Abstract
The t(11;17)(q23;q21) translocation is associated with a retinoic acid (RA)–insensitive form of acute promyelocytic leukemia (APL), involving the production of reciprocal fusion proteins, promyelocytic leukemia zinc finger–retinoic acid receptor α (PLZF-RARα) and RARα-PLZF. Using a combination of chromatin immunoprecipitation promotor arrays (ChIP-chip) and gene expression profiling, we identify novel, direct target genes of PLZF-RARα that tend to be repressed in APL compared with other myeloid leukemias, supporting the role of PLZF-RARα as an aberrant repressor in APL. In primary murine hematopoietic progenitors, PLZF-RARα promotes cell growth, and represses Dusp6 and Cdkn2d, while inducing c-Myc expression, consistent with its role in leukemogenesis. PLZF-RARα binds to a region of the c-MYC promoter overlapping a functional PLZF site and antagonizes PLZF-mediated repression, suggesting that PLZF-RARα may act as a dominant-negative version of PLZF by affecting the regulation of shared targets. RA induced the differentiation of PLZF-RARα–transformed murine hematopoietic cells and reduced the frequency of clonogenic progenitors, concomitant with c-Myc down-regulation. Surviving RA-treated cells retained the ability to be replated and this was associated with sustained c-Myc expression and repression of Dusp6, suggesting a role for these genes in maintaining a self-renewal pathway triggered by PLZF-RARα.
Introduction
Acute promyelocytic leukemia (APL) is characterized by the excessive production of immature myeloid cells and is associated with gene rearrangements that fuse various proteins (N) to the retinoic acid receptor α (RARα) encoded on chromosome 17 (q21). These chimeric (N-RARα) fusion proteins include the DNA and ligand binding domains of the RARα protein (regions B-F) and can disrupt normal retinoid signaling by acting as aberrant RARα receptors. The majority of APL involves the t(15;17)(q22;q21) translocation, which fuses the promyelocytic leukemia (PML) gene on chromosome 15 to the RARα gene, to yield the PML-RARα fusion, however 6 alternative partner genes have since been identified.1-7 Treatment of APL with pharmacologic levels of all-trans retinoic acid (ATRA) leads to differentiation of leukemic blasts and complete remission, however the t(11;17)(q23;q21) translocation, which yields the promyelocytic leukemia zinc finger (PLZF)–RARα and reciprocal RARα-PLZF fusion, is less responsive to ATRA and patients have a poorer prognosis.8,9
PLZF-RARα can block myeloid differentiation in human cell lines and in murine models of hematopoiesis.10-12 As a dominant negative RARα, PLZF-RARα may affect normal RARα/RXR signaling by regulating target promoters either as a homodimer or as a heterocomplex with RXR or by sequestering RXR from active RARα/RXR complexes. PLZF-RARα may also interfere with endogenous PLZF activity via interactions mediated through its BTB domain.13,14 The relative insensitivity of t(11;17) patients to ATRA differentiation therapy has been hypothesized to be due to the ability of PLZF-RARα to tightly bind corepressors even in the presence of ATRA. In vitro, PLZF-RARα fails to transactivate target promoters in the presence of ATRA.11,15,16 In cell and animal models, the presence of PLZF-RARα has been associated with relative retinoic acid resistance and the failure of ATRA to eradicate disease in mice.10,11,17,18 Recent data suggest that the reciprocal RARα-PLZF fusion may also be a critical factor affecting the response of leukemic cells to ATRA.19 This hypothesis is supported by the observation that a patient expressing only the PLZF-RARα fusion as a result of cryptic insertion event underwent complete hematologic remission upon treatment with ATRA and these patient blasts were also sensitive to ATRA-induced differentiation in vitro.20,21
In a transgenic murine model of t(11;17) APL, where PLZF-RARα is expressed under the cathespin G promoter, all mice developed leukemia and died within 1.5 years, whereas only 15% of mice harboring PML-RARα develop disease in this time period.11 This suggests that the PLZF-RARα fusion is highly oncogenic, and likely affects many important cellular processes in addition to blocking myeloid differentiation. Hence, identification of transcriptional and signaling networks perturbed by PLZF-RARα will yield important insights into disease pathogenesis and resistance to treatment.
Using chromatin immunoprecipitation promotor arrays (ChIP-chip) and gene expression profiling, we have identified novel, direct targets of PLZF-RARα with disease-specific relevance. We demonstrate that in addition to blocking myeloid differentiation, PLZF-RARα also promotes proliferation/self-renewal via the aberrant regulation of cell cycle–associated genes such as c-Myc, providing a basis for studying the aberrant response of this leukemia subtype to retinoic acid.
Methods
Cell lines and culture reagents
The U937T:PLZF-RARα–inducible cell line is based on the U937T autoregulatory tet-off system and cells were cultured as previously described.22 PLZF-RARα expression was induced by washing cells 3 times with phosphate-buffered saline and culturing without tetracycline. Cells were cultured in the presence of 10nM ATRA (Sigma-Aldrich) for 24 hours before PLZF-RARα induction as well as after protein induction. The 293 cells were cultured in Dulbecco modified Eagle medium (DMEM) containing 10% (vol/vol) fetal bovine serum, 50 U/mL penicillin, and 50 μg/mL streptomycin. Platinum E (PLATE) retroviral producer cells were cultured in DMEM containing 10% (vol/vol) fetal bovine serum, 50 U/mL penicillin, 50 μg/mL streptomycin, 1 μg/mL puromycin, and 10 μg/mL blasticidin.
RNA extraction and microarray analysis
U937T:PLZF-RARα cells were cultured in triplicate in the presence or absence of tetracycline for 48 hours. RNA was extracted using QIAGEN RNeasy, quantified using a NanoDrop ND-1000 (NanoDrop Technologies), and biotin labeled using the MessageAmpII kit (Ambion). After quality of biotinylated cRNA was determined using an Agilent Bioanalyzer, 10 μg was hybridized to U133 Plus 2.0 arrays (Affymetrix). Data were analyzed using ArrayAssist Version 5.2.2 (Stratagene), and normalization was performed using the Gene Chip Robust Multiarray Average (GC-RMA) algorithm. Microarray data have been deposited to GEO with the accession number GSE18476.23
cDNA preparation and qRT-PCR
cDNA was synthesized from 1 μg of total RNA using the Iscript cDNA Synthesis Kit (BioRad). Quantitative reverse-transcription polymerase chain reactions (qRT-PCR) were performed using the Quantitect SYBR Green PCR Kit (QIAGEN) using the Mx3000P Real-Time PCR System (Stratagene, Agilent Technologies). Primer sequences used for quantitative PCR are in supplemental Table 1 (available on the Blood website; see the Supplemental Materials link at the top of the online article).
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) was performed with 107 U937T:PLZF-RARα cells 48 hours after tetracycline removal using 6 μg of PLZF (2A9) monoclonal antibody (Calbiochem) or unrelated immunoglobulin G2a monoclonal antibody (Millipore; supplemental Methods). A subset of 12 genes was selected for validation by loci-specific qPCR (primer sequences are located in supplemental Table 2). DNA fragments enriched by ChIP were calculated as a percentage of input DNA. Significant fold changes in enrichment were calculated by comparing PLZF-RARα–expressing cells relative to either uninduced cells or negative control antibody for biologic triplicate ChIPs (supplemental Methods). Binding to previously identified RARα target promoters (RARβ2, NFE2) was used as a positive control for enrichment, and an exonic region of Bcl6 was used as a negative control.
ChIP-chip
PLZF-RARα and immunoglobulin G2a ChIP products and corresponding input chromatin were amplified by ligation-mediated PCR as described previously (http://genomics.ucdavis.edu/farnham; supplemental Methods). qPCR of positive and negative control loci using equivalent quantities of ChIP and input DNA (70 ng) was performed to ensure the conservation of enrichment after amplification. Biologic triplicate PLZF-RARα ChIP amplicons were labeled with cyanine 5 (Cy5) and total input amplicons were labeled with Cy3 by NimbleGen Systems Inc and cohybridized to the HG18 RefSeq Promoter Array. Enriched peaks were identified using NimbleScan24,25 and visualized using SignalMap (supplementary Methods). Peaks with a false discovery rate (FDR) less than 0.2 and enriched in at least two-thirds of biologic replicates were considered for further analysis.
