Abstract
Transcription factor CCAAT enhancer binding protein α (C/EBPα) is essential for granulopoiesis and its function is deregulated in leukemia. Inhibition of E2F1, the master regulator of cell-cycle progression, by C/EBPα is pivotal for granulopoiesis. Recent studies show microRNA-223 (miR-223), a transcriptional target of C/EBPα, as a critical player during granulopoiesis. In this report, we demonstrate that during granulopoiesis microRNA-223 targets E2F1. E2F1 protein was up-regulated in miR-223 null mice. We show that miR-223 blocks cell-cycle progression in myeloid cells. miR-223 is down-regulated in different subtypes of acute myeloid leukemia (AML). We further show that E2F1 binds to the miR-223 promoter in AML blast cells and inhibits miR-223 transcription, suggesting that E2F1 is a transcriptional repressor of the miR-223 gene in AML. Our study supports a molecular network involving miR-223, C/EBPα, and E2F1 as major components of the granulocyte differentiation program, which is deregulated in AML.
Introduction
CCAAT enhancer binding protein α (C/EBPα) functions as a key mediator of granulopoiesis.1 Conditional silencing of C/EBPα in mice shows a selective block in the differentiation of granulocytes.2 In acute myeloid leukemia (AML), C/EBPα is deregulated by various mechanisms including its own mutations.3 Inhibition of E2F activity by C/EBPα is a key step for the antimitotic activity of C/EBPα during granulopoiesis.1 Targeted disruption of the domains of C/EBPα needed for E2F interaction results in block of granulopoietic differentiation in mice.4 In addition, mice carrying a germline mutation that disrupts C/EBPα-mediated E2F repression develop AML.5 Interestingly, E2F1 is able to inhibit granulopoiesis and induce myeloid cell-cycle progression.6 However, to our knowledge there has been no definitive mechanism demonstrated for C/EBPα-mediated E2F1 repression in granulopoiesis.
MicroRNAs (miRNAs) are a novel group of gene regulators and play important roles in biologic processes such as cell proliferation and differentiation, development, and apoptosis, all of which are frequently affected in cancer. Growing number of studies demonstrate that the deregulation of microRNAs is associated with the development of many cancers including leukemia.7,8 In granulopoiesis, microRNA-223 (miR-223) is one of the most critical microRNAs.9,10 miR-223 has been shown to be regulated by myeloid transcription factors such as C/EBPs and PU.1. The crucial role of miR-223 in granulopoiesis was shown by a recent finding that mice lacking miR-223 display defects in granulopoiesis.11 Interestingly, miR-223 is inactivated by the oncoprotein AML1/ETO in AML.12
In this study, we explored the function of microRNA-223 in granulopoiesis and in AML in connection with C/EBPα-mediated inhibition of E2F1. Here we show that C/EBPα regulates miR-223, which in turn targets E2F1 via translational inhibition. We also report that miR-223 is down-regulated in human AML. Moreover, E2F1 binds to the miR-223 promoter and inhibits miR-223 transcription, thus generating a negative feedback loop. Our study demonstrates that granulopoiesis is regulated by C/EBPα–miR-223–E2F1 network, whereby miR-223 functions as a key regulator of myeloid cell proliferation interlinked with E2F1 in a mutual negative feedback loop.
Methods
Patient samples
AML patient samples were obtained from the Leukemia Diagnostics laboratories at University Hospital of Munich (Klinikum Grosshadern) and University Hospital of Münster. All samples were karyotyped and molecular genetics analysis was performed for mutations. The study was approved by the institutional review boards from University Hospital of Munich and University Hospital of Münster. Human cord blood samples were collected after full-term delivery with informed consent of the mothers from University Hospital of Halle. Hematopoietic CD34+ cells were isolated from cord blood samples using the CD34+ selection kit (Miltenyi Biotec).
Cells, reagents, and transfections
K562-C/EBPα-p42-ER, K562-C/EBPα-p30-ER, K562-C/EBPα-BRM2-ER, and K562-ER cells were maintained in RPMI 1640 without phenol red supplemented with 10% charcoal-treated fetal bovine serum, 1% penicillin-streptomycin, and 2 μg/mL puromycin.13 K562 cells are multipotential cells that can undergo granulocytic differentiation during C/EBPα induction13 or erythrocytic differentiation upon dimethyl sulfoxide treatment.14 U937 cells were cultured in RPMI 1640 supplemented with 10% heat inactivated fetal bovine serum and 1% penicillin-streptomycin. U937 cells are multipotential cells that can differentiate to granulocytic lineage during retinoic acid or C/EBPα induction and monocytic lineage during tetradecanoyl phorbol acetate (TPA).14
Human embryonic kidney 293T cells were maintained in Dulbecco modified Eagle medium supplemented with 10% fetal bovine serum and 1% penicillin-streptomycin.
For differentiation of K562-C/EBPα-p42ER cells, cells (106) were induced with 5μM β-estradiol (Sigma) dissolved in ethanol. NB4 cells (106) were induced for differentiation by the addition of 1μM retinoic acid (Sigma) dissolved in dimethyl sulfoxide.
293T cells (2 × 104 cells) were transfected with the Lipofectamine Plus reagent (Invitrogen) according to the manufacturer's instruction. Transfection of U937 was performed with the Nucleofector kit (AMAXA) and Effectene reagent (QIAGEN) as described by the manufacturers. DNA plasmid (2 μg) was used for each transfection and the transfection efficiency was analyzed using a plasmid with enhanced green fluorescent protein marker. U937 cells were transfected with nucleofection program V-01. Transfection efficiency of approximately 55% to 70% was observed in this cell line.
miRNA oligonucleotides (mimic) were obtained from Dharmacon. miRNA mimic was transfected in U937 cells using Lipofectamine 2000 (Invitrogen) and in 293T cells using Lipofectamine Plus (Invitrogen). The transfection efficiency was checked by fluorescence-activated cell sorting (FACS) analysis of cells transfected with siGloRed reagent (Dharmacon) and found to be approximately 64% to 67%.
Locked nucleic acid (LNA) oligonucleotides were obtained from Exiqon. LNA (160 nM) was transfected in U937 cell line using AMAXA transfection method using the program V-01. The transfection efficiency was checked by FACS analysis of cells transfected with fluorescein isothiocyanate (FITC)–labeled LNA and found to be approximately 75% to 89%.
