic mutation status was designated as unmutated if there were < 2% mutations (> 98% homology), or as mutated if there were ≥ 2% mutations (≤ 98% homology) compared with germline sequences.14
Assessment of ZAP70 protein expression
Expression of ZAP70 protein was assessed either by immunohistochemistry or flow cytometry. Immunohistochemical staining was performed using routinely fixed and processed paraffin-embedded tissue sections of bone marrow core biopsy and/or clot specimens and a specific monoclonal antibody (Upstate Cell Signaling Systems), as described previously.15,16 The flow cytometry assay for ZAP70 protein was performed by the Chronic Lymphocytic Leukemia Research Consortium laboratory, as described previously.17
LDOC1 mRNA expression by RT-PCR assay
For the RT-PCR assay, 2 primer pairs (AB and AC) were used. Primer pair AB was designed to amplify the entire LDOC1 coding region and yield a 464-bp product.7 We designed a second reverse primer, C (5′-AGCAGGTAACTGGAGCGCTA-3′), to bind within the 3′ untranslated region (3′ UTR). The AC primer pair was expected to yield a 649-bp product. The cDNA (80 ng) was amplified in the presence of primers, reaction buffer, deoxynucleotide triphosphates (2.5μM each), and HotStar Taq DNA polymerase. After incubation at 95°C for 10 minutes, the cDNA was amplified for 35 cycles of at 95°C for 15 seconds, 55°C for 30 seconds, and 72°C for 45 seconds, followed by a final extension at 72°C for 7 minutes. We performed 2 independent PCRs for each sample. The sequence of the gel-purified PCR products was determined directly using the forward and reverse primers, and an ABI 3700 or 3730 DNA Analyzer (Applied Biosystems). Sequences were aligned (SPLIGN and LaserGene Version 7.2; DNASTAR) to the LDOC1 reference sequence (GenBank RefSeq NM_012317). We also amplified β-actin using forward (5′-GATCATGTTTGAGACCTTCAAC-3′) and reverse (5′-TCTTTGCGGATGTCCACGTC-3′) primers, using the same conditions as for the LDOC1 RT-PCR assay.
Total LDOC1 mRNA expression by quantitative RT-PCR assays
We used 2 different quantitative RT-PCR (QRT-PCR) assays to detect total LDOC1 mRNA expression: a high-throughput assay using microfluidics cards (MF-QRT-PCR) and a standard QRT-PCR assay. Both assays use the same LDOC1 TaqMan probe and primer set; the primers bind to sequences in the 3′ UTR that are present in wild-type LDOC1 and its splice variant, LDOC1S (TaqMan Assay, Hs00273392_s1; Applied Biosystems). In the first assay, we printed custom microfluidics cards to detect 88 candidate mRNA biomarkers of somatic mutation status, including LDOC1 and ZAP70, and 5 endogenous control genes (18S rRNA, GAPD, PGK1, GUSB, and ECE-1), and performed the MF-QRT-PCR assays, as described previously.6
In the standard QRT-PCR assay for LDOC1 mRNA, the PCRs were carried out in 25-μL reaction volumes that contained 5 μL of cDNA, which was reverse transcribed from 3 μg of RNA and diluted 20-fold to achieve a final cDNA concentration of 10 ng/μL. In addition, 1× TaqMan Universal PCR Master Mix without AmpErase UNG, unlabeled LDOC1-specific PCR primers, and a 6-carboxy fluorescein (FAM)–labeled TaqMan minor groove binder (MGB) probe were added. Amplification of 18S ribosomal RNA (rRNA) was performed in all cases to normalize the LDOC1 values. The probe for 18S rRNA is labeled with VIC (part number: 4319413E, Pre-Developed TaqMan Assay Reagents; Applied Biosystems). After incubation at 95°C for 10 minutes, the cDNA was amplified for 45 cycles of denaturation at 95°C for 15 seconds and combined annealing/extension at 60°C for 1 minute. Each sample was analyzed in triplicate in a PRISM 7000 Sequence Detector (Applied Biosystems). We used the 7500 Fast System Version 1.4.0 software (Applied Biosystems) to analyze the fluorescence emission data after QRT-PCR. The threshold cycle (Ct) value represents the cycle number at which fluorescence passes a fixed threshold. The ΔCt for LDOC1 mRNA was obtained by subtracting the Ct value of 18S rRNA from the Ct value of LDOC1 mRNA for each sample. The LDOC1 mRNA expression levels in test samples are presented as relative quantities (RQ) of 2−ΔΔCt = 2−(ΔCt test − ΔCt calibrator), where the calibrator represents an equal mixture of cDNA obtained from GA-10 and Jurkat cells.
LDOC1 mRNA isoform expression by QRT-PCR assays
To assess the contribution of each of the LDOC1 mRNA isoforms to the total LDOC1 expression, we designed 2 specific TaqMan probe and primer sets, which distinguish between the wild-type LDOC1 and splice variant LDOC1S mRNA isoforms (Primer Express software; Applied Biosystems). One probe and primer set recognizes sequences restricted to the wild-type LDOC1 transcript: the 5′ primer anneals to sequences 5′-TGGTGCCCTACATCGAGATG-3′, the 3′ primer anneals to sequences 5′-CGAGGAAGGCCCGGTAA-3′, and the TaqMan probe anneals to sequences 5′-ATAGCCCCATCCTAGGTG-3′. The other probe and primer set specifically detects the splice variant LDOC1S transcript. The TaqMan probe targets the junction sequence located between nucleotides 183 to 233 and 718 to 785: the 5′ primer anneals to sequences 5′-TTCCAAGCACTTCCGAGTGA-3′, the 3′ primer anneals to sequences 5′-ATGGAACAGCTGCGGCTG-3′, and the TaqMan probe anneals to sequences 5′-CTATTCCTGGCGCAGCAG-3′.
