Abstract
Recent genome-wide association data have implicated genetic variation at 7p12.2 (IKZF1), 10q21.2 (ARIDB5), and 14q11.2 (CEBPE) in the etiology of B-cell childhood acute lymphoblastic leukemia (ALL). To verify and further examine the relationship between these variants and ALL risk, we genotyped 1384 cases of precursor B-cell childhood ALL and 1877 controls from Germany and the United Kingdom. The combined data provided statistically significant support for an association between genotype at each of these loci and ALL risk; odds ratios (OR), 1.69 (P = 7.51 ×10−22), 1.80 (P = 5.90 × 10−28), and 1.27 (P = 4.90 × 10−6), respectively. Furthermore, the risk of ALL increases with an increasing numbers of variant alleles for the 3 loci (ORper-allele = 1.53, 95% confidence interval, 1.44-1.62; Ptrend = 3.49 × 10−42), consistent with a polygenic model of disease susceptibility. These data provide unambiguous evidence for the role of these variants in defining ALL risk underscoring approximately 64% of cases.
Introduction
Two genome-wide association (GWA) studies provided evidence that common germline variation influences the risk of precursor B-cell childhood acute lymphoblastic leukemia (ALL).1,2 The strongest association was attained at 7p12.2 with the single nucleotide polymorphism (SNP) rs4132601 located 3′ to the ikaros family zinc finger 1 (IKZF1) gene. Ikaros proteins are master regulators of lymphocyte development and differentiation directing the CD4 versus CD8 T-cell lineage commitment.3 Mutant mice expressing only non–DNA-binding Ikaros isoforms develop aggressive lymphoblastic leukemia.4,5 IKZF1 somatic deletions are common at diagnosis in high-risk/poor prognosis B-cell precursor ALL and in ALL with BCR-ABL1 fusions.6,7 Functional basis for the association between rs4132601 and ALL was provided by the correlation between reduced IKZF1 expression and risk genotype in lymphocytes.1 The second locus identified indentified was at 10q21.2 with rs7089424 located in intron 3 of the AT-rich interactive domain 5B (ARIDB5) gene. ARIDB5 is a member of the AT-rich interaction domain family of transcription factors, important in embryogenesis and growth regulation.8,9 The third association seen in only one of the GWA studies was at 14q11.2 with rs2239633 in the vicinity of CEBPE, encoding CCAAT/enhancer-binding protein, ϵ a regulator of myelopoiesis.
Association studies are often capricious; therefore, replication is highly desirable. Replication across populations can also augment generalization of findings. Estimates of the impact of risk loci derived from GWA studies have a tendency to be overinflated because of “winner's curse.” To verify and further explore the relationship between the 7p12.2, 10q21.2, and 14q11.2 variants and ALL risk, we genotyped 2 case-control series from Germany and the United Kingdom totaling 1384 patients and 1877 controls.
Methods
The German case-control study comprised 1193 patients (663 male and 530 female; mean age, 6 years) with childhood ALL ascertained through the Berlin-Frankfurt-Münster trials between 1993 and 2004. A total of 1516 controls (762 male and 754 female; mean age, 58 years) were ethnically matched healthy persons of German origin recruited in 2004 at the Institute of Transfusion Medicine, Mannheim, Germany.10 The United Kingdom series was composed of 191 patients (101 male and 90 female) not previously analyzed as part of our GWA study: 114 prevalent cases from the United Kingdom Childhood Cancer Study, an epidemiologic study of childhood malignancies conducted between 1991 and 1998, 24 incident cases from the United Kingdom MRC ALL 97 (99) trial, and 53 cases from Royal Marsden NHS Hospitals Trust. Mean age at diagnosis of patients was 6 years. Controls were healthy persons collected through a national study of colorectal cancer being conducted in the United Kingdom (117 male and 244 female; mean age, 59 years).11
Collection of blood samples and clinicopathologic information from patients and controls was undertaken with informed consent and relevant ethical review board approval from the Institute for Cancer Research in accordance with the Declaration of Helsinki. DNA extractions were performed using standard methodologies. SNP genotyping was conducted using the Kaspar allele-specific polymerase chain reaction (KBiosciences) and analyzed in an Applied Biosystem ABI 7900HT system. Primer sequences and polymerase chain reaction conditions are available on request. Quality control was monitored through blind genotyping of 15 DNA samples with 100% concordance and inclusion of 6% of DNA samples as replicates in each round of genotyping.