RNA isolation and miRNA detection
Total RNA from cells was extracted using the Trizol method. For isolation of RNA from patient samples, the RNeasy mini kit (QIAGEN) was used. The miRNA quantification was done with the TaqMan miRNA Detection Kit (Applied Biosystems) using 100 ng of RNA in a Rotor-Gene RG-3000 cycler (Corbett Research Australia) by the comparative threshold cycle method using U6 expression for normalization. Corresponding reverse-transcription (RT) and polymerase chain reaction (PCR) primers for miRNA-223 and U6 were obtained from Applied Biosystems. All reactions were performed in triplicate.
Real-time RT-PCR
Total RNA was isolated from cells using Trizol reagent (Invitrogen). RNA (750 ng) was used to synthesize cDNA by reverse transcription. Equal amounts of cDNA were taken for a subsequent quantitative real-time PCR (Q-RT-PCR) using the SYBR Green PCR kit (QIAGEN) in a Rotor-Gene RG-3000 cycler (Corbett Research Australia). The relative quantity of E2F1 mRNA was determined by the comparative threshold cycle method using glyceraldehyde-3-phosphate dehydrogenase (GAPDH) expression for normalization. All reactions were performed in triplicate. The following primers were used: E2F1 forward primer, 5′-GGG GAG AAG TCA CGC TAT GA-3′; E2F1 reverse primer, 5′-CTC AGG GCA CAG GAA AAC AT-3′; GAPDH forward primer, 5′-ACC ACA GTC CAT GCC ATC AC-3′; GAPDH reverse primer, 5′-TCC ACC ACC CTG TTG CTG TA-3′.
Immunoblot analysis
Total cell extracts (30-50 μg) were resolved on a 10% sodium dodecyl sulfate polyacrylamide gel, electroblotted to a nitrocellulose membrane, and reacted with the mouse or rabbit polyclonal antibody anti-E2F1 (Santa Cruz Biotechnology). Monoclonal mouse anti–β-actin and polyclonal rabbit anti–β-tubulin antibodies were used to normalize the E2F1 protein. The immunodetection was performed with an enhanced chemiluminescence reagent (Amersham Biosciences). The band intensities were quantified using ImageJ software (National Institutes of Health).
DNA constructs, cloning, and mutagenesis
The luciferase vector containing a 647–base pair fragment of the 3′ untranslated region (UTR) of E2F1 in a pGL3 vector was published before.15 Site-directed mutagenesis of the pGL3 E2F1 3′UTR wild-type vector was performed using the QuikChange Site-Directed Mutagenesis Kit (Stratagene) according to the manufacturer's instructions using the following primers: E2F1 3′UTR mut forward primer, 5 ′-TGA GGG AGG GAG ACA GCC CGC CTG ACA GCC ATG-3′; reverse primer, 5′-CAT GGC TGT CAG GCG GGC TGT CTC CCT CCC TC-3′.
A DNA fragment −1097 bases and +108 relative to the transcription start site of the human ppri-miR-223 was PCR amplified from genomic DNA using the forward primer 5′-TAG GGT ACC TCA TTA GCC TGA AGG-3′ and the reverse primer 5′-GTC TCG AGG CAA ATG GAT ACC ATA C-3′ and cloned into KpnI and XhoI digested pGL3 basic vector (ppri-miR-223 promoter-1). ppri-miR-223 promoter-2 was published before.10
A DNA fragment +1910 bases and +3409 relative to the transcription start site of the human ppri-miR-223 was PCR amplified from genomic DNA using the forward primer 5′-TAG GGT ACC GCT GAA TTG GGT AGG-3′ and the reverse primer 5′-GTC TCG AGC CAA GAG CTT CTG TGG-3′ and cloned into KpnI and XhoI digested pGL3 basic vector (pre-miR-223 promoter).
Luciferase assays
To test whether miR-223 directly targets E2F1, 293T cells were transiently cotransfected with 0.1 μg of each of the reporter constructs (wild-type and mutant E2F1 3′UTR, and pGL3 basic vector), 0.02 μg of Renilla construct, and 10μM synthetic miRNA-223 oligonucleotides (mimic-223) or control oligonucleotides (mimic-control) using the Lipofectamine reagent (Invitrogen). Reporter assays in U937 cells were carried out by transfecting 0.1 μg of wild-type E2F1 3′UTR reporter construct and LNA oligonucleotides (160nM) using the AMAXA transfection method. Firefly luciferase activities from the reporter constructs and Renilla luciferase activity from the internal control plasmid were determined 24 hours after transfection using the Dual-Luciferase Reporter Assay System (Promega). Values were normalized using Renilla luciferase.
To find the regulation of miR-223 promoter by E2F1 and C/EBPα, U937 cells were transiently transfected with 0.7 μg of the miR-223 promoter constructs (ppri-miR-223 promoter-1, 2 and pre-miR-223-promoter), 0.1 μg of Renilla construct, 0.2 μg of C/EBPα, and 0.2 μg of E2F1 vectors by Effectene transfection reagent (QIAGEN) as described by the manufacturer. Firefly luciferase activities from the promoter constructs and Renilla luciferase activity from the internal control plasmid were determined 24 hours after transfection using the Dual-Luciferase Reporter Assay System (Promega). Values were normalized using Renilla luciferase.
Lentiviral transfections
Lentiviral transfections were done essentially as reported before.9
Cell-growth assays
For proliferation analysis, U937 cells were transfected with control or miR-223 lentiviral vectors. Viable cells were counted on different days using trypan blue exclusion method.
Cell-cycle analysis
For cell-cycle analysis, U937 cells were transfected with control or miR-223 lentiviral vectors. Two days later, cells were fixed in cold ethanol, washed, resuspended in phosphate-buffered saline containing 50 μg/mL propidium iodide (PI) and 50 μg/mL RNAase A, and analyzed by flow cytometry on a FACScan (Becton Dickinson).17
BrdU incorporation assay
5-Bromodeoxyuridine (BrdU) incorporation assay was done with the FITC BrdU Flow kit (BD Pharmingen). U937 cells were transfected with control or miR-223 lentiviral vectors. Two days later, cells were labeled with BrdU for 45 minutes. Cells were then fixed, stained with FITC-labeled anti-BrdU antibody and PI, and analyzed by flow cytometry to determine the cell-cycle distribution on a FACScan (Becton Dickinson).