To determine the assay specificity, that is, whether the TaqMan assays designed against the sequences of either LDOC1 or LDOC1S mRNA recognize their own sequence specifically, and discriminate efficiently between the two, we prepared synthetic templates that consist of the amplicons between the forward and reverse primer sequences, inclusive: the wild-type template is 5′-TGGTGCCCTACATCGAGATGATAGCCCCATCCTAGGTGTTACCGGGCCTTCCTCG-3′, and the splice variant template is 5′-TTCCAAGCACTTCCGAGTGACTATTCCTGGCGCAGCAGCAGCCGCAGCTGTTCCAT-3′. We used 33 000 molecules of each primer as the cDNA template. We determined that the assay for the wild-type LDOC1 mRNA isoform recognizes the wild-type template with 14 × 106-fold specificity compared with the splice variant template, as determined by RQ of 2−ΔCt = 2−(Ct target − Ct homolog). The assay for LDOC1S mRNA isoform recognizes the splice variant template with 43 × 103-fold specificity compared with the wild-type template (data not shown). Because a specificity of > 30 × 103-fold is the standard used for the commercially available TaqMan assays, the assays we designed are highly specific. To account for the different efficiencies, we used the data from all samples to fit a model expressing total LDOC1 mRNA as a weighted linear combination of wild-type (WT) and splice variant (SV) contributions. For normalized mRNA levels, we found the optimal model to be: Total = 0.67 × WT + 0.24 × SV.
Detection of genomic gains and losses by single nucleotide polymorphism genotyping
Genotypic analysis was performed on DNA obtained from 100 of the CLL samples using the Illumina HumanHap610 chip, according to the manufacturer's instructions. Background subtraction and normalization were performed using the default settings in the Illumina BeadStudio software. Log R ratios (LRR), B-allele frequencies (BAF), and genotype calls were exported from BeadStudio for analysis in the R statistical programming environment (Version 2.8.1). Segments of constant copy number in the LRR data were identified by applying the circular binary segmentation (CBS) algorithm, as implemented in the DNAcopy package (Version 1.16.0).18 Segments with mean LRR < −0.15 and 2 bands in the BAF plot were called “deleted”; segments with mean LRR > 0.15 and 4 bands in the BAF plot were called “gained.” The data were evaluated to detect common abnormalities associated with CLL, that is, deletions of 6q21, 11q22.3, 13q14.3, and 17p13.1, and trisomy 12; we also evaluated the DNA for gains or deletions of LDOC1.
Survival analysis
Time-to-event (“survival”) analysis was performed using Cox proportional hazards models. Significance was assessed using the log-rank (score) test. To assess multivariate models, we used a forward-backward stepwise algorithm to eliminate redundant factors and optimize the Akaike Information Criterion (AIC).19 All computations were performed using the survival package (Version 2.35-8) in the R statistical programming environment (Version 2.11.1).
Results
LDOC1 mRNA expression distinguishes mutated from unmutated CLL cases
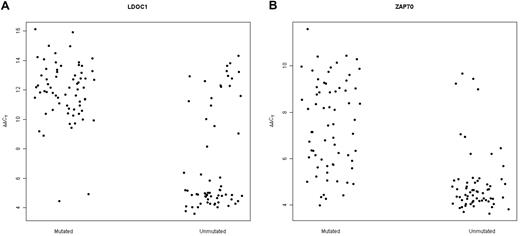
We expanded our previous analysis of 49 samples6 to a total of 131 samples obtained from untreated CLL patients. We found that LDOC1 expression was bimodally distributed, with no patients exhibiting ΔCt values between 7 and 8 cycles (data not shown). Thus, we defined samples to be LDOC1-positive if ΔCt ≤ 7.5 and LDOC1-negative if ΔCt > 7.5. Patient characteristics are summarized in Table 1. With respect to age, sex, Rai stage, white blood cell count, and serum β-2 microglobulin levels at the time the sample was obtained, there were no statistically significant differences between patients whose cells expressed LDOC1 mRNA and those whose cells were negative. Because we originally identified LDOC1 mRNA as a biomarker of somatic mutation status,6 its expression strongly correlated with the somatic mutation status, as expected (Fisher exact test; P = 2.20 × 10−16). For each case, the somatic mutation status, IGHV family, percent homology to the germline sequence, LDOC1 mRNA expression, ZAP70 protein expression, and cytogenetic findings, are listed in supplemental Table 1 (available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Of a total of 131 cases, 65 of 67 mutated cases (97%) were negative for LDOC1 mRNA, and 43 of 63 unmutated cases (68%) were positive; the somatic mutation status was unavailable for 1 case (Figure 1A; supplemental Table 1). Equivalently, 43 of 45 (96%) LDOC1-positive cases were unmutated and 65 of 85 (76%) LDOC1-negative cases were mutated. Four cases (3%) used the VH3-21 gene, a marker of poor prognosis independent of somatic mutation status.20,21