Bioinformatic analysis
Gene ontology was assessed using the PANTHER (http://www.pantherdb.org).26 Gene Biofunctions and network analysis was performed using Ingenuity Pathway Analysis (IPA; Ingenuity Systems). The frequency of direct repeats associated with natural retinoic response elements (RAREs; 5′-PuG(G/T)TCA) within PLZF-RARα–enriched peak sequences (∼ 0.3-1 kB) was determined using a pattern-finding algorithm. A consensus of sequence motifs found in PLZF-RARα–enriched peaks from a filtered list of PLZF-RARα targets (2/3 biologic replicates, FDR < 0.2, and differentially expressed > 1.5-fold; P < .05; 1672 genes) was analyzed using the MATRIXReduce algorithm27 with affinity logos generated using STAMP (http://www.benoslab.pitt.edu/stamp).28 Consensus motifs were compared with transcription factor binding sites in the JASPAR V3 database.29 Transcription factor binding sites in the c-MYC promoter were identified using PATCH Public 1.0 (http://www.gene-regulation.com).
For comparison of PLZF-RARα targets with different acute myeloid leukemia (AML) subtypes, Affymetrix U133A.CEL data files from leukemic blasts of 4 PLZF-RARα–expressing19 and 18 PML-RARα–expressing30 specimens and 99 representing other AML subtypes (selected at random from 267 patient samples30 ) were imported into ArrayAssist Version 5.2.2 and probe intensities were summarized and normalized using GC-RMA. Gene Set Enrichment Analysis (GSEA; V2.0) was used to generate a ranked list of genes, using the signal-to-noise ratio ranking metric, such that the final position of a gene in the list is dependent on the strength of the gene to discriminate between the 2 phenotypes (APL vs AML). This ranked gene list was divided into deciles where D1 represents genes that are highly expressed in APL versus AML and D10 represents genes that are expressed at a low level in APL compared with other AMLs. PLZF-RARα direct, transcriptional targets (1672 genes) were located within the context of this ranked list and the number of genes in each decile was calculated.
Production of retrovirus
Retroviral constructs used for human cord blood and murine hematopoietic progenitor assays used a 2-promoter MSCV-PGK-EGFP retroviral vector (MPG). MPG-PLZF-RARα contains the full-length human PLZF(B)-RARα,31 whereas MPG-PLZF contains the full-length PLZF cDNA. These plasmids were transfected into the PLATE packaging cell line (Cell Biolabs) using a 3:1 ratio of Fugene6:DNA (Roche). After 24 hours, transfection reagent was removed and replaced with Expansion Stock (1:1 RPMI-1640/DMEM, 0.075% NaHCO3, 50 U/mL penicillin, and 50 μg/mL streptomycin) plus 1× Nutridoma-SP (Roche) and incubated at 32°C for 48 hours. Supernatants were pooled and filtered through a 0.45-μm syringe filter for immediate use.
Murine hematopoietic assays
Bone marrow from 8- to 12-week-old C57BL/6 mice was lineage depleted with the EasySep Mouse Kit (StemCell Technologies). Cells were prestimulated for 24 hours in Expansion Stock supplemented with murine interleukin-3 (mIL-3; 10 ng/mL), mIL-6 (10 ng/mL), stem cell factor (SCF; 50 ng/mL; all from Peprotech), and 0.1μM of cortisol. For infections, approximately 0.5 × 106 cells were combined 1:1 with retroviral supernatant supplemented with the prestimulation cytokine cocktail and 4 μg/mL of polybrene in a final volume of 1 mL and centrifuged for 50 minutes at 1010g at 25°C. After 48 hours of growth, GFP-positive cells were collected by cell sorting using a DakoCytomation MoFlo. For colony assays, 5000 or 10 000 cells were seeded into 3 mL of Methocult medium (StemCell Technologies) supplemented with mIL-3, mIL-6, and mSCF with IL-3 (10 ng/mL), IL-6 (10 ng/mL), mCSF (50 ng/mL), and GMCSF (10 ng/mL), along with granulocyte-macrophage colony-stimulating factor (10 ng/mL) and duplicate 1.2-mL aliquots were plated into 35-mm dishes. Colonies were scored after 7 days in culture. After 8 days, cells were recovered from methylcellulose and either analyzed or replated (5000 or 10 000 cells per well). For differentiation assays, cells were plated with dimethyl sulfoxide or 1μM of ATRA. Details of c-Myc knockdown in PLZF-RARα–transformed cells can be found in supplemental Methods.
Human cord blood assays
Lin− cord blood (CB) cells were prestimulated and transduced in X-VIVO 10 (Cambrex) medium with 1% bovine serum albumin, 2mM l-glutamine, 100 U/mL penicillin-streptomycin, plus SCF (100 ng/mL), FMS-like tyrosine kinase 3 ligand (100 ng/mL), thrombopoietin (50 ng/mL), and IL-6 (20 ng/mL; all from Amgen) for 24 hours. For viral infections, approximately 106 cells were transduced 4 times with changes of 1 to 5 × 107 viral particles, each for 48 hours. Wells were precoated with CH-296 fibronectin (4 μg/cm2; Takara Bio Inc). For liquid cultures, 105 infected cells were grown in Iscove modified Dulbecco medium plus 20% BIT 9500 Serum Substitute (StemCell Technologies), 2mM l-glutamine, plus SCF (100 ng/mL), FMS-like tyrosine kinase 3 ligand (100 ng/mL), IL-6 (20 ng/mL), granulocyte colony-stimulating factor (20 ng/mL), and granulocyte-macrophage colony-stimulating factor (20 ng/mL; Amgen). Cultures were maintained at a density of approximately 106 cells/mL for the duration of cumulative cell counts.
In vivo mouse studies
Animal experiments were performed according to the guidelines of institutional animal care committees, using protocols approved by the Comité Régional d'Ethique Expérimentation Animale no. 4. PML-RARα and PLZF-RARα/RARα-PLZF APL transplantation models were previously described.17 Mice were treated with retinoic acid by subcutaneous implantation of release pellets containing 10 mg of retinoic acid for 24 or 48 hours (Innovative Research of America). RNA was extracted from untreated and RA-treated bone marrow using RNeasy kits (QIAGEN). qRT-PCR for Cebpϵ, Id1, Csf2ra, c-Myc, Dusp6, and Bcl2 were performed as described earlier using the Ywhaz gene as a normalization control. Primer sequences can be found in supplemental Table 1.
Immunophenotypic analysis
Immunophenotypic analysis of U937 cells was performed by staining with 1 μg of antibody/1 × 106 cells (Cd11b–allophycocyanin [APC], CD33-APC, CD14–fluorescein isothiocyanate, and CD116-phycoerythrin) or isotype controls (BD Biosciences or Caltag) for 15 minutes and analyzed on a BD LSRII flow cytometer (BD Biosciences). Flow cytometric data were analyzed using FlowJo (TreeStar) or FACSDiva (BD Biosciences). Immunophenotypic analysis of murine cells for Cd11b, Gr-1, c-kit, and Sca1 was performed as described.32
Cell-cycle analysis
U937 cell-cycle profiles were assessed using propidium iodide.33 For primary murine hematopoietic progenitors, the cell cycle was assessed between days 7 and 8 using the APC bromodeoxyuridine (BrdU) Flow Kit (BD Biosciences). BrdU was added directly to colonies in methylcellulose (100μM final) and after gentle resuspension, cells were incubated at 37°C for 3 hours and BrdU assays were performed on approximately 106 cells according to the manufacturer's instructions. Cell-cycle profiles were analyzed on a BD LSRII flow cytometer.