Chromatin immunoprecipitation
The cross-linking of proteins to DNA was accomplished by the addition of 1% formaldehyde for 10 minutes to cultured cells at 37°C. After sonication, the chromatin was immunoprecipitated with 5 μg of anti-E2F1, C/EBPα, and anti–immunoglobulin G (Santa Cruz Biotechnology) antibodies at 4°C overnight. The following primers were used: oligo 1 (forward), 5′-TTG TAC AGC TTC ACA GGG CTC-3′; oligo 1 (reverse), 5′-CCA TGG ACT CCA AAT CAC ATG-3′; oligo 2 (forward), 5′-GCC CTC TTT GTT GAT GTG TC-3′; oligo 2 (reverse), 5′-GGC AGC TAT TAA AGT GCC CT-3′; oligo 3 (forward), 5′-CAT CAC CTT CTC ATC TTC TGC-3′; oligo 3 (reverse), 5′-CTT GAA CAA TCA CAC ACA GCC-3′.
Statistical analysis
We used the Student t test to determine the statistical significance of experimental results. A P value of .05 or less was considered significant. The results were represented as the mean plus or minus SD from 3 independent experiments.
Results
C/EBPα-p42 up-regulates miR-223, and C/EBPα mutants fail to up-regulate miR-223
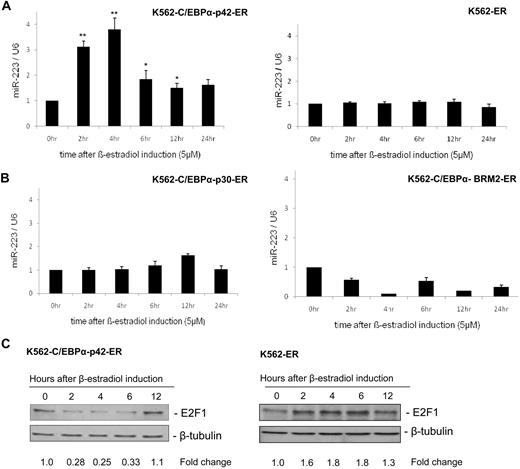
A previous study has shown that miR-223 is up-regulated upon retinoic acid induction.9 Retinoic acid up-regulates myeloid transcription factors including C/EBPα, C/EBPβ, PU.1, and Oct-1, which have been shown to have distinct functions in granulopoiesis.14,18 To understand the regulation of miR-223 by C/EBPα in granulopoiesis, we used a stable cell line in which C/EBPα protein was introduced into K562 cells upon β-estradiol induction.13 We selected this cell line because these cells do not have endogenous C/EBPα.13 Our finding shows that C/EBPα (42-kDa isoform) regulates miR-223 during granulopoiesis (Figure 1A left). A control cell line, which does not express C/EBPα, did not show any up-regulation of miR-223 (Figure 1A right). Next, we analyzed the ability of C/EBPα mutant proteins—C/EBPα with N-terminal mutation (C/EBPα-p30) and basic domain mutant (C/EBPα-BRM2)—in regulating miR-223. These mutants fail to induce granulocytic differentiation13,19 and have been observed in AML (supplemental Figure 1, available on the Blood website; see the Supplemental Materials link at the top of the online article).20 Our data show that these mutants failed to up-regulate miR-223 (Figure 1B). This suggests the induction of miR-223 as a specific function of C/EBPα (42-kDa isoform).
C/EBPα-p42 up-regulates miR-223 and leads to down-regulation of E2F1 protein expression. (A-B) K562-C/EBPα-p42-ER cells and K562-ER cells (A) and K562-C/EBPα-p30-ER cells and K562-C/EBPα-BRM2-ER cells (B) were induced with β-estradiol (5μM) for respective time points. Total RNA was analyzed by quantitative real-time RT-PCR with oligos for miR-223. Data are represented as mean ± SD from 3 independent experiments. *P < .05; **P < .01. (C) K562-C/EBPα-p42-ER cells and K562-ER cells were induced with β-estradiol (5μM) for respective time points. Total protein was analyzed by Western blot analysis with anti-E2F1 antibody. Values below the gel image indicate the down-regulation (fold) of E2F1 protein level normalized to β-tubulin.
C/EBPα-p42 up-regulates miR-223 and leads to down-regulation of E2F1 protein expression. (A-B) K562-C/EBPα-p42-ER cells and K562-ER cells (A) and K562-C/EBPα-p30-ER cells and K562-C/EBPα-BRM2-ER cells (B) were induced with β-estradiol (5μM) for respective time points. Total RNA was analyzed by quantitative real-time RT-PCR with oligos for miR-223. Data are represented as mean ± SD from 3 independent experiments. *P < .05; **P < .01. (C) K562-C/EBPα-p42-ER cells and K562-ER cells were induced with β-estradiol (5μM) for respective time points. Total protein was analyzed by Western blot analysis with anti-E2F1 antibody. Values below the gel image indicate the down-regulation (fold) of E2F1 protein level normalized to β-tubulin.
C/EBPα down-regulates E2F1 protein during granulopoiesis
Because it is known that C/EBPα inhibits E2F1 during granulopoiesis, we analyzed the E2F1 mRNA and protein levels during C/EBPα induction. C/EBPα down-regulated E2F1 protein during granulopoiesis (Figure 1C left). A control cell line did not show any down-regulation of E2F1 protein levels (Figure 1C right). Meanwhile the mRNA level of E2F1 remained unaffected (data not shown). We also analyzed the ability of C/EBPα to down-regulate E2F1 in NB4 cells upon retinoic acid induction. Our data showed that retinoic acid induction in NB4 cells led to up-regulation of miR-223 and down-regulation of E2F1 protein (supplemental Figure 2).