Clinical and laboratory features
| . | All patients, n = 131 . | LDOC1 positive, n = 46 . | LDOC1 negative, n = 85 . | P* . |
|---|---|---|---|---|
| Median age, y (range) | 59 (27-82) | 60 (27-82) | 59 (27-81) | .6796 |
| Sex | .7066 | |||
| Male, n (%) | 81 (62%) | 27 (59%) | 54 (64%) | |
| Female, n (%) | 50 (38%) | 19 (41%) | 31 (36%) | |
| Rai stage | (n = 131) | (n = 46) | (n = 85) | .826 |
| 0-2, n (%) | 102 (78%) | 35 (76%) | 67 (79%) | |
| 3-4, n (%) | 29 (22%) | 11 (24%) | 18 (21%) | |
| WBC count | (n = 131) | (n = 46) | (n = 85) | .0621 |
| ≤ 150 × 109/L, n (%) | 118 (90%) | 38 (83%) | 80 (94%) | |
| > 150 × 109/L, n (%) | 13 (10%) | 8 (17%) | 5 (6%) | |
| Serum β2 microglobulin† | (n = 130) | (n = 46) | (n = 84) | .1384 |
| < 4, n (%) | 98 (75%) | 31 (67%) | 67 (80%) | |
| ≥ 4, n (%) | 32 (25%) | 15 (33%) | 17 (20%) | |
| IGHV somatic mutation status† | (n = 130) | (n = 45) | (n = 85) | 2.20 × 10−16 |
| Mutated, n (%) | 67 (52%) | 2 (4%) | 65 (76%) | |
| Unmutated, n (%) | 63 (48%) | 43 (96%) | 20 (24%) | |
| ZAP70 protein status† | (n = 113) | (n = 39) | (n = 74) | 1.06 × 10−6 |
| Positive, n (%) | 51 (45%) | 30 (77%) | 21 (28%) | |
| Negative, n (%) | 62 (55%) | 9 (23%) | 53 (72%) | |
| Cytogenetic changes† | (n = 100) | (n = 39) | (n = 61) | .0005834 |
| None, n (%) | 27 (27%) | 14 (36%) | 14 (23%) | |
| Isolated del(13q), n (%) | 36 (36%) | 5 (13%) | 30 (49%) | |
| del(6q), del(11q), del(17p), +12, or del(13q) with other abnormalities, n (%) | 37 (37%) | 20 (51%) | 17 (28%) |
| . | All patients, n = 131 . | LDOC1 positive, n = 46 . | LDOC1 negative, n = 85 . | P* . |
|---|---|---|---|---|
| Median age, y (range) | 59 (27-82) | 60 (27-82) | 59 (27-81) | .6796 |
| Sex | .7066 | |||
| Male, n (%) | 81 (62%) | 27 (59%) | 54 (64%) | |
| Female, n (%) | 50 (38%) | 19 (41%) | 31 (36%) | |
| Rai stage | (n = 131) | (n = 46) | (n = 85) | .826 |
| 0-2, n (%) | 102 (78%) | 35 (76%) | 67 (79%) | |
| 3-4, n (%) | 29 (22%) | 11 (24%) | 18 (21%) | |
| WBC count | (n = 131) | (n = 46) | (n = 85) | .0621 |
| ≤ 150 × 109/L, n (%) | 118 (90%) | 38 (83%) | 80 (94%) | |
| > 150 × 109/L, n (%) | 13 (10%) | 8 (17%) | 5 (6%) | |
| Serum β2 microglobulin† | (n = 130) | (n = 46) | (n = 84) | .1384 |
| < 4, n (%) | 98 (75%) | 31 (67%) | 67 (80%) | |
| ≥ 4, n (%) | 32 (25%) | 15 (33%) | 17 (20%) | |
| IGHV somatic mutation status† | (n = 130) | (n = 45) | (n = 85) | 2.20 × 10−16 |
| Mutated, n (%) | 67 (52%) | 2 (4%) | 65 (76%) | |
| Unmutated, n (%) | 63 (48%) | 43 (96%) | 20 (24%) | |
| ZAP70 protein status† | (n = 113) | (n = 39) | (n = 74) | 1.06 × 10−6 |
| Positive, n (%) | 51 (45%) | 30 (77%) | 21 (28%) | |
| Negative, n (%) | 62 (55%) | 9 (23%) | 53 (72%) | |
| Cytogenetic changes† | (n = 100) | (n = 39) | (n = 61) | .0005834 |
| None, n (%) | 27 (27%) | 14 (36%) | 14 (23%) | |
| Isolated del(13q), n (%) | 36 (36%) | 5 (13%) | 30 (49%) | |
| del(6q), del(11q), del(17p), +12, or del(13q) with other abnormalities, n (%) | 37 (37%) | 20 (51%) | 17 (28%) |
Age, Rai stage, WBC count, and serum β2 microglobulin values are reported for the time the sample was obtained for LDOC1 mRNA expression; IGHV somatic mutation status, ZAP70 protein status, and cytogenetic changes were determined on samples obtained before treatment.
All P values were calculated using the 2-sided Fisher exact test except for age in years, which was calculated using the 2-sided t test.
For serum β2 microglobulin and IGHV somatic mutation status, 1 value was unavailable; for ZAP70 protein, 18 values were unavailable; and for genomic abnormalities, 31 values were unavailable.