Luciferase reporter assays
The pXP2:c-myc2.5kB promoter luciferase reporter construct containing −2451 to +49 bp of the human c-Myc gene34 was coexpressed with MPG, MPG-PLZF and MPG-PLZF-RARα, MSCV-puro-PLZF-RARα containing the full-length human PLZF-RARα fusion transcript, MSCV-hygro-PLZF-RARαDMD35N/R49Q, which loses repression and homodimerization ability, and MSCV-hygro-FKBP-PLZF-RARαDMD35N/R49Q mutant, which restores oligomerization through a FKBP module.35 For luciferase assays, reporter and effector plasmids were used in a 1:4 ratio. The 293 cells were transfected in triplicate with Fugene6 (Roche) and lysates were harvested after 48 hours and assayed for luciferase activity using the Dual Luciferase Kit (Promega). Raw values were normalized for cell number by measuring the protein concentration of individual lysates by Bradford assay (BioRad).
Results
ChIP-chip and gene expression profiling identifies novel PLZF-RARα targets
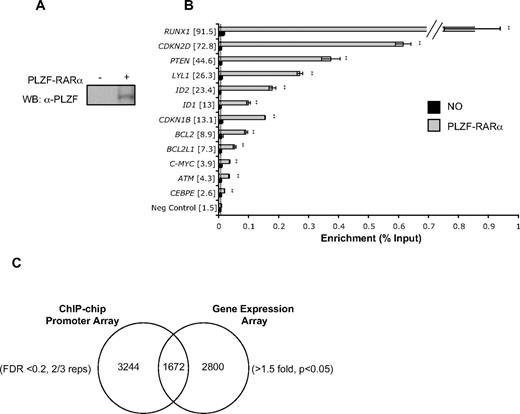
ChIP-chip was performed in the U937 myelomonocytic cell line, used previously to identify transcriptional and biologic networks regulated by PLZF and PLZF-RARα.10,33,36 PLZF-RARα was induced by tetracycline withdrawal for 48 hours and ChIP was performed using an anti-PLZF monoclonal antibody37 (Figure 1A). Immunoprecipitated DNA fragments and corresponding inputs were amplified and labeled, and biologic triplicates were cohybridized in triplicate to NimbleGen HG18RefSeq arrays, representing 24 659 human promoters. Potential sites of PLZF-RARα binding were identified at 4916 gene promoters, and these included known transcriptional targets such as IL-8 and S100P38 (FDR < 0.2, 2/3 biologic replicates; supplemental Table 3). We confirmed binding of PLZF-RARα to 12 putative target promoters, selected on the basis that they were either (1) bound and transcriptionally regulated by PLZF-RARα in U937 cells, and involved in myeloid differentiation/cell cycle/apoptosis (RUNX1, CDKN2D, PTEN, LYL1, ID1, CDKN1B, BCL2L1, c-MYC, CEBPϵ) or (2) bound by both PLZF-RARα and PLZF in ChIP-chip screens and may therefore be potentially regulated by both proteins (RUNX1, ID2, BCL2, c-MYC, and ATM; Figure 1B, supplemental Figure 1). All 12 genes showed binding by PLZF-RARα in U937 cells only upon PLZF-RARα induction, with fold enrichment ranging from between approximately 2.5- and 92-fold. No significant binding was seen at these loci when PLZF-RARα was absent and these promoters were not enriched by IP with an unrelated antibody (data not shown).
Identification of PLZF-RARα direct target genes by ChIP-chip and gene expression arrays. (A) U937T:PLZF-RARα cells were withdrawn from tetracycline for 48 hours and PLZF-RARα expression was assessed by Western blot of whole-cell lysate using PLZF monoclonal antibody (2A9). (B) Loci-specific qPCR using unamplified ChIP samples confirmed PLZF-RARα binding to 100% of putative target promoters compared with uninduced controls (fold enrichment in brackets), a negative control exon region (—). Data are expressed as mean ± SEM of 3 independent experiments (**P < .01). (C) Comparison of PLZF-RARα ChIP dataset (FDR < 0.2, 2/3 replicates; 4916 genes) with gene expression dataset (> 1.5-fold, P < .05; 4472 genes) identified 1672 genes bound and transcriptionally regulated by PLZF-RARα.
Identification of PLZF-RARα direct target genes by ChIP-chip and gene expression arrays. (A) U937T:PLZF-RARα cells were withdrawn from tetracycline for 48 hours and PLZF-RARα expression was assessed by Western blot of whole-cell lysate using PLZF monoclonal antibody (2A9). (B) Loci-specific qPCR using unamplified ChIP samples confirmed PLZF-RARα binding to 100% of putative target promoters compared with uninduced controls (fold enrichment in brackets), a negative control exon region (—). Data are expressed as mean ± SEM of 3 independent experiments (**P < .01). (C) Comparison of PLZF-RARα ChIP dataset (FDR < 0.2, 2/3 replicates; 4916 genes) with gene expression dataset (> 1.5-fold, P < .05; 4472 genes) identified 1672 genes bound and transcriptionally regulated by PLZF-RARα.
To identify transcriptional pathways directly regulated by PLZF-RARα, gene expression profiling was performed under identical conditions used for ChIP-chip. We thus identified 4472 genes altered in expression in response to PLZF-RARα induction (> 1.5 fold, P < .05). A comparison of ChIP-chip and gene expression datasets revealed that a highly significant 34% of genes (P < .001 by χ2) bound by PLZF-RARα were also transcriptionally regulated (1672 genes; Figure 1C, supplemental Table 4). Of these, 56% of genes were up-regulated and 44% of genes were down-regulated, suggesting that PLZF-RARα functions as both a transcriptional activator and a repressor in the absence of exogenous ATRA. Ingenuity Pathway Analysis revealed that many of the 1672 putative direct targets were involved in cell death (362 genes; P < .001), cell cycle (191 genes; P < .001), and hematologic system development and function (214 genes; P < .001; Table 1).
PLZF-RARα target genes are differentially expressed in APL
To investigate the disease relevance of these direct, transcriptional targets, we determined whether such genes were associated with an APL-specific gene expression pattern. Gene expression data from 22 patients with APL (4 PLZF-RARα, 18 PML-RARα specimens) and 99 patients with other forms of AML (supplemental Table 5) were used to generate a ranked gene list that could discriminate APL and AML phenotypes (Figure 2). There was an overrepresentation of PLZF-RARα direct target genes among those down-regulated in APL. By contrast, a set of 1672 randomly selected genes was not significantly overrepresented in any gene expression decile. Ontologic analyses of the top 100 genes down-regulated in APL compared with AML revealed an enrichment of genes involved in major histocompatibility complex II–mediated immunity (P < .001), immunity and defense (P < .001), amino acid catabolism (P < .001), and cell proliferation (P < .001; Table 2). PLZF-RARα directly bound targets were specifically associated with immunity and defense and cell proliferation and differentiation, and included the dual specificity phosphatase 6 (DUSP6) gene, which encodes a negative regulator of mitogen-activated protein kinase signaling and has been implicated in control of hematopoietic precursor cell expansion derived from embryoid bodies.39 Conversely, PLZF-RARα direct targets identified in the top 100 leading-edge genes up-regulated in APL compared with AML were associated with protein metabolism and modification (Table 2).
PLZF-RARα direct transcriptional target genes are associated with an APL-specific phenotype. Gene expression profiles of 22 APL (PLZF-RARα [n = 4] and PML-RARα [n = 18]) and 99 AML patients were normalized by GC-RMA, and GSEA was used to generate a ranked list of genes distinguishing these phenotypes (red = up-regulated in APL vs AML blue = down-regulated in APL vs AML). The distribution of PLZF-RARα direct targets (1672 genes) within this ranked gene list was calculated and expressed as number of genes/decile. PLZF-RARα–bound and regulated genes are highly enriched in deciles 8 to 10, representing genes down-regulated in APL versus AML (●, P < .001), whereas genes from a randomly selected dataset were not significantly enriched in these quartiles (○, .01 < P < .1).
PLZF-RARα direct transcriptional target genes are associated with an APL-specific phenotype. Gene expression profiles of 22 APL (PLZF-RARα [n = 4] and PML-RARα [n = 18]) and 99 AML patients were normalized by GC-RMA, and GSEA was used to generate a ranked list of genes distinguishing these phenotypes (red = up-regulated in APL vs AML blue = down-regulated in APL vs AML). The distribution of PLZF-RARα direct targets (1672 genes) within this ranked gene list was calculated and expressed as number of genes/decile. PLZF-RARα–bound and regulated genes are highly enriched in deciles 8 to 10, representing genes down-regulated in APL versus AML (●, P < .001), whereas genes from a randomly selected dataset were not significantly enriched in these quartiles (○, .01 < P < .1).