miR-223 targets E2F1 via translational repression
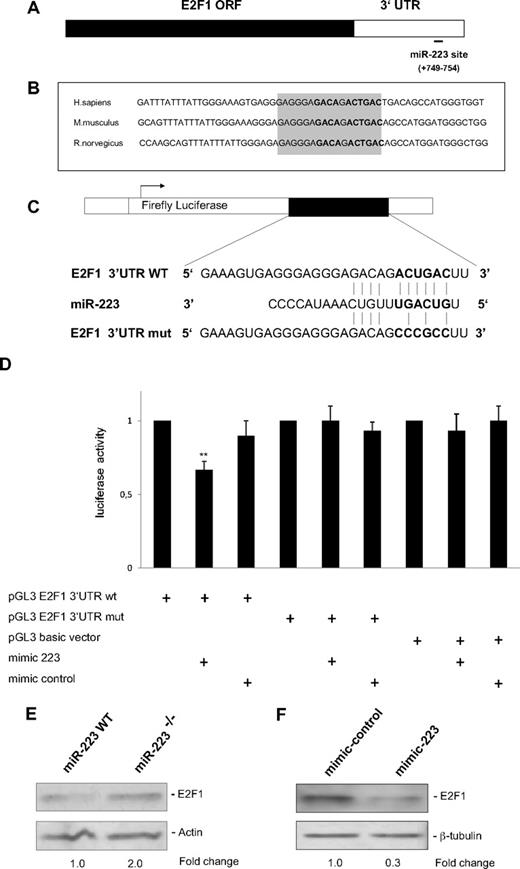
Because the majority of microRNAs function by translational repression, we hypothesized that microRNAs involved in granulopoiesis could have a role in the repression of E2F1 protein by C/EBPα (Figure 1C). By computational analysis using miRNA target prediction programs such as Target Scan (http://www.targetscan.org) and miRanda (http://www.microrna.org), we found that E2F1 is a putative target of miR-223 (Figure 2A). Conserved miRNA target sites exert a stronger effect than nonconserved target sites.21 We found that the E2F1 3′UTR predicted for miR-223 binding is conserved in humans, mice, and rats (Figure 2B).
E2F1 is a direct target of miR-223. (A) Schematic representation of the E2F1 transcript. Predicted miR-223 binding site is depicted. The numbers (+749 to +754) represent the nucleotides (relative to the E2F1 termination codon) that are predicted to base pair with the miR-223 seed sequence. (B) Sequences of the predicted miRNA-223 binding sites in human, mouse, and rat genomes. Highly conserved nucleotides are shown in gray. (C) Schematic representation of luciferase constructs used for reporter assays. (D) Luciferase assays in 293T cells transfected with vectors shown in panel C and miR-223 and control oligonucleotides (mimic). Bars represent luciferase activity for the corresponding vectors. Data are represented as mean ± SD from 3 independent experiments. **P < .01. (E-F) Western blot analysis of E2F1 on whole-cell lysates from sorted Ly6Gpos bone marrow neutrophils from wild-type (WT) and miR-223 knockout (KO) animals11 (E), on whole-cell lysates from U937 cells treated with 20 nm of control and miR-223 oligonucleotides (mimic; F).
E2F1 is a direct target of miR-223. (A) Schematic representation of the E2F1 transcript. Predicted miR-223 binding site is depicted. The numbers (+749 to +754) represent the nucleotides (relative to the E2F1 termination codon) that are predicted to base pair with the miR-223 seed sequence. (B) Sequences of the predicted miRNA-223 binding sites in human, mouse, and rat genomes. Highly conserved nucleotides are shown in gray. (C) Schematic representation of luciferase constructs used for reporter assays. (D) Luciferase assays in 293T cells transfected with vectors shown in panel C and miR-223 and control oligonucleotides (mimic). Bars represent luciferase activity for the corresponding vectors. Data are represented as mean ± SD from 3 independent experiments. **P < .01. (E-F) Western blot analysis of E2F1 on whole-cell lysates from sorted Ly6Gpos bone marrow neutrophils from wild-type (WT) and miR-223 knockout (KO) animals11 (E), on whole-cell lysates from U937 cells treated with 20 nm of control and miR-223 oligonucleotides (mimic; F).
To verify E2F1 as a target of miR-223, we performed luciferase reporter assays with 3′UTR of the E2F1 gene with putative miR-223 binding site (E2F1 3′UTR WT and E2F1 3′UTR mut, Figure 2C). Our data indicated a miR-223–specific regulation of reporter gene expression (Figure 2D). The ability of miR-223 to target E2F1 3′UTR was underlined by the finding that silencing of miR-223 by locked nucleic acid (LNA) oligonucleotides in U937 cells results in up-regulation of luciferase activity of E2F1 3′UTR by 60% (supplemental Figure 3). Because C/EBPα is able to up-regulate miR-223 (Figure 1A) and miR-223 target E2F1 (Figure 2D), we analyzed the effect of C/EBPα on translational repression of E2F1. C/EBPα down-regulated the reporter gene expression of E2F1 3′UTR (supplemental Figure 4). The inhibition of E2F1 by C/EBPα was relieved when miR-223 was blocked with LNA specific for miR-223 (supplemental Figure 4). This suggests that inhibition of E2F1 by miR-223 could be a possible mechanism for the E2F1 inhibition by C/EBPα. However, we cannot rule out the possibility of other microRNAs regulated by C/EBPα being involved in E2F1 inhibition.
To validate our finding that E2F1 is a target of miR-223, we analyzed the E2F1 protein levels in bone marrow neutrophils from miR-223 null mice.11 Western blot analysis showed a 2.0-fold increase in E2F1 protein levels in miR-223 null bone marrow neutrophils (Figure 2E), indicating that endogenous levels of miR-223 control E2F1 expression. Although the change in E2F1 protein level is subtle upon inhibition of miR-223, this could still be significant in regulating E2F1 protein levels in the myeloid cell.21,22 We also tested the ability of miR-223 in down-regulating E2F1 protein levels. Overexpression of miR-223 resulted in a 70% decrease in E2F1 protein levels (Figure 2F). Altogether, these results suggest that E2F1 is a potential target of miR-223.