Expression of LDOC1 and ZAP70 mRNAs measured by MF-QRT-PCR assay distinguishes between mutated and unmutated cases of CLL. (A) Dot-plot for LDOC1 mRNA expression. For the mutated cases, 65 of 67 mutated cases were unambiguously negative for LDOC1 mRNA (higher ΔΔCt values) and 2 were positive (lower ΔΔCt values). For the unmutated cases, 43 of 63 unmutated cases were positive for LDOC1 mRNA and 20 were negative. (B) Dot-plot for ZAP70 mRNA expression. For the unmutated cases, 53 of 63 unmutated cases were positive for ZAP70 mRNA and 10 were negative. Eleven of 67 mutated cases were positive for ZAP70 mRNA and 56 were negative.
Expression of LDOC1 and ZAP70 mRNAs measured by MF-QRT-PCR assay distinguishes between mutated and unmutated cases of CLL. (A) Dot-plot for LDOC1 mRNA expression. For the mutated cases, 65 of 67 mutated cases were unambiguously negative for LDOC1 mRNA (higher ΔΔCt values) and 2 were positive (lower ΔΔCt values). For the unmutated cases, 43 of 63 unmutated cases were positive for LDOC1 mRNA and 20 were negative. (B) Dot-plot for ZAP70 mRNA expression. For the unmutated cases, 53 of 63 unmutated cases were positive for ZAP70 mRNA and 10 were negative. Eleven of 67 mutated cases were positive for ZAP70 mRNA and 56 were negative.
Twenty-two cases (17%) showed discordance between LDOC1 mRNA expression and somatic mutation status. We were unable to identify any features with respect to somatic mutation status or family that would have allowed us to predict the discordant cases. In contrast, for ZAP70 mRNA expression, most unmutated CLL samples were either positive or negative, whereas mutated cases showed a continuous gradient of expression (Figure 1B).
LDOC1 mRNA expression by RT-PCR in NBC, CLL samples, and cell lines
Little is known about the expression of LDOC1 mRNA in NBCs or in lymphoid neoplasms other than CLL. Thus, we assessed LDOC1 expression in NBCs, a variety of primary B-cell lymphoma samples, and carcinoma cell lines, using 2 primer pairs, AB and AC (Figure 2). Primer pair AB was designed to amplify the entire LDOC1 coding region and yield a product of 464 bp.7 The reverse primer, C, in primer pair AC was designed to bind within the 3′ UTR and yield a product of 649 bp. Using primer pair AB, we detected strong expression of the expected WT 464 bp product in 7 unmutated CLL cases (CLL 42, 46, 49, 51, 53, 54, 60), 2 NBC samples (NBC 4, 6), and the MCF-7 breast carcinoma cell line (Figure 3). We detected faint or no RT-PCR products in 7 mutated CLL cases (CLL 58, 62, 12, 37, 61, 67, 99), or in the GA-10 Burkitt lymphoma and Jurkat T-cell lymphoblastic lymphoma cell lines. However, primer pair AC yielded two distinct bands in all positive samples, the expected 649-bp product and an unexpected 165-bp product. Three of 4 additional NBC samples, as well as normal peripheral blood T cells and HeLa cervical carcinoma cells were also positive with both primer pairs (data not shown).
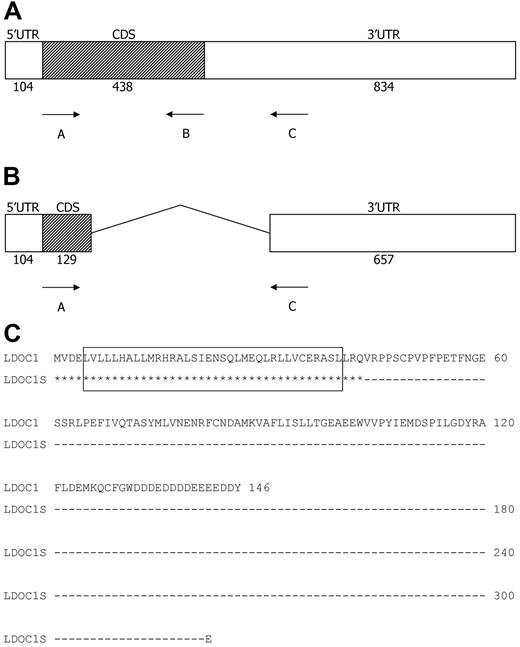
Structure of the LDOC1 wild-type and splice variant mRNAs, and translated proteins. (A-B) Structure of the LDOC1 and LDOC1S mRNAs. The LDOC1 gene is an intronless gene that spans 1376 bp. Open boxes represent 5′ and 3′ untranslated regions (UTR) and closed boxes represent the coding sequences (CDS); the number of nucleotides is indicated below. (C) Alignment of the amino acid sequences of wild-type LDOC1 protein and the putative splice variant protein. The wild-type mRNA encodes a protein composed of 146 amino acids. The splice variant mRNA, if translated, would yield a truncated protein of 44 amino acids that corresponds mainly to the leucine zipper region of the wild-type LDOC1. Identical residues are indicated by asterisks; the dashes indicate nucleotides that have been removed from the LDOC1S mRNA by alternative splicing. The leucine zipper domain (amino acids 5-40) in the wild-type protein, indicated by an open box, would be preserved in the splice variant.