PLZF-RARα binds an extended repertoire of RAREs
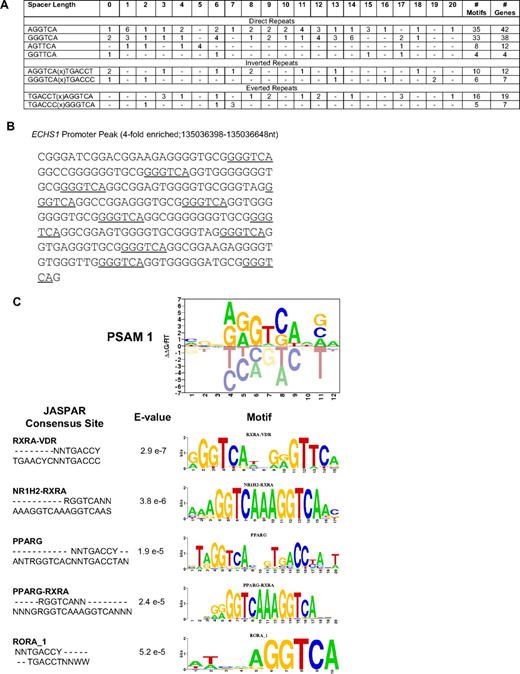
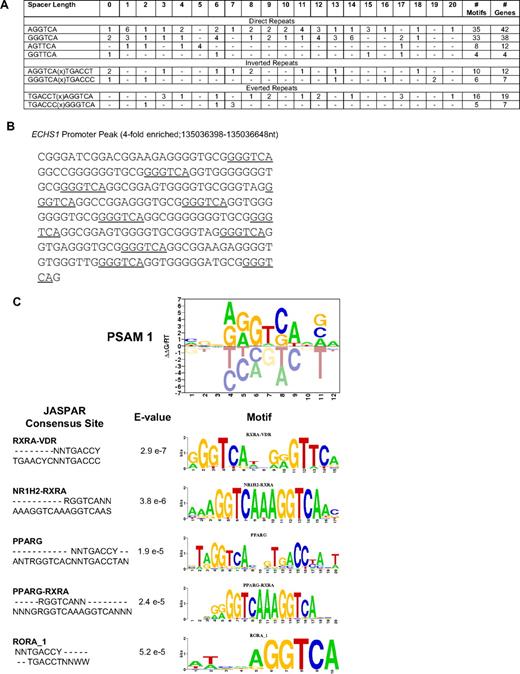
To test the hypothesis that PLZF-RARα may bind and regulate RARα targets, we calculated the frequency of RAREs in PLZF-RARα–bound promoters (Figure 3A). Only 2% of PLZF-RARα–bound promoters contained a classical putative RARE. Of these, one-third of genes were regulated by PLZF-RARα in U937 cells, with 47% down-regulated and 53% up-regulated, again suggesting that PLZF-RARα can both activate and repress transcription in the absence of exogenous ATRA (supplemental Table 6). Whereas conventional RAREs most commonly contain a closely spaced core motif RG(G/T)TCA separated by 2 or 5 bp, we observed RARE direct repeats separated by up to 20 bp (Figure 3A). The majority of promoters contained a singular direct repeat; however, 3 promoters contained multiple GGGTCA combinations. For example, the ECHS1 promoter contains up to 6 different combinations of GGGTCA(0-20)GGGTCA repeats (Figure 3B), and the MESDC2 promoter contains 3 different combinations. We also observed various inverted and everted configurations of the -PuGGTCA- RARE half site (Figure 3A). In an unbiased approach, we also used the MATRIXReduce algorithm27 to identify overrepresented motifs in the 1672 genes bound and regulated in response to PLZF-RARα. The top scoring motif from these analyses (CGGAGGTCAAGA) also ranked highest in terms of similarity to known transcription factor binding sites, largely represented by nuclear hormone receptor sites (Figure 3C). Given the low frequency of classical RARE half site direct repeats in PLZF-RARα–bound promoters, these results suggest that PLZF-RARα might bind to other nuclear receptor binding sites such as the vitamin D receptor and the peroxisome proliferator-activated receptor γ, potentially disrupting these signaling pathways.
PLZF-RARα binds an extended repertoire of RAREs. (A) The frequency of consensus RAREs (PuG(G/T)TCA)(0-20 bp)(PuG(G/T)TCA) in PLZF-RARα–bound promoters (4916 genes) was calculated. The total number of RAREs and genes containing RAREs are shown to the far right. The frequency of spacer lengths corresponding to each RARE was also calculated. (B) The ECHS1 promoter peak contains multiple RAREs (underlined). (C) MATRIXReduce was used to identify consensus motifs in PLZF-RARα–enriched peak sequences (2/3 biologic replicates, FDR < 0.2, differentially expressed > 1.5-fold, P < .05; 1672 genes). The top-scoring motif identified in PLZF-RARα–bound promoters was most similar to nuclear hormone receptor binding sites containing the RARE (A/G)GGTCA half site.
PLZF-RARα binds an extended repertoire of RAREs. (A) The frequency of consensus RAREs (PuG(G/T)TCA)(0-20 bp)(PuG(G/T)TCA) in PLZF-RARα–bound promoters (4916 genes) was calculated. The total number of RAREs and genes containing RAREs are shown to the far right. The frequency of spacer lengths corresponding to each RARE was also calculated. (B) The ECHS1 promoter peak contains multiple RAREs (underlined). (C) MATRIXReduce was used to identify consensus motifs in PLZF-RARα–enriched peak sequences (2/3 biologic replicates, FDR < 0.2, differentially expressed > 1.5-fold, P < .05; 1672 genes). The top-scoring motif identified in PLZF-RARα–bound promoters was most similar to nuclear hormone receptor binding sites containing the RARE (A/G)GGTCA half site.
PLZF-RARα–mediated myeloid block is responsive to ATRA
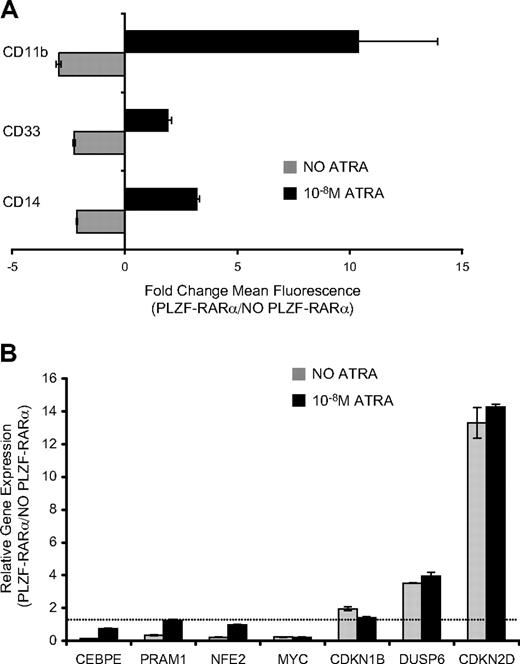
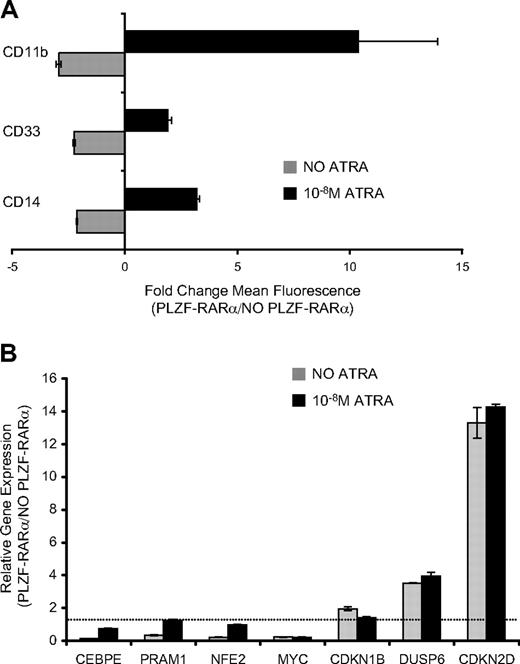
Induction of PLZF-RARα in U937 cells for 5 days inhibited expression of differentiation-associated myeloid markers CD11b, CD33, CD14 (Figure 4A), and CSF2RA (data not shown). In keeping with this biology, PLZF-RARα repressed transcriptional regulators of myeloid differentiation including CEBPϵ, NFE2, PRAM1, ITGB2, and CSF2RA (Table 1, Figure 4B). However, in the presence of physiologic concentrations of ATRA (10nM), PLZF-RARα enhanced myeloid differentiation, and repression of several myeloid-specific genes was relieved (Figure 4A-B). This indicates that the PLZF-RARα–induced differentiation block is responsive to ATRA.