miR-223 is down-regulated in human acute myeloid leukemia samples
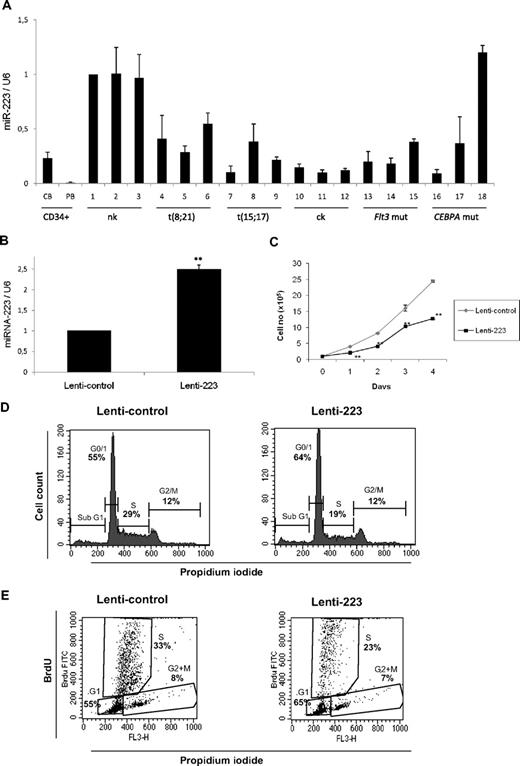
To address the role of miR-223 in AML, we examined its expression in CD34+ cells and diagnostic samples from AML patients. The genetic features of the different AML samples are shown in supplemental Table 1. We observed lowest expression level of miR-223 in CD34+ cells (Figure 3A), as reported before.12 We used AML samples with normal karyotype (nk) as control group (samples 1-3), because this subtype has not been shown to have C/EBPα deregulation. We found miR-223 was down-regulated in most of the samples with different subtypes of AML (Figure 3A samples 4-17). Our finding that miR-223 is down-regulated in different subtypes of AML, in which C/EBPα function is also deregulated, suggests the critical regulation of miR-223 by C/EBPα in normal granulopoiesis. However, one sample with CEBPA mutation showed no significant change in miR-223 expression, suggesting additional mechanisms could be relevant in this subtype.
miR-223 functions as a tumor suppressor in acute myeloid leukemia. (A) Quantitative real-time RT-PCR for miR-223 was carried out using bone marrow cells derived from AML patients. Values were normalized with U6. Cord blood (CB) and peripheral blood (PB) and AML with normal karyotype (nk), with complex karyotype (ck), with FLT3 mutation (FLT3 mut), and with CEBPA mutation (CEBPA mut). Data are represented as mean from 3 experiments. (B) U937 cells were transfected with lentiviral control or lenti-223 vectors. Two days later, total RNA was isolated and analyzed for miR-223 by quantitative real-time RT-PCR. Data are represented as mean ± SD from 3 independent experiments. **P < .01. (C) Growth curve of U937 cells transfected with lentiviral control or lenti-223 vectors. Data are represented as mean ± SD from 3 independent experiments. **P < .01. (D) Analysis of cell-cycle distribution during miR-223 lentiviral overexpression by flow cytometry. Representative FACS data from U937 cells transfected for 2 days with control and miR-223 lentiviral vector. (E) Flow cytometry of BrdU/PI-stained U937 cells transfected with lentiviral control or lenti-223 vectors from a representative experiment. Two days after lentiviral transfection, cells were labeled with BrdU for 45 minutes. Cells were then fixed, stained with FITC-labeled anti-BrdU antibody and PI, and analyzed by flow cytometry to determine the cell-cycle distribution. Bivariate analysis of the incorporation of BrdU (BrdU FITC, vertical axis) and the DNA content (FL3, PI staining, horizontal axis) was performed.
miR-223 functions as a tumor suppressor in acute myeloid leukemia. (A) Quantitative real-time RT-PCR for miR-223 was carried out using bone marrow cells derived from AML patients. Values were normalized with U6. Cord blood (CB) and peripheral blood (PB) and AML with normal karyotype (nk), with complex karyotype (ck), with FLT3 mutation (FLT3 mut), and with CEBPA mutation (CEBPA mut). Data are represented as mean from 3 experiments. (B) U937 cells were transfected with lentiviral control or lenti-223 vectors. Two days later, total RNA was isolated and analyzed for miR-223 by quantitative real-time RT-PCR. Data are represented as mean ± SD from 3 independent experiments. **P < .01. (C) Growth curve of U937 cells transfected with lentiviral control or lenti-223 vectors. Data are represented as mean ± SD from 3 independent experiments. **P < .01. (D) Analysis of cell-cycle distribution during miR-223 lentiviral overexpression by flow cytometry. Representative FACS data from U937 cells transfected for 2 days with control and miR-223 lentiviral vector. (E) Flow cytometry of BrdU/PI-stained U937 cells transfected with lentiviral control or lenti-223 vectors from a representative experiment. Two days after lentiviral transfection, cells were labeled with BrdU for 45 minutes. Cells were then fixed, stained with FITC-labeled anti-BrdU antibody and PI, and analyzed by flow cytometry to determine the cell-cycle distribution. Bivariate analysis of the incorporation of BrdU (BrdU FITC, vertical axis) and the DNA content (FL3, PI staining, horizontal axis) was performed.
miR-223 blocks myeloid cell proliferation
Given the importance of E2F1 in the regulation of cell-cycle progression, we decided to address the role of miR-223 in myeloid cell proliferation. We elected U937 cells for further experiments because they express C/EBPα endogenously and have been reported as a good model for granulocytic differentiation in the context of C/EBPα.14 U937 cells were infected with lentiviral vector expressing miR-223 (lenti-223) as reported before.9 We found a 2-fold expression of miR-223 upon lenti-223 infection in U937 cells compared with the empty vector–infected cells (Figure 3B). Overexpression of miR-223 by lentiviral vector significantly down-regulated the proliferation rate of myeloid U937 cells (Figure 3C). To better understand the regulation of myeloid cell proliferation by miR-223, we examined cell-cycle profiles 2 days after overexpression of miR-223 lentiviral vector. U937 cells transfected with miR-223 lentiviral vector displayed a significant increase in the number of cells in G0/G1 and significant decrease in the number of cells in S phase (Figure 3D). This indicates that lentiviral overexpression of miR-223 is able to block cell-cycle progression by inhibiting G1 to S phase transition of cell cycle. To gain further insight about myeloid cell-cycle regulation by miR-223, we performed 5-bromodeoxyuridine (BrdU) assay in cells transfected with miR-223 lentiviral vector. Our data show that overexpression of miR-223 leads to a decrease in the number of myeloid cells in the S phase (Figure 3E). The finding from miR-223 null animals together with our finding that miR-223 targets E2F1 (Figure 2) and inhibits myeloid cell-cycle progression (Figure 3B-E) suggest that down-regulation of E2F1 by miR-223 could be a critical step in granulocytic differentiation.