Structure of the LDOC1 wild-type and splice variant mRNAs, and translated proteins. (A-B) Structure of the LDOC1 and LDOC1S mRNAs. The LDOC1 gene is an intronless gene that spans 1376 bp. Open boxes represent 5′ and 3′ untranslated regions (UTR) and closed boxes represent the coding sequences (CDS); the number of nucleotides is indicated below. (C) Alignment of the amino acid sequences of wild-type LDOC1 protein and the putative splice variant protein. The wild-type mRNA encodes a protein composed of 146 amino acids. The splice variant mRNA, if translated, would yield a truncated protein of 44 amino acids that corresponds mainly to the leucine zipper region of the wild-type LDOC1. Identical residues are indicated by asterisks; the dashes indicate nucleotides that have been removed from the LDOC1S mRNA by alternative splicing. The leucine zipper domain (amino acids 5-40) in the wild-type protein, indicated by an open box, would be preserved in the splice variant.
Expression of LDOC1 and LDOC1S mRNAs assessed by RT-PCR. (A) Expression in cell lines, normal peripheral blood B cells, and CLL cells. Wild-type LDOC1 (464 and 649 bp) and LDOC1S (165 bp) mRNAs were detected in the MCF7 breast carcinoma cell line, 2 normal peripheral blood B-cell (NBC) samples, and 2 unmutated CLL samples. Little or no LDOC1 or LDOC1S mRNA were detected in the GA10 Burkitt lymphoma cell line, the Jurkat T-cell lymphoblastic lymphoma cell line, or 2 mutated CLL samples. (B) Expression in CLL cells. Wild-type LDOC1 (464 and 649 bp) and LDOC1S (165 bp) mRNAs were detected in 5 additional unmutated CLL samples. Little or no LDOC1 or LDOC1S mRNA were detected in 5 additional mutated CLL samples. The amount of cDNA amplified for each sample was comparable, as shown by the β-actin signal.
Expression of LDOC1 and LDOC1S mRNAs assessed by RT-PCR. (A) Expression in cell lines, normal peripheral blood B cells, and CLL cells. Wild-type LDOC1 (464 and 649 bp) and LDOC1S (165 bp) mRNAs were detected in the MCF7 breast carcinoma cell line, 2 normal peripheral blood B-cell (NBC) samples, and 2 unmutated CLL samples. Little or no LDOC1 or LDOC1S mRNA were detected in the GA10 Burkitt lymphoma cell line, the Jurkat T-cell lymphoblastic lymphoma cell line, or 2 mutated CLL samples. (B) Expression in CLL cells. Wild-type LDOC1 (464 and 649 bp) and LDOC1S (165 bp) mRNAs were detected in 5 additional unmutated CLL samples. Little or no LDOC1 or LDOC1S mRNA were detected in 5 additional mutated CLL samples. The amount of cDNA amplified for each sample was comparable, as shown by the β-actin signal.
Sequence analysis of the LDOC1 mRNA RT-PCR products
We determined the sequences of the 464, 165, and 649-bp products in a subset of CLL cases that expressed LDOC1 mRNA (CLL 46, 49, 53). We also determined the sequences of the 464- and 649-bp products in a subset of NBC cases (NBC 1, 3, 5). The sequences of the 464- and 649-bp products were identical to the published LDOC1 reference sequence (GenBank RefSeq NM_012317). Sequence analysis of the 165-bp product revealed that it was a splice variant (Figure 2B). The LDOC1 gene is an intronless gene that encodes an mRNA of 1376 bp.7 The mRNAs for LDOC1 and its splice variant, LDOC1S (GenBank HQ343285), have identical start sites, with a sequence similar to that described by Kozak.22 The splice variant contains canonical splice donor (AG|GTACGT at nucleotide 232) and acceptor sequences (TGTCTTTGTTCCAG|G at nucleotide 704), as well as a branch sequence (TTCAT at nucleotide 685; Alex's Splice Site Finder Version 0.5). Thus, in the splice variant, approximately the first third of the amino acid–coding region is joined with the 3′ UTR at nucleotide 718. After one codon (GAA, glutamic acid), the coding sequence is terminated by a stop codon (TAG), followed by a 3′ UTR that is identical to the WT sequence. If translated, the 165-bp splice variant would produce a truncated protein of 44 amino acids that contains the leucine zipper region of the WT protein; it would lack the proline-rich region (amino acids 46-65) and the remainder of the coding region (Figure 2C).
Total LDOC1 mRNA expression by QRT-PCR assay in NBC and B-cell subsets, CLL and primary B-cell lymphoma samples, and cell lines
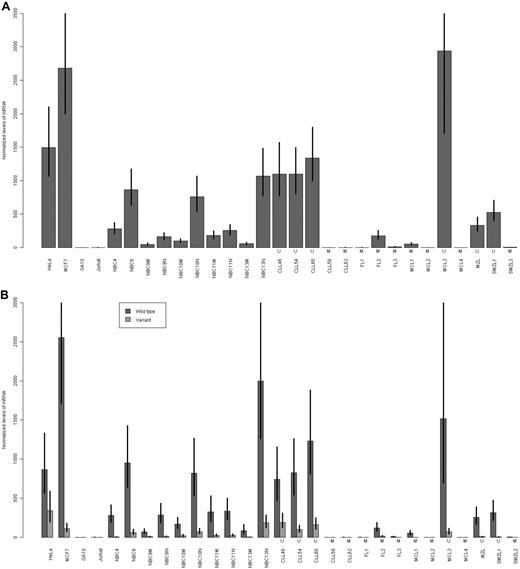
Using a more sensitive QRT-PCR assay, we assessed the expression of LDOC1 mRNA in unfractionated NBC samples and in NBC samples that had been enriched for naive (CD27−) or memory (CD27+) B cells. We also assessed its expression in 10 primary B-cell lymphoma samples, for which we determined the somatic mutation status. The primary lymphoma samples included 3 follicular (FL1, FL2, FL3, all mutated), 4 mantle cell (MCL1, MCL2, and MCL4, mutated; MCL3 unmutated), 1 extranodal marginal zone B-cell (MZL, unmutated), and 2 splenic marginal zone (SMZL1, unmutated; SMZL2, mutated) lymphoma samples. We used the commercially available QRT-PCR assay, which detects total LDOC1 mRNA; it does not distinguish between the isoforms. The results for unfractionated NBC and CLL samples, and lymphoma and carcinoma cell lines were consistent with the results of the RT-PCR assay (Figure 4A). For NBC samples, the fraction enriched for naive B cells expressed higher levels of LDOC1 than the fraction enriched for memory B cells. Primary B-cell lymphoma samples also expressed LDOC1. Although the sample size is insufficiently large for a statistical analysis of individual lymphoma subtypes, there was a trend for the unmutated lymphoma samples to express higher levels of LDOC1 than mutated samples.