PLZF-RARα differentiation block in U937 cells is responsive to retinoic acid. (A) U937T:PLZF-RARα cells were induced by tetracycline withdrawal for 5 days and the cell-surface expression of myeloid/monocytic differentiation markers (CD11b, CD33, CD14) was assessed by flow cytometry. Expression was calculated as the fold change in mean fluorescence intensity between PLZF-RARα–expressing and nonexpressing cells. In the absence of ligand ( ), PLZF-RARα inhibited the expression of CD11b, CD33, and CD14; however, in the presence of 10nM ATRA (■), PLZF-RARα enhances the expression of these markers. (B) Transcriptional response of PLZF-RARα target genes to low-dose ATRA (10nM). Data are expressed as mean ± SEM of 3 independent experiments.
), PLZF-RARα inhibited the expression of CD11b, CD33, and CD14; however, in the presence of 10nM ATRA (■), PLZF-RARα enhances the expression of these markers. (B) Transcriptional response of PLZF-RARα target genes to low-dose ATRA (10nM). Data are expressed as mean ± SEM of 3 independent experiments.
PLZF-RARα differentiation block in U937 cells is responsive to retinoic acid. (A) U937T:PLZF-RARα cells were induced by tetracycline withdrawal for 5 days and the cell-surface expression of myeloid/monocytic differentiation markers (CD11b, CD33, CD14) was assessed by flow cytometry. Expression was calculated as the fold change in mean fluorescence intensity between PLZF-RARα–expressing and nonexpressing cells. In the absence of ligand ( ), PLZF-RARα inhibited the expression of CD11b, CD33, and CD14; however, in the presence of 10nM ATRA (■), PLZF-RARα enhances the expression of these markers. (B) Transcriptional response of PLZF-RARα target genes to low-dose ATRA (10nM). Data are expressed as mean ± SEM of 3 independent experiments.
), PLZF-RARα inhibited the expression of CD11b, CD33, and CD14; however, in the presence of 10nM ATRA (■), PLZF-RARα enhances the expression of these markers. (B) Transcriptional response of PLZF-RARα target genes to low-dose ATRA (10nM). Data are expressed as mean ± SEM of 3 independent experiments.
PLZF-RARα stimulates proliferation of primary hematopoietic progenitors
Overexpression of PLZF-RARα in U937 cells was associated with decreased total viable cells over 5 days, a decrease in S and G2/M populations, and the appearance of a sub-G1 apoptotic population of cells (supplemental Figure 2). Accordingly, PLZF-RARα directly up-regulated several genes that negatively affect G1/S transition and S phase, including cell-cycle inhibitors CDKN2D and CDKN1B (Table 1). In keeping with the inhibitory effect of PLZF-RARα on cell proliferation, PLZF-RARα also up-regulated DUSP6 and repressed the c-MYC proto-oncogene (Figure 4B, Table 1). Whereas myeloid genes were de-repressed after ATRA treatment, PLZF-RARα target genes involved in cell proliferation were unaffected by treatment with ATRA (Figure 4B), suggesting that ATRA may not be capable of reversing all of the effects of PLZF-RARα.
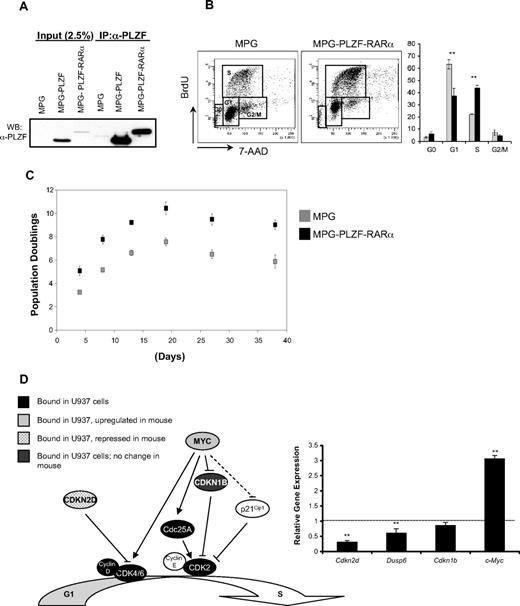
To investigate the effect of PLZF-RARα on cell proliferation in nontransformed cells, we used a retroviral expression system to overexpress PLZF-RARα in 2 primary hematopoietic systems: lineage-depleted (Lin−) murine hematopoietic progenitors and Lin− human cord blood (CB) cells. The MPG:PLZF-RARα vector yielded a protein of the expected size (Figure 5A). Constitutive expression of PLZF-RARα in lineage-depleted murine bone marrow cells inhibited myeloid development as assessed by the 2-fold reduction in myeloid colonies and decreased expression of the myeloid cell-surface marker, Cd11b, in accordance with a prior report40 (supplemental Figure 3). PLZF-RARα–expressing murine progenitors also displayed enhanced serial replating compared with control vector and expressed higher levels of the tyrosine kinase receptor Kit (c-kit), which plays a role in the maintenance and survival of hematopoietic stem cells (supplemental Figure 3B-C).
PLZF-RARα increases the proliferative capacity of hematopoietic progenitors. (A) PLATE 293T retroviral producer cells were transfected with MPG, MPG-PLZF, and MPG-PLZF-RARα retroviral vectors, and whole-cell lysates were Western blotted with PLZF (2A9) antibody. (B) Murine lin− hematopoietic progenitors were infected with MPG or MPG-PLZF-RARα retrovirus and plated in methylcellulose under myeloid-promoting conditions. Cell-cycle profiles of day-8 colonies were assessed after a 3-hour BrdU pulse. Cells were stained with anti–BrdU-APC antibody and 7-amino-actinomycin D and analyzed by flow cytometry. (C) Human lineage-depleted (Lin−) CD34+ progenitors from cord blood were infected with MPG or MPG-PLZF-RARα retrovirus and maintained in liquid culture. Cell counts were performed over a period of 40 days. (D) PLZF-RARα directly binds to the promoters of several key regulators of G1/S transition. RNA was extracted from MPG or MPG-PLZF-RARα–expressing murine colonies at day 8 and the expression of PLZF-RARα proliferation target genes (c-Myc, Dusp6, Cdkn2d, and Cdkn1b) was analyzed by qRT-PCR. Data are expressed as mean ± SEM of 3 independent experiments (**P < .05).
PLZF-RARα increases the proliferative capacity of hematopoietic progenitors. (A) PLATE 293T retroviral producer cells were transfected with MPG, MPG-PLZF, and MPG-PLZF-RARα retroviral vectors, and whole-cell lysates were Western blotted with PLZF (2A9) antibody. (B) Murine lin− hematopoietic progenitors were infected with MPG or MPG-PLZF-RARα retrovirus and plated in methylcellulose under myeloid-promoting conditions. Cell-cycle profiles of day-8 colonies were assessed after a 3-hour BrdU pulse. Cells were stained with anti–BrdU-APC antibody and 7-amino-actinomycin D and analyzed by flow cytometry. (C) Human lineage-depleted (Lin−) CD34+ progenitors from cord blood were infected with MPG or MPG-PLZF-RARα retrovirus and maintained in liquid culture. Cell counts were performed over a period of 40 days. (D) PLZF-RARα directly binds to the promoters of several key regulators of G1/S transition. RNA was extracted from MPG or MPG-PLZF-RARα–expressing murine colonies at day 8 and the expression of PLZF-RARα proliferation target genes (c-Myc, Dusp6, Cdkn2d, and Cdkn1b) was analyzed by qRT-PCR. Data are expressed as mean ± SEM of 3 independent experiments (**P < .05).