E2F1 functions as a repressor of the miR-223 gene
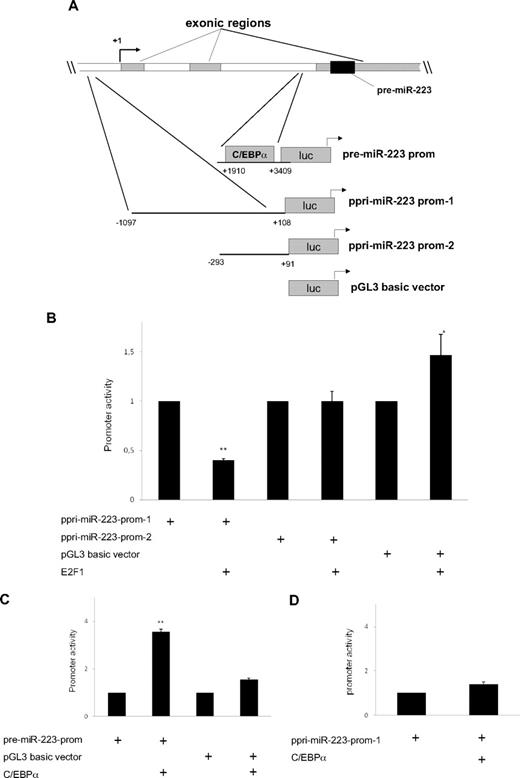
Recent studies show the ability of E2F1 to regulate microRNAs and how this could be significant in the pathogenesis of certain cancers.23 E2F1 has been shown to interfere with myeloid differentiation and promote proliferation of myeloid progenitors.6 Based on these findings, we sought to evaluate whether miR-223 is a direct target of E2F1. To assess the regulation of miR-223 by E2F1, we did promoter assays with different promoter constructs for miR-223 (Figure 4A). Our data suggest that E2F1 is able to inhibit miR-223 transcription (Figure 4B) through an element located in between 1097 and 293 bases upstream of the transcription start site. We could not observe any repression of miR-223 by E2F1 in the miR-223 promoter region, which has been reported to be necessary for C/EBPα transactivation9 (supplemental Figure 5).
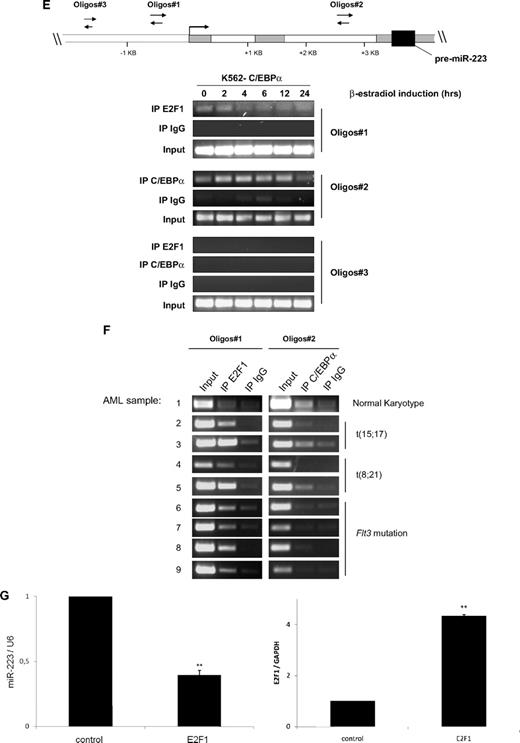
E2F1 functions as a repressor of the miR-223 gene. (A) Schematic representation of the different miR-223 promoter constructs used for reporter assays. (B-D) Luciferase reporter assays were performed in U937 cells using indicated reporters and E2F1 (B) and C/EBPα (C-D). Bars represent promoter activity for the corresponding vectors. Data are represented as mean ± SD from 3 independent experiments. *P < .05; **P < .01. (E-F) Chromatin derived from K562-C/EBPα-p42-ER cells (E) and AML blast cells (F) was immunoprecipitated with anti-E2F1, anti-C/EBPα, and immunoglobulin G antibodies. Recovered DNA was PCR amplified with primers specific for the E2F1 binding amplicon (oligo 1) and C/EBPα binding amplicon (oligo 2). (G) U937 cells were transfected with control or E2F1 vector. Total RNA was analyzed by quantitative RT-PCR with oligos for miR-223 (top) and with oligos for E2F1 (bottom). Data are represented as mean ± SD from 3 independent experiments. *P < .05; **P < .01.
E2F1 functions as a repressor of the miR-223 gene. (A) Schematic representation of the different miR-223 promoter constructs used for reporter assays. (B-D) Luciferase reporter assays were performed in U937 cells using indicated reporters and E2F1 (B) and C/EBPα (C-D). Bars represent promoter activity for the corresponding vectors. Data are represented as mean ± SD from 3 independent experiments. *P < .05; **P < .01. (E-F) Chromatin derived from K562-C/EBPα-p42-ER cells (E) and AML blast cells (F) was immunoprecipitated with anti-E2F1, anti-C/EBPα, and immunoglobulin G antibodies. Recovered DNA was PCR amplified with primers specific for the E2F1 binding amplicon (oligo 1) and C/EBPα binding amplicon (oligo 2). (G) U937 cells were transfected with control or E2F1 vector. Total RNA was analyzed by quantitative RT-PCR with oligos for miR-223 (top) and with oligos for E2F1 (bottom). Data are represented as mean ± SD from 3 independent experiments. *P < .05; **P < .01.
We observed that C/EBPα is able to transactivate miR-223 promoter (Figure 4C), as reported before.9 However, we did not observe any transactivation of miR-223 by C/EBPα in the promoter region, which has been shown as necessary for the regulation by PU.1 and C/EBPβ10 (Figure 4D).
Because we observed that E2F1 represses miR-223 transcription, we next examined putative E2F1 binding sites in the miR-223 genomic sequence, which is 1 kilobase upstream of the transcription start site, using TransFac software (BIOBASE Biological Databases, http://www.gene-regulation.com). We could not find any consensus E2F1 binding site in this region (data not shown). E2F1 can bind to target genes through sites that are different from consensus binding sites.24,25 To find the binding of E2F1 to the miR-223 promoter, we performed chromatin immunoprecipitation assays in K562-C/EBPα-p42-ER cells. E2F1 binds to the miR-223 promoter in the uninduced state and this binding is decreased during C/EBPα-induced granulocytic differentiation (Figure 4E). One possible explanation for the decrease in E2F1 binding to the miR-223 promoter during granulocytic differentiation could be tethering of E2F1 by C/EBPα via direct protein-protein interaction reported before.13,26 This explains why C/EBPα with mutations in the basic region (C/EBPα-BRM2), which can bind and transactivate target genes19 but do not interact with E2F1, fails to up-regulate miR-223 (Figure 1B right).