Expression of LDOC1 mRNAs measured by QRT-PCR assay. (A) Expression of total LDOC1 mRNA. We used a commercially available assay that does not distinguish between the LDOC1 and LDOC1S mRNA isoforms. (B) Expression of wild-type LDOC1 and LDOC1S mRNA isoforms. We designed an assay that distinguishes between the isoforms. In both experiments, expression was measured in carcinoma (HeLa, MCF7) and lymphoma (GA10, Jurkat) cell lines, unfractionated normal peripheral blood B cells (NBC4, NBC6), normal peripheral blood B cells enriched for memory B cells (NBC9M, NBC10M, NBC11M, NBC13M), normal peripheral blood B cells enriched for naive B cells (NBC9N, NBC10N, NBC11N, NBC13N), and unmutated (U) or mutated (M) CLL and primary B-cell lymphoma samples, including follicular (FL), mantle cell (MCL), marginal zone (MZL), or splenic marginal zone (SMZL) lymphoma samples. Error bars represent the SE of the ΔΔCt values.
Expression of LDOC1 mRNAs measured by QRT-PCR assay. (A) Expression of total LDOC1 mRNA. We used a commercially available assay that does not distinguish between the LDOC1 and LDOC1S mRNA isoforms. (B) Expression of wild-type LDOC1 and LDOC1S mRNA isoforms. We designed an assay that distinguishes between the isoforms. In both experiments, expression was measured in carcinoma (HeLa, MCF7) and lymphoma (GA10, Jurkat) cell lines, unfractionated normal peripheral blood B cells (NBC4, NBC6), normal peripheral blood B cells enriched for memory B cells (NBC9M, NBC10M, NBC11M, NBC13M), normal peripheral blood B cells enriched for naive B cells (NBC9N, NBC10N, NBC11N, NBC13N), and unmutated (U) or mutated (M) CLL and primary B-cell lymphoma samples, including follicular (FL), mantle cell (MCL), marginal zone (MZL), or splenic marginal zone (SMZL) lymphoma samples. Error bars represent the SE of the ΔΔCt values.
LDOC1 and LDOC1S mRNA isoform expression by QRT-PCR assay
Because the commercially available QRT-PCR assay fails to distinguish between the LDOC1 and LDOC1S mRNA isoforms, we were unable to assess the contribution of each isoform to the total LDOC1 levels that we observed using the RT-PCR and commercially available QRT-PCR assays. Thus, we designed a QRT-PCR assay that distinguishes between the isoforms, and reassessed the previously tested samples described previously (Figure 4B). In general, for both benign and malignant cells, cells that expressed the wild-type LDOC1 also expressed the splice variant, LDOC1S, but the WT isoform was predominant.
LDOC1 mRNA expression more strongly predicts IGHV somatic mutation status than does ZAP70 protein expression
Because ZAP70 protein has been considered a surrogate biomarker for somatic mutation status,23,24 we evaluated the association between somatic mutation status and ZAP70 protein measured by immunohistochemistry or flow cytometry (IHC/Flow). We detected a positive association between ZAP70 positivity by IHC/Flow and somatic mutation status (Fisher exact test; P = 1.42 × 10−9). Of the 112 cases for which ZAP70 protein data were available, 24 (21%) showed discordance between somatic mutation status and ZAP70, consistent with the results of previous studies.3,15-17,23,24 Eleven of 59 mutated cases (19%) were ZAP70 positive; 13 of 53 unmutated cases (25%) were ZAP70 negative (supplemental Table 1). Thus, LDOC1 mRNA expression was more strongly associated with somatic mutation status than was ZAP70 protein.
LDOC1 mRNA expression is associated with cytogenetic markers of prognosis
We found that expression of LDOC1 mRNA correlated with cytogenetic markers of prognosis (Table 1, Fisher exact test; P = .0005834) supplemental Table 1). Cases that were negative for LDOC1 were more likely to harbor isolated deletions in chromosome 13q14.3, a marker of good prognosis, compared with samples that were positive for LDOC1. In contrast, cases that were positive for LDOC1 mRNA were more likely to harbor genomic abnormalities associated with poor prognosis, that is, del(6)(q21), del(11)(q22.3), del(17)(p13.1), +12, or del(13)(q14.3) with another cytogenetic marker of poor prognosis, than cases that were negative for LDOC1.