PLZF-RARα–transduced cells yielded larger colonies in methylcellulose at day 8 compared with vector control (data not shown), suggesting that these cells had increased proliferative capacity. PLZF-RARα–expressing progenitor cells exhibited a 2-fold increase in S-phase cells compared with vector control (Figure 5B). Consistent with these observations, PLZF-RARα enhanced the proliferation of human CD34+ (Lin−) cells in liquid culture, with a 7-fold increase in total cells after 38 days compared with vector control (Figure 5C). Accordingly, we observed a 3-fold increase in c-Myc, a 3-fold decrease in Cdkn2d, and a 1.6-fold decrease in Dusp6 expression in PLZF-RARα–expressing day-8 colonies compared with vector controls (P < .05; Figure 5D, supplemental Figure 4).
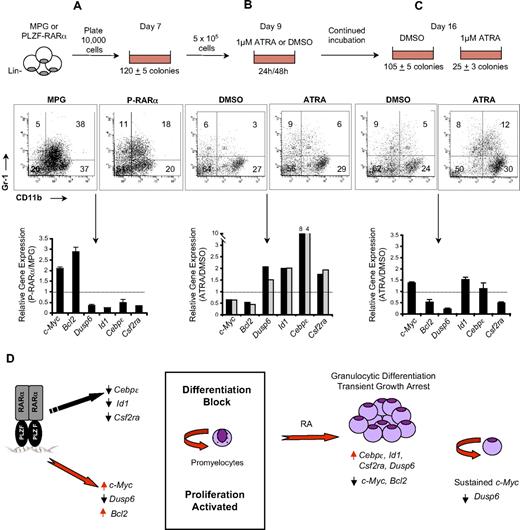
Sustained c-Myc expression is associated with survival of retinoic acid–treated PLZF-RARα–transformed murine progenitors
The response of U937-PLZF-RARα cells to retinoic acid may not reflect the response in APL cells, therefore we assessed target gene expression in APL transplantation models derived from mice expressing either the Tg(PML-RARα)935Kog or the PLZF-RARα/RARα-PLZF human transgenes.17 Treatment of mice with retinoic acid (10 mg) led to the up-regulation of Cebpϵ in both mice, consistent with the induction of granulocytic differentiation (supplemental Figure 5). Likewise, c-Myc was down-regulated and Dusp6 was activated in response to retinoic acid for both APLs.
Similarly, ex vivo treatment of PLZF-RARα–transformed murine progenitors with ATRA (1μM) for 24 and 48 hours led to the up-regulation of Csf2ra, Cebpϵ, Id1, and Dusp6, and repression of c-Myc and Bcl2, and this was associated with granulocytic differentiation (Figure 6A-B). After 8 days of ATRA treatment, we observed a 4-fold reduction in secondary colony formation. Importantly, these cells expressed sustained c-Myc levels and reduced Dusp6 expression compared with untreated cells (Figure 6C). Furthermore, as previously reported,17 these ATRA-treated cells were capable of being replated (data not shown). Knockdown of c-Myc in PLZF-RARα–transformed cells led to a 2-fold decrease in the frequency of clonogenic cells (supplemental Figure 6). Taken together, these results suggest that the ability of PLZF-RARα cells to survive and proliferate, even after prolonged RA treatment, may be linked to the ability of PLZF-RARα to sustain c-Myc expression and inhibit Dusp6 (Figure 6D).
Sustained c-Myc expression is associated with survival of retinoic acid–treated PLZF-RARα–transformed murine progenitors. (A) Murine lin− hematopoietic progenitors were infected with MPG or MPG-PLZF-RARα retrovirus and plated in methylcellulose under myeloid-promoting conditions for 7 days. Granulocytic differentiation was assessed by flow cytometry using CD11b-Pe-Cy7 and Gr-1-APC antibodies and gene expression was assessed by qRT-PCR. (B) Day-7 colonies were disaggregated and replated in the presence of 1μM ATRA (or dimethyl sulfoxide control). After 24 or 48 hours, several colonies were picked and disaggregated and differentiation was measured by flow cytometry as in panel A and changes in gene expression were measured by qRT-PCR. (C) Replated colonies treated with ATRA or control were harvested after 9 days of ATRA treatment, and differentiation was measured by flow cytometry as in panel A. Changes in gene expression were measured by qRT-PCR. Data are expressed as mean; error bars represent the maximum deviation of duplicate experiments. (D) PLZF-RARα blocks differentiation and activates proliferation by directly affecting genes involved in granulocyte differentiation and cell-cycle control. RA induces differentiation and transient growth arrest. The persistence of c-Myc expression and down-regulation of Dusp6 is associated with the continued survival of RA-treated PLZF-RARα–expressing cells.
Sustained c-Myc expression is associated with survival of retinoic acid–treated PLZF-RARα–transformed murine progenitors. (A) Murine lin− hematopoietic progenitors were infected with MPG or MPG-PLZF-RARα retrovirus and plated in methylcellulose under myeloid-promoting conditions for 7 days. Granulocytic differentiation was assessed by flow cytometry using CD11b-Pe-Cy7 and Gr-1-APC antibodies and gene expression was assessed by qRT-PCR. (B) Day-7 colonies were disaggregated and replated in the presence of 1μM ATRA (or dimethyl sulfoxide control). After 24 or 48 hours, several colonies were picked and disaggregated and differentiation was measured by flow cytometry as in panel A and changes in gene expression were measured by qRT-PCR. (C) Replated colonies treated with ATRA or control were harvested after 9 days of ATRA treatment, and differentiation was measured by flow cytometry as in panel A. Changes in gene expression were measured by qRT-PCR. Data are expressed as mean; error bars represent the maximum deviation of duplicate experiments. (D) PLZF-RARα blocks differentiation and activates proliferation by directly affecting genes involved in granulocyte differentiation and cell-cycle control. RA induces differentiation and transient growth arrest. The persistence of c-Myc expression and down-regulation of Dusp6 is associated with the continued survival of RA-treated PLZF-RARα–expressing cells.
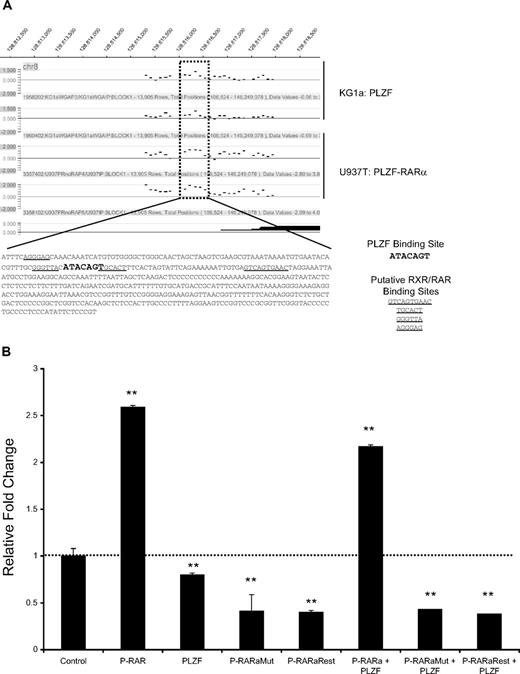
PLZF-RARα interferes with PLZF-mediated repression at the c-MYC promoter
The ability of PLZF-RARα to directly regulate c-MYC was intriguing, since c-MYC was previously identified as a direct target gene of PLZF and mediates the growth suppressive effects of PLZF in U937 cells.33 We performed ChIP-chip for PLZF targets in KG1a myeloblastic cells (K.L.R. and J.D.L., unpublished observations, May 2007) that revealed that both PLZF and PLZF-RARα bind to an overlapping region of the c-MYC promoter that contains a known PLZF binding site33 (Figure 7A) approximately 1.5-kB upstream of the major P1 transcriptional start site.41 Consistent with previous reports, PLZF modestly repressed expression from a 2.5-kB c-MYC promoter construct33 (Figure 7B). By contrast, PLZF-RARα activated the c-MYC promoter and antagonized PLZF-mediated repression. Activation was dependent on a functionally intact BTB domain, since point mutations that disrupt the ability of PLZF-RARα to repress transcription and homodimerize affected the ability of PLZF-RARα to activate transcription. Rescue of the oligomerization defect via a synthetic peptide (PLZF-RARα FKBP) did not restore the ability of PLZF-RARα mutant to activate transcription, suggesting that in addition to mediating self-association, the BTB domain is required for other activities mediated by PLZF-RARα.