C/EBPα has been shown to be associated with miR-223 promoter during granulopoiesis.9 As shown in Figure 4E, C/EBPα associates with miR-223 promoter in a time-dependent manner, supporting our finding that C/EBPα regulates miR-223 levels during granulopoiesis.
To further validate our data in vivo, we analyzed binding of E2F1 and C/EBPα to miR-223 promoter in diagnostic samples from AML patients by chromatin immunoprecipitation. The genetic features of the AML samples are shown in supplemental Table 2. We used AML sample with normal karyotype (nk) as control. We found weak binding of E2F1 to the miR-223 promoter in this sample (Figure 4F sample 1, oligos 1). However, we found a strong binding of E2F1 to the miR-223 promoter in AML samples having AML1/ETO, PML/RARα, and Flt3 mutation (Figure 4F samples 2-9, oligo 1). This indicates that E2F1 negatively regulates miR-223 in AML. Interestingly, the binding of C/EBPα to the miR-223 promoter was found to be weak in AML samples (Figure 4F samples 2-9, oligos 2). These data explain our previous finding (Figure 3A), which showed lower miR-223 expression in those AML samples in which C/EBPα function is blocked.
E2F1 binding to genes that do not have a consensus E2F1 binding site has been reported for several genes including hydroxysteroid sulfotransferase, thioether-S-methyltransferase, and carboxylesterase.24 Different mechanisms have been proposed for E2F1 binding to a nonconsensus motif.25 Because C/EBPα and E2F1 can bind directly by protein-protein interaction, we investigated the possibility that E2F1 is being recruited to the miR-223 promoter via protein-protein interaction. We found no binding of C/EBPα to the miR-223 promoter in the E2F1 binding element (oligos 1) (supplemental Figure 6). Therefore, we assume that E2F1 is recruited to miR-223 promoter via a mechanism distinct from interaction with C/EBPα. Because we could observe that E2F1 binds and represses the miR-223 promoter activity (Figure 4B,4E-F), we hypothesized that E2F1 could down-regulate miR-223 levels. We found a 60% down-regulation of miR-223 level during E2F1 overexpression (Figure 4G top). Taken together, these findings demonstrate that E2F1 functions as a transcriptional repressor of the miR-223 gene.
Discussion
C/EBPα, the first leucine zipper domain transcription factor discovered, depends on different pathways for mediating growth arrest in various tissues.27,28 Inhibition of E2F activity has been found as a unique mechanism for the antimitotic activity of C/EBPα in granulopoiesis.1,20 Previous studies have shown the significance of microRNAs in regulating cell-cycle progression.23 It is becoming clear that miRNAs act in the context of regulatory feedback loops to inhibit E2F1. As shown in this study, inhibition of E2F1, the master regulator of cell-cycle progression, could be one of the most likely potential functions of miR-223 in granulopoiesis, because deregulation of cell-cycle machinery has been demonstrated to be crucial in the development of AML.
Our findings give new insights to the C/EBPα-mediated inhibition of myeloid cell-cycle progression, which is a key step disrupted in different subtypes of AML. In this report, we show that C/EBPα regulates miR-223, and miR-223 blocks myeloid cell-cycle progression by targeting E2F1. We cannot rule out the possibility of miR-223 targeting other cell-cycle regulators during granulopoiesis. To our knowledge, this is the first report that shows the mechanism for E2F1 inhibition by C/EBPα during granulopoiesis. The mutant form of C/EBPα, which cannot interact with E2F1 (C/EBPα-BRM2), fails to induce miR-223 (Figure 1B right). This mutant form is defective in inducing granulocytic differentiation.4 Our data suggest that E2F1 is bound to miR-223 promoter in the uninduced state and this binding is decreased during C/EBPα induction (Figure 4E). This suggests tethering of E2F1 by C/EBPα from the miR-223 promoter is essential for the up-regulation of miR-223 and for granulopoiesis. The inability of the transactivation domain mutant of C/EBPα (C/EBPα-p30) in up-regulating miR-223 (Figure 1B left) suggests transactivation domain of C/EBPα is also necessary for the induction of miR-223. Altogether, these data demonstrate that E2F1 interaction domain, as well as N-terminal transactivation domain, is critical for proper regulation of miR-223 in granulopoiesis by C/EBPα. Even though different microRNAs including miR-17-5p, miR-20a, miR-93, and miR-106b have been reported to target E2F1,15,23,29,30 these microRNAs are not reported to be expressed in granulopoiesis. Because miR-223 is the most widely expressed microRNA in granulopoiesis, we propose E2F1 inhibition by miR-223 as a major pathway through which C/EBPα mediates cell-cycle inhibition in granulopoiesis. However, we cannot rule out the possibility of other microRNAs regulated by C/EBPα targeting E2F1 during granulopoiesis.
Myeloid master regulator C/EBPα has been shown to be deregulated by different oncoproteins (AML1/ETO, BCR/ABL, PML/RARα, Flt3 mutant protein).3 In addition, CEBPA mutations are reported in approximately 10% of AML patient samples.20,31 This suggests deregulation of C/EBPα as one of the major steps in AML. Our observation that miR-223 is down-regulated in different subtypes of AML (Figure 3A), in which C/EBPα function is also blocked, proposes miR-223 as a key component of tumor suppressor function of C/EBPα in AML. Down-regulation of miR-223 pathway could be one of the critical steps in leukemogenesis during C/EBPα deregulation.