Total LDOC1 mRNA expression is a better predictor of overall survival than either IGHV somatic mutation status or ZAP70 protein expression
Because LDOC1 mRNA expression was associated with somatic mutation status and ZAP70 protein, both strong predictors of prognosis in CLL patients, we sought to determine whether LDOC1 expression could also serve as a biomarker of prognosis. Thus, we analyzed the relationship between LDOC1 and overall survival. We found that patients whose cells were negative for LDOC1 had a significantly longer median survival than patients whose cells were positive, regardless of whether overall survival was measured from the time of diagnosis (Figure 5; log-rank test, P = .009581) or from the time the sample was obtained (log-rank test, P = .02294; data not shown). The median survival for the LDOC1-negative patients was not reached, whereas the median survival for LDOC1-positive patients was 164 months. Furthermore, we applied the Akaike Information Criterion to a multivariate model that incorporated LDOC1, ZAP70 protein, and somatic mutation status. The optimal model retained only LDOC1 (AIC = 167.93), eliminating ZAP70 protein (AIC = 169.2) and mutation status (AIC = 171.02). (Lower values of AIC identify better models.19 ) Thus, in this sample set, LDOC1 mRNA expression was a better predictor of overall survival than either somatic mutation status or ZAP70 protein.
LDOC1 mRNA expression predicts overall survival. Overall survival was measured from the time of CLL diagnosis. The median survival for LDOC1 mRNA-positive patients was 164 months; the median survival for the LDOC1 mRNA-negative patients was not reached.
LDOC1 mRNA expression predicts overall survival. Overall survival was measured from the time of CLL diagnosis. The median survival for LDOC1 mRNA-positive patients was 164 months; the median survival for the LDOC1 mRNA-negative patients was not reached.
Discussion
The physiologic functions of LDOC1 in normal B-cell development are unknown, and we have found no published studies that have investigated LDOC1 expression during the course of human lymphocyte development. A recent study that evaluated the gene expression profiles of human cord blood subpopulations identified LDOC1 mRNA as up-regulated in the CD34+/CD133+ subpopulation, which contains hematopoietic stem cells and progenitor cells, compared with the more mature CD34−/CD133− subpopulation.25 Taken together, these findings suggest that during the course of normal B-cell development LDOC1 mRNA levels may vary with maturational stage and state of activation. An assessment of B cells obtained from different compartments and subjected to a variety of different stimuli is required to address this question.
The presence or absence of somatic mutations in the IGHV genes separates patients into 2 prognostic subsets.1,26 Gene expression profiling studies demonstrated that the majority of unmutated CLL cases express ZAP70 mRNA.27 Subsequently, others showed that expression of ZAP70 protein correlates with somatic mutation status and clinical outcome.3,28 Recent studies suggest that ZAP70 protein expression may be a better predictor of time-to-treatment than somatic mutation status.15,17 However, the relationship between somatic mutation status and prognosis is not absolute. Patients whose CLL cells use the VH3-21 gene and contain somatic mutations have a short overall survival, similar to patients with unmutated VH genes.20,21 In our study, LDOC1 mRNA expression, somatic mutation status, and ZAP70 protein expression all predicted time-to-treatment in univariate analyses. In multivariate analyses, somatic mutation status performed marginally better than ZAP70 protein, which performed marginally better than LDOC1 (data not shown). However, our data suggest that expression of LDOC1 mRNA may predict overall survival better than either somatic mutation status or ZAP70 protein in previously untreated CLL patients.
Chromosomal abnormalities, predominantly gains and deletions (del), are strong independent predictors of prognosis in CLL. The most common abnormality is del(13)(q14.3), followed by del(11)(q22.3), trisomy 12, del(6)(6q21-q23), and del(17)(p13.1). In clinical practice, these abnormalities are usually assessed using a panel of FISH probes.29-31 As the sole abnormality, del(13)(q14.3) is associated with a good prognosis. In contrast, del(6)(q21-q23), del(11)(q22.3), del(17)(p13.1), del(13)(q14.3) with other abnormalities, and trisomy 12 are associated with more rapid disease progression and inferior survival. The abnormalities del(17)(p13.1), the site of the TP53 gene, and del(11)(q22.3), the site of the ATM gene, are the most important independent cytogenetic markers of poor prognosis. Deletion of (17)(p13.1) is associated with resistance to therapy with purine analogs, such as fludarabine, and short survival.32-34 Somatic mutation status is associated with cytogenetic abnormalities detected by FISH analysis.35-37 For example, del(13)(q14.3) is found more often in mutated than unmutated cases. Our results indicate that LDOC1 mRNA expression is also associated with cytogenetic markers of prognosis. Cases that were LDOC1-negative were more likely to contain isolated del(13)(q14.3), while LDOC1-positive cases were more likely to demonstrate cytogenetic markers of poor prognosis. By SNP genotyping, no case showed loss of the LDOC1 gene. Three mutated cases showed a gain in the LDOC1 copy number, but were negative for LDOC1 mRNA (data not shown). Furthermore, we found no mutations in LDOC1 mRNA in the subset of CLL cases that we subjected to sequence analysis. Thus, the differences that we observed in levels of LDOC1 mRNA expression in mutated compared with unmutated CLL cases appear to result neither from copy number variation in the gene, nor from mutations in the coding regions.