For the 3 SNPs, risk of ALL was quantified by calculating odds ratios (OR) with 95% confidence intervals using the STATA statistical package (Version 10.0; Stata Corporation). All statistical tests were 2-sided. To provide increased power to demonstrate a relationship between SNP genotype and risk, we pooled data from the 2 case-control series. Meta-analysis was performed under a fixed effects model, estimating Cochran's Q statistic to test for heterogeneity and the I2 statistic to quantify the proportion of the total variation between studies. Combined effect of pairs of the 3 loci with risk was investigated by logistic regression modeling, and evidence for interactive effects between SNPs was assessed by a likelihood ratio test. The population attributable fraction was estimated from 1 − Π(1 − (x − 1)/x), where x = (1 − p)2 + 2p(1 − p)OR1 + p2OR2, p is the population allele frequency, and OR1 and OR2 are the ORs associated with heterozygosity and homozygosity, respectively.
Results and discussion
Genotypes were obtained for 3572 of 3579 for DNA samples from the German cases (99.8%) and 4519 of 4548 controls (99.4%). The call rate for United Kingdom series was 555 of 578 (96.0%) in the cases and 1068 of 1086 (98.3%) in the controls. SNP call rates were not significantly different between all cases and controls or between each of the 2 case-control studies. Population stratification was ruled out as the genotype distribution satisfied the criterion for Hardy-Weinberg equilibrium in both control series. The genotype frequencies for each of the 3 SNPs in both case-control series showed a strong relationship with ALL risk in the German series (Table 1). Although a similar relationship between genotype and risk was observed in the United Kingdom series, the association was not statistically significant for rs2239633 (Table 1). In both datasets, the pattern of risk associated with each SNP was compatible with an additive model of disease susceptibility.
Association between SNPs and ALL risk
| SNP (gene) . | Population . | Genotype . | Cases (%) . | Controls (%) . | OR (95% CI) . | P . |
|---|---|---|---|---|---|---|
| rs4132601 (IKZF1) | German | AA | 471 (39.6) | 811 (54.0) | 1.0 (referent) | |
| AC | 552 (46.4) | 574 (38.2) | 1.7 (1.4-2.0) | 1.2 × 10−9 | ||
| CC | 166 (14.0) | 116 (7.8) | 2.5 (1.9-3.2) | 7.6 × 10−12 | ||
| United Kingdom | AA | 60 (31.9) | 206 (57.2) | 1.0 (referent) | ||
| AC | 96 (51.1) | 133 (36.9) | 2.5 (1.7-3.7) | 3.8 × 10−6 | ||
| CC | 32 (17.0) | 21 (5.8) | 5.2 (2.7-10.2) | 2.9 × 10−8 | ||
| Overall | AA | 531 (38.6) | 1017 (54.6) | 1.0 (referent) | ||
| AC | 648 (47.0) | 707 (38.0) | 1.8 (1.5-2.0) | 1.4 × 10−13 | ||
| CC | 198 (14.4) | 137 (7.4) | 2.8 (2.2-3.6) | 2.9 × 10−17 | ||
| rs7089424 (ARIDB5) | German | AA | 346 (29.0) | 683 (45.3) | 1.0 (referent) | |
| AC | 595 (50.0) | 673 (44.6) | 1.8 (1.5-2.1) | 1.1 × 10−10 | ||
| CC | 249 (21.0) | 152 (10.1) | 3.2 (2.5-4.1) | 1.0 × 10−22 | ||
| United Kingdom | AA | 46 (25.0) | 153 (43.0) | 1.0 (referent) | ||
| AC | 99 (53.8) | 165 (46.3) | 2.0 (1.3-3.0) | 9.5 × 10−4 | ||
| CC | 39 (21.2) | 38 (10.7) | 3.4 (1.9-6.1) | 8.8 × 10−6 | ||
| Overall | AA | 392 (28.5) | 836 (44.9) | 1.0 (referent) | ||