C/EBPα has been shown to inhibit cell-cycle progression.32,33 Loss of C/EBPα-mediated cell-cycle arrest is pivotal in the development of AML. C/EBPα-mediated E2F inhibition is important in controlling the proliferative capacity of early myeloid progenitors.34 Conditional knockout animals for C/EBPα show a selective block in the common myeloid progenitor to granulocyte macrophage progenitor (GMP) stage of granulopoiesis.2 Mice with targeted disruption of domains necessary for E2F inhibition display an increase in GMP population.4 Mice homozygous for a C/EBPα knock-in mutation that impairs E2F repression at the N-terminus show accumulation of committed myeloid progenitors and develop AML.5 All these findings demonstrate that effective E2F inhibition by C/EBPα is essential for myeloid lineage commitment. A recent study shows that miR-223 null animals display increase in the GMP population of myeloid progenitors.11 These data are in correlation with data from different C/EBPα mice models mentioned above, which show accumulation of myeloid progenitors. The finding from miR-223 null animals together with our finding that miR-223 targets E2F1 (Figure 2) and inhibits myeloid cell-cycle progression (Figure 3B-E) suggest that down-regulation of E2F1 by miR-223 could be a critical step in granulocytic differentiation.
Our study also gives new dimensions for E2F1 function during transcriptional control during granulopoiesis. E2F1 is widely considered as a transcriptional activator. However, emerging studies propose that E2F1 can function as transcriptional activator or repressor, based on the molecular context.35 E2F1 has been reported to act as a transcriptional repressor for several genes including androgen receptor,16 the antiapoptotic protein Mcl-1,36 plasminogen activator inhibitor-1 (PAI-1),37 human telomerase reverse transcriptase (hTERT),38 and urokinase-type plasminogen activator (uPA).39 The mechanisms for E2F1-mediated negative regulation are still not well understood. Our data suggest that E2F1 functions as a repressor of the miR-223 gene during transcription and this is mediated by an element located within several hundred base pairs of the transcription start site of miR-223 (Figure 4).
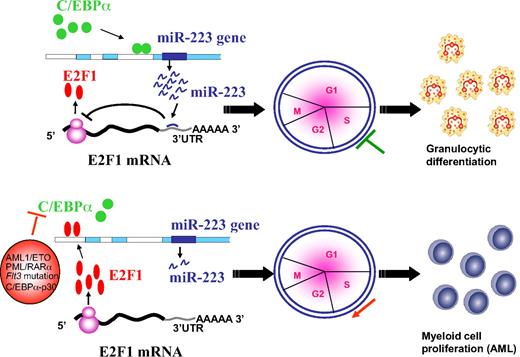
Our data suggest that during C/EBPα induction, miR-223 is induced, which in turn blocks the translation of E2F1 leading to inhibition of cell-cycle progression followed by myeloid differentiation (Figure 5 top). Our finding suggests that deregulation of C/EBPα by various mechanisms in AML results in weak binding of C/EBPα to the miR-223 promoter, which results in fewer miR-223 molecules in the cell to inhibit E2F1 translation (Figure 5 bottom). The overexpressed E2F1 could bind to the miR-223 promoter and in turn lead to a further decrease in miR-223 expression through a negative feedback loop followed by myeloid cell-cycle progression at the expense of differentiation. Overexpression of E2F1 has been shown to be an oncogenic event that predisposes cells to transformation.40 This fits with the recent finding from a mouse model that show lack of C/EBPα-mediated E2F repression is an important component of the transformation process during leukemogenesis.5 Taken together, our findings support a regulatory network existing between miR-223 and transcription factors C/EBPα and E2F1 during myeloid cell proliferation and differentiation. Changing the balance from one to the other may result in the shift from differentiation to proliferation (ie, from normal granulocytic differentiation to differentiation block) which results in AML.
Schematic representation of a model for the role of miR-223, C/EBPα, and E2F1 in normal granulopoiesis and in AML. During granulopoiesis (top panel), C/EBPα binds and transactivates miR-223 promoter, which in turn leads to E2F1 repression and inhibition of cell-cycle progression resulting in myeloid differentiation. When C/EBPα is deregulated by various mechanisms in AML (bottom panel), transactivation of miR-223 is inhibited, which results in accumulation of E2F1. Overexpressed E2F1 binds to miR-223 promoter and inhibits miR-223 transcription through a negative feedback loop resulting in myeloid cell-cycle progression and block of differentiation.
Schematic representation of a model for the role of miR-223, C/EBPα, and E2F1 in normal granulopoiesis and in AML. During granulopoiesis (top panel), C/EBPα binds and transactivates miR-223 promoter, which in turn leads to E2F1 repression and inhibition of cell-cycle progression resulting in myeloid differentiation. When C/EBPα is deregulated by various mechanisms in AML (bottom panel), transactivation of miR-223 is inhibited, which results in accumulation of E2F1. Overexpressed E2F1 binds to miR-223 promoter and inhibits miR-223 transcription through a negative feedback loop resulting in myeloid cell-cycle progression and block of differentiation.
In summary, our study provides evidence that E2F1 repression by miR-223 is a crucial event in the antimitotic function of C/EBPα during granulopoiesis. Our data suggest microRNA-223 as a molecular switch regulated by C/EBPα and E2F1 in granulopoiesis and in AML, respectively. Manipulation of miR-223 could be therapeutically relevant in AML subtypes in which E2F1 inhibition is deregulated.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank F. D. Camargo for providing bone marrow cells from miR-223 null mice, C. Nervi for lenti-223 vector and valuable discussions, A. D. Friedman for valuable discussions, T. Fukao for miR-223 promoter constructs, J. T. Mendell for E2F1 3′UTR construct, M. L. Day for E2F1 construct, D. Eick and Z. Weng for discussions, and A. Zubaiga for E2F1-null mice bone marrow cells. We also thank K. Nerger and J. Luetzkendorf for assistance with FACS analysis and J. Dittmer for help with luciferase assays.
This work was supported by National Institutes of Health grants HL-56745 and CA-118316 (D.G.T.) and Deutsche Forschungsgemeinschaft and Krebshilfe grants (G.B.).
National Institutes of Health
Authorship
Contribution: J.A.P., V.D., and P.S.P. performed experiments and analyzed the data; S.K.B. and C.M.-T. provided AML patient samples; D.G.T. provided K562-ER cell lines; A.A.P.Z., C.M.-T., and G.B. commented on the paper; J.A.P. and V.D. wrote the paper; J.A.P. designed the research; and G.B. supervised the work.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Gerhard Behre, Department of Internal Medicine IV–Oncology and Hematology, Martin Luther University Halle-Wittenberg, Ernst Grube Str 40, Halle 06120, Germany; e-mail: gerhard.behre@medizin.uni-halle.de.
References
Author notes
J.A.P. and V.D. contributed equally to this work.