PLZF-RARα functionally antagonizes PLZF-mediated repression at the c-MYC promoter. (A). PLZF and PLZF-RARα bind to an overlapping region of the c-MYC promoter, highlighted by the dotted box. This region lies upstream of the P0 promoter, approximately 1.5-kB upstream of the major transcriptional start site initiated from promoter P1. PLZF endogenous ChIP-chip was performed in the KG1a cell line and compared with PLZF-RARα ChIP-chip target genes identified in the U937:PLZF-RARα–inducible cell line. The major c-MYC transcripts initiated from this promoter region are indicated by solid black lines. Sequence analysis of the enriched peak using PATCH Public 1.0 identified putative RXR and RAR binding sites (underline) that may bind PLZF-RARα, overlapping with a previously characterized PLZF binding site (bold, enlarged font). (B) The pXP2-MYC2.5-kB luciferase reporter vector contains a 2.5-kB region of the c-MYC promoter harboring the PLZF-RARα–enriched peak. The c-Myc reporter (200 ng) was cotransfected with the indicated PLZF, PLZF-RARα, or PLZF-RARα mutant plasmids (800 ng) in 293 cells and lysates were harvested at 48 hours for luciferase activity. Data are expressed as mean ± SEM of 3 independent experiments (**P < .05).
PLZF-RARα functionally antagonizes PLZF-mediated repression at the c-MYC promoter. (A). PLZF and PLZF-RARα bind to an overlapping region of the c-MYC promoter, highlighted by the dotted box. This region lies upstream of the P0 promoter, approximately 1.5-kB upstream of the major transcriptional start site initiated from promoter P1. PLZF endogenous ChIP-chip was performed in the KG1a cell line and compared with PLZF-RARα ChIP-chip target genes identified in the U937:PLZF-RARα–inducible cell line. The major c-MYC transcripts initiated from this promoter region are indicated by solid black lines. Sequence analysis of the enriched peak using PATCH Public 1.0 identified putative RXR and RAR binding sites (underline) that may bind PLZF-RARα, overlapping with a previously characterized PLZF binding site (bold, enlarged font). (B) The pXP2-MYC2.5-kB luciferase reporter vector contains a 2.5-kB region of the c-MYC promoter harboring the PLZF-RARα–enriched peak. The c-Myc reporter (200 ng) was cotransfected with the indicated PLZF, PLZF-RARα, or PLZF-RARα mutant plasmids (800 ng) in 293 cells and lysates were harvested at 48 hours for luciferase activity. Data are expressed as mean ± SEM of 3 independent experiments (**P < .05).
Discussion
To date, our knowledge of PLZF-RARα target genes has been restricted to gene expression profiling, which although informative, provides limited information regarding the mechanisms by which PLZF-RARα directly affects target gene activity.10,36,37 To address this, we used a genome-wide approach combining ChIP-chip, gene expression arrays, patient samples, and primary hematopoietic models to identify PLZF-RARα target genes that contribute to the pathogenesis of APL. Using a gain-of-function cell line system, we identified 1672 direct, transcriptional targets of PLZF-RARα involved in crucial cellular processes such as myeloid differentiation, proliferation, apoptosis, and cellular metabolism. These genes are highly enriched in the repressed genes that differentiate APL from other myeloid leukemias, confirming the hypothesis that PLZF-RARα acts primarily as a transcriptional repressor in APL.
APL is characterized by the uncontrolled proliferation of cells blocked at the promyelocyte stage of differentiation. In addition to directly repressing genes involved in myeloid cell differentiation and function, such as CEBPϵ, PRAM1, ITGB2, and CSF2RA, our studies show that PLZF-RARα also directly controls genes controlling cell cycle and proliferation. These include c-MYC, a critical regulator of cell proliferation, differentiation, and apoptosis, DUSP6, a negative regulator of extracellular signal-related kinase (ERK) activity, and CDKN2D, a CDK inhibitor controlling G1 progression. In primary hematopoietic cells, PLZF-RARα repressed Id1, Cebpϵ, Csf2ra, Dusp6, and Cdkn2d and robustly induced c-Myc expression, correlating with a block in myeloid differentiation and increased proliferative potential.
The role of c-MYC in controlling proliferation, differentiation, and apoptosis is well established and overexpression of c-MYC is associated with hematologic malignancies.42,43 Terminal myeloid differentiation is characterized by the exit from cell cycle and a reduction in translation and protein synthesis and requires a down-regulation of c-MYC. Enforced expression of c-MYC blocks differentiation and induces myeloid leukemia in mice.44 Previous studies showed that AML-associated translocation products (AML1-ETO, PML-RARα, and PLZF-RARα) activate c-MYC expression indirectly through the Wnt signaling pathway.45 Bioinformatic analysis of PLZF-RARα target genes in U937 cells identified c-MYC at the center of a regulatory network that included genes involved in cell cycle, cancer, skeletal, and muscle disorders (Ingenuity Pathway Analysis, data not shown). Thus, our studies provide evidence that c-MYC and associated downstream pathways are directly regulated by PLZF-RARα. DUSP6 is a negative feedback regulator of fibroblast growth factor signaling, which is capable of cross-talk with other signaling pathways. Previous studies have shown that the expansion of stem cell ligand–expressing hematopoietic progenitors from embryonic stem cells is dependent on vascular endothelial-derived growth factor activation of Erk1/2, which may potentially be inhibited by DUSP6.46 Repression of DUSP6 may therefore lead to the expansion of hematopoietic progenitors and, indeed, we observed an increase in c-KIT+/Sca+ cells in PLZF-RARα–expressing murine progenitors. In support of this hypothesis, overexpression of HOXB4 in embryoid bodies led to down-regulation of Dusp6 and expansion of embryonic stem cells as measured by increased colony formation.39 Intriguingly, DUSP6 expression is down-regulated in APL compared with other AMLs, suggesting that dysregulated ERK signaling may constitute a mechanism for leukemic cell proliferation in this subtype of AML.
In the unliganded state, RAR/RXR heterodimers repress transcription. Upon ligand binding, a conformational change is induced, leading to the release of corepressor molecules such as NcoR47 and SMRT,48 and associated histone deacetylases, and the recruitment of coactivator molecules such as CBP/p300,49 TIF-2/Grip1,50 and ACTR/RAC3/p/CIP leading to gene transcription. The ability of PLZF-RARα to tightly bind corepressors even in the presence of 1μM of ATRA constitutes one possible mechanism for the ATRA insensitivity of t(11;17) patients.11 Our studies indicate that retinoic acid differentiates PLZF-RARα–induced differentiation block in 3 different hematopoietic models and is associated with the activation of PLZF-RARα direct targets previously shown to be involved in granulocytic differentiation. This finding is supported by recent studies showing that treatment of PLZF-RARα/RARα-PLZF APL with RA induces differentiation, although interestingly, normal hematopoiesis was not restored in treated mice.17 Taken together, these data strengthen the argument that differentiation block may not be the sole factor affecting the clinical response of t(11;17) APL patients to ATRA. PLZF-RARα–transformed progenitors were capable of self-renewal even after ATRA treatment compared with vector transduced cells. Knockdown of c-Myc prevented the self-renewal of these cells. This suggests that the ability of PLZF-RARα to bind and regulate a specific set of targets underlies its ability to transform cells and allow leukemia to persist even after therapy.