| AC | 694 (50.5) | 838 (44.9) | 1.8 (1.5-2.1) | 8.7 × 10−13 | ||
| CC | 288 (21.0) | 190 (10.2) | 3.2 (2.6-4.0) | 7.1 × 10−27 | ||
| rs2239633 (CEBPE) | German | AA | 197 (16.5) | 307 (20.3) | 1.0 (referent) | |
| AG | 559 (46.9) | 773 (51.2) | 1.1 (0.9-1.4) | 2.6 × 10−1 | ||
| GG | 437 (36.6) | 430 (28.5) | 1.6 (1.3-2.0) | 5.1 × 10−5 | ||
| United Kingdom | AA | 26 (14.2) | 64 (18.2) | 1.0 (referent) | ||
| AG | 95 (51.9) | 183 (52.0) | 1.3 (0.7-2.2) | 3.5 × 10−1 | ||
| GG | 62 (33.9) | 105 (29.8) | 1.5 (0.8-2.6) | 1.8 × 10−1 | ||
| Overall | AA | 223 (16.2) | 371 (20.0) | 1.0 (referent) | ||
| AG | 654 (47.5) | 956 (51.3) | 1.1 (0.9-1.4) | 1.9 × 10−1 | ||
| GG | 499 (36.3) | 535 (28.7) | 1.6 (1.3-1.9) | 2.8 × 10−5 |
| SNP (gene) . | Population . | Genotype . | Cases (%) . | Controls (%) . | OR (95% CI) . | P . |
|---|---|---|---|---|---|---|
| rs4132601 (IKZF1) | German | AA | 471 (39.6) | 811 (54.0) | 1.0 (referent) | |
| AC | 552 (46.4) | 574 (38.2) | 1.7 (1.4-2.0) | 1.2 × 10−9 | ||
| CC | 166 (14.0) | 116 (7.8) | 2.5 (1.9-3.2) | 7.6 × 10−12 | ||
| United Kingdom | AA | 60 (31.9) | 206 (57.2) | 1.0 (referent) | ||
| AC | 96 (51.1) | 133 (36.9) | 2.5 (1.7-3.7) | 3.8 × 10−6 | ||
| CC | 32 (17.0) | 21 (5.8) | 5.2 (2.7-10.2) | 2.9 × 10−8 | ||
| Overall | AA | 531 (38.6) | 1017 (54.6) | 1.0 (referent) | ||
| AC | 648 (47.0) | 707 (38.0) | 1.8 (1.5-2.0) | 1.4 × 10−13 | ||
| CC | 198 (14.4) | 137 (7.4) | 2.8 (2.2-3.6) | 2.9 × 10−17 | ||
| rs7089424 (ARIDB5) | German | AA | 346 (29.0) | 683 (45.3) | 1.0 (referent) | |
| AC | 595 (50.0) | 673 (44.6) | 1.8 (1.5-2.1) | 1.1 × 10−10 | ||
| CC | 249 (21.0) | 152 (10.1) | 3.2 (2.5-4.1) | 1.0 × 10−22 | ||
| United Kingdom | AA | 46 (25.0) | 153 (43.0) | 1.0 (referent) | ||
| AC | 99 (53.8) | 165 (46.3) | 2.0 (1.3-3.0) | 9.5 × 10−4 | ||
| CC | 39 (21.2) | 38 (10.7) | 3.4 (1.9-6.1) | 8.8 × 10−6 | ||
| Overall | AA | 392 (28.5) | 836 (44.9) | 1.0 (referent) | ||
| AC | 694 (50.5) | 838 (44.9) | 1.8 (1.5-2.1) | 8.7 × 10−13 | ||
| CC | 288 (21.0) | 190 (10.2) | 3.2 (2.6-4.0) | 7.1 × 10−27 | ||
| rs2239633 (CEBPE) | German | AA | 197 (16.5) | 307 (20.3) | 1.0 (referent) | |
| AG | 559 (46.9) | 773 (51.2) | 1.1 (0.9-1.4) | 2.6 × 10−1 | ||
| GG | 437 (36.6) | 430 (28.5) | 1.6 (1.3-2.0) | 5.1 × 10−5 | ||
| United Kingdom | AA | 26 (14.2) | 64 (18.2) | 1.0 (referent) | ||
| AG | 95 (51.9) | 183 (52.0) | 1.3 (0.7-2.2) | 3.5 × 10−1 | ||
| GG | 62 (33.9) | 105 (29.8) | 1.5 (0.8-2.6) | 1.8 × 10−1 | ||
| Overall | AA | 223 (16.2) | 371 (20.0) | 1.0 (referent) | ||
| AG | 654 (47.5) | 956 (51.3) | 1.1 (0.9-1.4) | 1.9 × 10−1 | ||
| GG | 499 (36.3) | 535 (28.7) | 1.6 (1.3-1.9) | 2.8 × 10−5 |
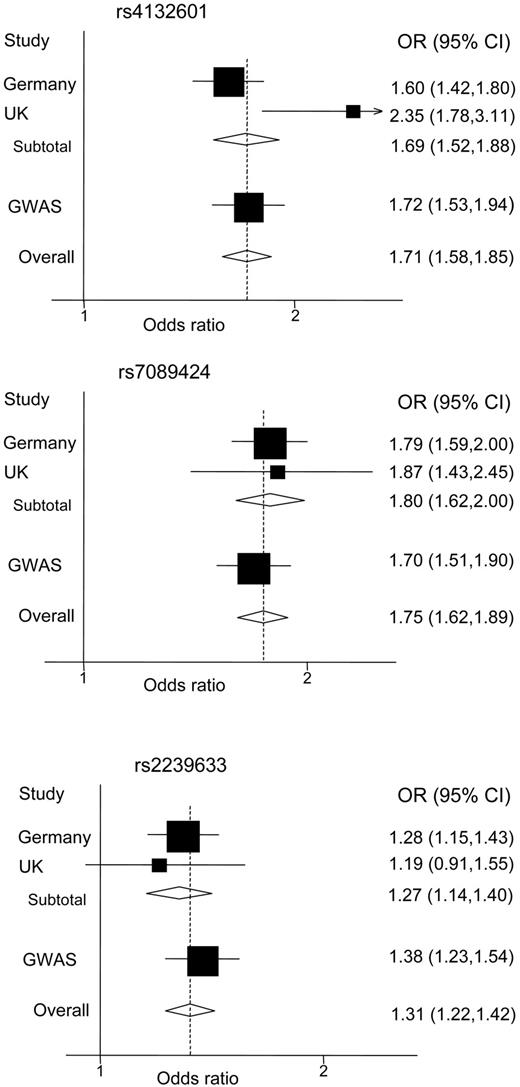
The per allele OR of ALL associated with rs4132601, rs7089424, and rs2239633 was 1.69 (P = 7.51 × 10−22), 1.80 (P = 5.90 × 10−28), and 1.27 (P = 4.90 × 10−6), respectively (Figure 1). The associated measures of heterogeneity for the same SNPs were Phet = .026, I2 = 83%; Phet = .76, I2 = 0%; and Phet = .63, I2 = 0%, respectively. Further pooling with data from the United Kingdom GWA study provided unambiguous evidence for a relationship between variations at these loci and ALL risk1 (Figure 1).