If LDOC1 functions as a transcription factor, as hypothesized, then small alterations in its level of expression could profoundly affect other genes that it regulates, and could promote or inhibit tumor formation, depending on the context. We are the first to report LDOC1S, a new splice variant of LDOC1. In cancer cells, altered expression of mRNA splice variants may result from the generation of new mRNA variants or from changes in the tissue-specific ratio of normal mRNA isoforms. An example of the former is Ikaros, a zinc finger DNA binding protein that is critical for normal lymphocyte development. Alternative splicing of Ikaros pre-mRNA yields 8 different isoforms, each with a different DNA binding capacity and differential expression in normal and neoplastic lymphocytes.38 Alterations in the tissue-specific ratio of normal mRNA isoforms may also be associated with tumorigenesis. The interferon regulatory factor-1 (IRF-1) is a transcriptional activator that may function as a tumor suppressor gene. IRF-1 has 5 splice variants that lack various combinations of exons 7, 8, and/or 9. Although the variants are expressed in both normal and malignant cervical cells, they are found more abundantly in malignant cells. Most of the variants have been shown to inhibit the transcriptional activity of the wild type IRF-1.39
If the LDOC1S mRNA splice variant were translated into protein, it would contain the leucine zipper motif of the wild-type protein, but would lack the proline-rich region, which contains an SH3-binding consensus sequence, and the acidic region in the C-terminus. Mizutani and coworkers40 constructed LDOC1 deletion mutants, and studied their localization and protein interactions after transfection into canine kidney cells. While full-length LDOC1 localized predominantly to the nucleus, the N-terminal mutant protein (the leucine zipper region) localized to both the nucleus and cytoplasm. They also identified WAVE3, a predominantly cytoplasmic protein, as a binding partner of LDOC1. Coexpression of full-length LDOC1 and WAVE3 shifted LDOC1 from the nucleus to the cytoplasm, and was associated with decreased apoptosis. Their results suggest that WAVE3 may inhibit the proapoptotic activity of LDOC1, either by sequestering it in the cytoplasm or by shuttling it from the nucleus to cytoplasm. Similarly, it is conceivable that LDOC1S binds to LDOC1 to form nonfunctional dimers that inhibit the proapoptotic and antiproliferative activities of LDOC1, possibly by sequestering LDOC1 in the cytoplasm. Alternatively, LDOC1S could form nonfunctional dimers with LDOC1 within the nucleus that compete with functional LDOC1 dimers for DNA binding sites and inhibit its transcriptional regulatory activity. This scenario would be similar to the inhibitory actions displayed by members of the Id protein family, which contain helix-loop-helix dimerization domains but lack DNA-binding domains.41 Id proteins act in a dominant-negative fashion and control critical events in cell differentiation, proliferation, and tumorigenesis. It is also possible that LDOC1S may form nonfunctional heterodimers with other proapoptotic and/or antiproliferative proteins. Either mechanism might contribute to the aggressive behavior of some tumor types that aberrantly express LDOC1 isoforms.
The relatively high expression of LDOC1 mRNA isoforms in unmutated CLL cases compared with mutated cases, and their expression in a variety of tumor cell lines suggest that LDOC1 may contribute to aggressive clinical behavior. Because we are a tertiary care center, many of our patients received the diagnosis of CLL months to years before seeking care at our hospital. Thus, we do not know the LDOC1 mRNA expression status of their CLL cells at the time of initial diagnosis. We also do not know if LDOC1 protein expression predicts overall survival in previously treated CLL patients. We have been unable to identify a sensitive and specific commercially available antibody for use in either a Western blot or flow cytometry–based assay. However, QRT-PCR assays for a variety of different mRNA transcripts, such as BCR/ABL1 fusion transcripts, are now performed routinely in clinical molecular diagnostics laboratories.42 Thus, the lack of a robust antibody does not preclude the use of LDOC1 mRNA as a clinically relevant biomarker of prognosis. Whether LDOC1 is stable in cases that have undergone clonal evolution over the disease course or after therapeutic interventions, and if it is truly a better predictor of overall survival than either IGHV somatic mutation status or ZAP70 protein expression, can only be answered by a larger longitudinal study.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Dr Jolanta Bondaruk for expert assistance with the SNP genotyping. We also thank Dr Gilbert Cote for helpful discussions and advice.
This work was supported in part by grants from the National Center For Research Resources (H.D., TL1RR024147), the Business and Professional Women's Club of Texas (L.V.A.), the CLL Global Research Foundation (L.V.A.), and the National Cancer Institute (L.V.A., R01CA123252-3).
National Institutes of Health
Authorship
Contribution: H.D. designed and performed experiments, collected and analyzed data, and wrote the manuscript; C.D.S. designed and performed experiments, and ensured the integrity of the clinical data; K.R.C. performed the statistical analyses; L.L.B. performed experiments, collected data, and maintained the laboratory tissue bank and database; A.F., S.O., W.G.W., E.JF., and M.J.K. provided patient samples, clinical data, and clinical expertise; J.P. designed experiments and analyzed data; T.M. performed the SNP genotyping assays; B.A.C. provided laboratory resources and personnel to perform experiments; J.L.J. analyzed data; L.J.M. analyzed data and wrote the manuscript; L.V.A. designed the research, supervised the study, and wrote the manuscript; and all authors critically reviewed the final version of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Lynne V. Abruzzo, MD, PhD, Department of Hematopathology, The University of Texas M. D. Anderson Cancer Center, Box 350, 1515 Holcombe Blvd, Houston, TX 77030; e-mail: labruzzo@mdanderson.org.