RAR and RXR typically function as heterodimers by binding to a consensus sequence 5′(A/G)GGTCA-3′ arranged in a direct repeat with interspacing of 1 to 5 bp (DR+1-DR+5).51 Although the frequency of canonical RAREs in PLZF-RARα–bound promoters was low, these results may reflect PLZF-RARα binding to alternative elements, which may be related to its ability to homodimerize or form higher order oligomers.52,53 Given that PLZF-RARα–bound promoters tend to contain nuclear hormone receptor binding sites, one mechanism of action may be to interfere with RXR-nuclear hormone receptor or PPARγ activity. For example, PML-RARα can bind to a PPARα response element in the acyl–coenzyme A oxidase promoter, inhibiting PPARα-mediated transcription.54 The notion that PLZF-RARα may interfere with other nuclear receptors is supported by a recent genome-wide ChIP-chip tiling array study of the RAR in breast cancer cells that showed that that RAR binding overlapped with estrogen receptor binding and gene regulation, suggesting antagonistic regulation of shared targets.55
Our directed motif analyses suggest that PLZF-RARα, like PML-RARα, may acquire an extended repertoire of target genes by its ability to bind widely spaced RARE half sites in different spatial configurations.56 The biologic consequences of this gain of function are unclear, however given that differential spacing of RARE consensus half sites has been shown to affect corepressor/activator interactions and affect response to ligand, it is possible that PLZF-RARα may aberrantly regulate normal RAR/RXR as well as novel target genes. For example, we identified RAREs in PLZF-RARα–bound promoters with a spacer of DR+1 at the highest frequency. Previous studies have shown that the specific orientation of RAR/RXR heterodimers on RARE half sites may direct the response to activating ligand and is dependent on half site spacing. Thus, in the case of direct repeats spaced by 5 bp, RXR occupies the 5′ site and RAR binds the 3′ site and heterodimers activate transcription in response to ATRA. In the case of DR+1 elements, however, RAR occupies the 5′ site and RAR/RXR heterodimers do not release nuclear corepressors in the presence of ligand and constitutively repress transcription.47,57 The effect of PLZF-RARα at these specific RAREs and response to ligand may provide further insights into the mechanisms by which PLZF-RARα acts as an aberrant RAR.
An interesting aspect of this investigation was the finding that PLZF-RARα and PLZF shared a set of common target promoters. Up to 25% of PLZF targets identified in KG1a cells by ChIP-chip (K.L.R. and J.D.L., unpublished data, May 2007) were also bound by PLZF-RARα in our U937 gain-of-function system. Two such promoters included c-MYC, which contains a previously characterized PLZF responsive element,33 and DUSP6. In contrast to PLZF, which is a modest repressor of the c-MYC promoter in transient luciferase reporter assays, PLZF-RARα activated the c-MYC promoter and antagonized PLZF-mediated repression. Thus, in addition to acting as a dominant negative RARα31,53,58 or as an antagonist of normal PLZF function via protein-protein interactions,58 the dysregulation of a shared set of target genes may constitute a significant mechanism of action of the PLZF-RARα fusion.
In conclusion, we identified direct targets of PLZF-RARα involved in many important processes governing normal and malignant hematopoiesis. These studies provide the basis for understanding how chimeric RARs, such as PLZF-RARα, are capable of inducing a self-renewal phenotype, which coupled with the differentiation block gives rise to leukemia.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors are grateful to Julia Meyer for her contribution in optimizing the PLZF (2A9) antibody, Dr Ronald DePinho for providing the shMyc constructs, and Dr Simon Lin for bioinformatics support.
This work was supported by National Institutes of Health grant CA-59936, the Leukemia Research Foundation, and the Samuel Waxman Cancer Research Foundation. D.G. is supported by the Leukemia Research Fund of Great Britain. M.L. is supported by the Ramón Areces Foundation.
National Institutes of Health
Authorship
Contribution: K.L.R. and J.D.L. designed the research; K.L.R., I.H., S.D., C.A., M.L., and J.A. performed experiments; J.M.F. performed bioinformatics motif analyses; D.G. and K.I.M. characterized APL patient samples for gene expression profiling; M.J.M. created the U937T:PLZF-RARα cell line; J.E.D. contributed cord blood reagents; and K.L.R. and J.D.L. wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Jonathan D. Licht, Division of Hematology/Oncology, Northwestern University, Robert H. Lurie Comprehensive Cancer Center, 303 E Superior St, Lurie 5-123, Chicago, IL 60611; e-mail: j-licht@northwestern.edu.

![Figure 2. PLZF-RARα direct transcriptional target genes are associated with an APL-specific phenotype. Gene expression profiles of 22 APL (PLZF-RARα [n = 4] and PML-RARα [n = 18]) and 99 AML patients were normalized by GC-RMA, and GSEA was used to generate a ranked list of genes distinguishing these phenotypes (red = up-regulated in APL vs AML blue = down-regulated in APL vs AML). The distribution of PLZF-RARα direct targets (1672 genes) within this ranked gene list was calculated and expressed as number of genes/decile. PLZF-RARα–bound and regulated genes are highly enriched in deciles 8 to 10, representing genes down-regulated in APL versus AML (●, P < .001), whereas genes from a randomly selected dataset were not significantly enriched in these quartiles (○, .01 < P < .1).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/114/27/10.1182_blood-2009-03-206524/4/m_zh89990945990002.jpeg?Expires=1762792544&Signature=Yl-gHMAztfwvSiAgXycv~v6uItUeL23g0e73b6Do9r8B3ND55ioM7Fbq2L930kCm8jvfvh9jNSDSqzYtcGklByMFEkCt~7MgOIuyKMkEL1yzDTbVn-G8q-JZtAQs90vyExvXZ-a2ZqwW2b2Dz1-O2RNL4cwndw2azyRDjmynIcCH~NK2xklXqxagX0gD5D7r9P4T3lJOcmEOZq88ejlMWR7ZB3dtMSwnX8FMoae3mGF06LJEXSvGq7BFisn-IkTp15eu4b0EQkR-sUSo~8tuGqEsKn-ZuZkgTwrt-U-1pnpg9b60S1vvDL5sEO1lhUzYPX5oiIljhPL1569dcQ0XOw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)


![Figure 2. PLZF-RARα direct transcriptional target genes are associated with an APL-specific phenotype. Gene expression profiles of 22 APL (PLZF-RARα [n = 4] and PML-RARα [n = 18]) and 99 AML patients were normalized by GC-RMA, and GSEA was used to generate a ranked list of genes distinguishing these phenotypes (red = up-regulated in APL vs AML blue = down-regulated in APL vs AML). The distribution of PLZF-RARα direct targets (1672 genes) within this ranked gene list was calculated and expressed as number of genes/decile. PLZF-RARα–bound and regulated genes are highly enriched in deciles 8 to 10, representing genes down-regulated in APL versus AML (●, P < .001), whereas genes from a randomly selected dataset were not significantly enriched in these quartiles (○, .01 < P < .1).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/114/27/10.1182_blood-2009-03-206524/4/m_zh89990945990002.jpeg?Expires=1762792545&Signature=ng8pR-DKKoYII46FjP67ItSLrQuLmzLbsyJT0cwv73K4WW5ooGAS2pGsoLqj9HXA5aGMXy4~2Beuk7Y9ayauhqhokphW6FR2SFRIjT9lSTHdlexVjefrK-~YNXqwr-~DsxQItFGpzb4UfdJVGyCFqN0Pen0GadNYB2-r1YPX6aPtyVbZw03r116v0MyU74043cpFFzdEXLkJKrDawxEWn2z-Tl3dZ5TPftKoqbGNCn96e8xjf3Bl1zqSkG6nklhgXmtSDqayWJeIY9WKuv-CfHLWP~0tnv~XZzugMm5-eGoxfEyO9crYJcr-7~i9OmztLwdBh0D1lztKkXSD7xbxCw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
 ), PLZF-RARα inhibited the expression of CD11b, CD33, and CD14; however, in the presence of 10nM ATRA (■), PLZF-RARα enhances the expression of these markers. (B) Transcriptional response of PLZF-RARα target genes to low-dose ATRA (10nM). Data are expressed as mean ± SEM of 3 independent experiments.
), PLZF-RARα inhibited the expression of CD11b, CD33, and CD14; however, in the presence of 10nM ATRA (■), PLZF-RARα enhances the expression of these markers. (B) Transcriptional response of PLZF-RARα target genes to low-dose ATRA (10nM). Data are expressed as mean ± SEM of 3 independent experiments.