Forest plots of ORs of ALL for the 3 SNPs. Boxes represent allelic OR point estimates, their areas being proportional to the inverse variance weight of the estimate. Horizontal lines represent 95% confidence intervals. Diamond (and broken line) represents the summary OR computed under a fixed effects model, with 95% confidence interval given by its width. The unbroken vertical line is at the null value (OR = 1.0).
Forest plots of ORs of ALL for the 3 SNPs. Boxes represent allelic OR point estimates, their areas being proportional to the inverse variance weight of the estimate. Horizontal lines represent 95% confidence intervals. Diamond (and broken line) represents the summary OR computed under a fixed effects model, with 95% confidence interval given by its width. The unbroken vertical line is at the null value (OR = 1.0).
No evidence of interactive effects between any of the 3 pairs of loci (P > .05) was observed, suggesting that each locus has an independent role in defining the risk. Although the risks of ALL associated with the 3 variants are individually modest, the carrier frequencies of the risk alleles are high in the European population, and the loci make a major contribution to the development of ALL. The estimated population attributable fraction was compatible with approximately 64% of ALL being defined by these variants. Risk of ALL increases with an increasing number of risk alleles for the 3 loci, counting 2 for a homozygote and one for a heterozygote assuming equal weights (ORper-allele = 1.53, 95% confidence interval, 1.44-1.62; Ptrend = 3.49 × 10−42). Persons with 5 or more risk alleles have a more than 4-fold increase in risk compared with those with a median number of risk alleles. These ORs may be underestimates because the additive model assumes equal weighting across SNPs. The present data provide only estimates of the effect on susceptibility attributable to the variations. The effect of the actual causal variant responsible for the association is probably larger.
The assertion that ALL may have a genetic basis has long been pursued through association studies based on candidate genes without clear insight into the causal basis of ALL. Many of the studies have been based on small sample series with inconsistent findings. As with the previously published GWA studies, the strongest associations identified were for the SNPs mapping to IKZF1 and ARIDB5. However, our study provides independent evidence that variation in CEBPE plays a significant role in ALL. Collectively, these data provide robust evidence that variations in IKZF1, ARIDB5, and CEBPE influence the risk of ALL and that these variants collectively play a major role in the development of this hematologic malignancy.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank the participants for their involvement in the study.
This work was supported by Tumorzentrum Heidelberg-Mannheim, European Union (EU Food-CT-2005-016320 and Health-F3-2007-200767), Leukemia Research (United Kingdom), Kay Kendal Leukemia Fund, Cancer Research United Kingdom (C1298/A8362, supported by the Bobby Moore Fund), and Deutsche Krebshilfe.
Authorship
Contribution: R.B.P., J.V., E.P., A.G., and R. Koehler performed experiments; C.R.B., R.S.H., R. Kumar, and K.H. designed research; F.J.H. analyzed data; R. Koehler, M.G., E.S., S.E.K., T.L., E.R., M.T., K.P.-J., M. Stanulla, and M. Schrappe provided DNA for analysis; R.S.H., R. Kumar, and K.H. wrote the paper; M.G., K.P.-J., M. Stanulla, M. Schrappe, and C.R.B. contributed to the paper; and all authors approved the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Rajiv Kumar, Division of Molecular Genetic Epidemiology, German Cancer Research Center (DKFZ), Im Neuenheimer Feld 580, 69120 Heidelberg, Germany; e-mail: r.kumar@dkfz.de.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal