Abstract
Acute promyelocytic leukemia (APL) is characterized by a block in differentiation and accumulation of promyelocytes in the bone marrow and blood. The majority of APL patients harbor the t(15:17) translocation leading to expression of the fusion protein promyelocytic-retinoic acid receptor α. Treatment with retinoic acid leads to degradation of promyelocytic-retinoic acid receptor α protein and disappearance of leukemic cells; however, 30% of APL patients relapse after treatment. One potential mechanism for relapse is the persistence of cancer “stem” cells in hematopoietic organs after treatment. Using a novel sorting strategy we developed to isolate murine myeloid cells at distinct stages of differentiation, we identified a population of committed myeloid cells (CD34+, c-kit+, FcγRIII/II+, Gr1int) that accumulates in the spleen and bone marrow in a murine model of APL. We observed that these cells are capable of efficiently generating leukemia in recipient mice, demonstrating that this population represents the APL cancer–initiating cell. These cells down-regulate the transcription factor CCAAT/enhancer binding protein α (C/EBPα) possibly through a methylation-dependent mechanism, indicating that C/EBPα deregulation contributes to transformation of APL cancer–initiating cells. Our findings provide further understanding of the biology of APL by demonstrating that a committed transformed progenitor can initiate and propagate the disease.
Introduction
Acute myeloid leukemia (AML) is a hematologic disease characterized by accumulation in bone marrow and blood of immature blasts that fail to differentiate and invade the bone marrow and the peripheral blood. AMLs are characterized by a block in myeloid differentiation and can be classified into distinct subgroups; one well-defined subgroup is acute promyelocytic leukemia (APL). APL belongs to the M3 variant according to the French-American-British classification and is characterized by an accumulation of promyelocytes.1 All APL cases harbor a reciprocal chromosomal translocation involving the retinoic acid receptor α gene (RARα) on chromosome 17, and in 95% of the cases, the promyelocytic leukemia gene (PML) on chromosome 15.2 This t(15;17) results in the expression of the fusion protein promyelocytic-retinoic acid receptor α (PML-RARα) that has been shown to induce the myeloid differentiation arrest observed in APL.3-5 The oncogenic fusion protein PML-RARα can recruit corepressor complexes to promoters of specific genes. These corepressor complexes harbor both DNA methylase6 and histone deacetylase activity.7 In addition, PML-RARα can induce a repressed chromatin conformation on its direct genomic target genes.8
The treatment of APL differs from the therapy for other subtypes of AML in that APL cells differentiate in the presence of all-trans retinoic-acid (ATRA). ATRA induces a complete remission in the majority of APL cases; however, some patients relapse after ATRA treatment.9 The cancer “stem” cell hypothesis proposes that relapses in these and other cancers might be the result of inefficient therapeutic targeting of a small fraction of cells responsible for initiating and/or maintaining the bulk population of cancer cells, the cancer-initiating cell population, or cancer stem cell population.10 So far, the exact identity of APL cancer–initiating cell has not been characterized. In contrast to other subtypes of AML, transplantation of CD34+ CD38− cells (defining immature blood cells in human bone marrow) isolated from APL patients into SCID mice failed to generate leukemia.11 This result suggests that the compartment containing the APL cancer–initiating cells may differ from other AML subtypes.
As AML is characterized by a block of myeloid differentiation, a number of studies have investigated the role of myeloid-specific transcription factors in this process. Changes in expression of myeloid transcription factors have been implicated in human AML,12-14 and down-regulation of myeloid transcription factor expression in experimental models leads to disruption of normal hematopoiesis with subsequent development of leukemia.15-17 In APL, expression of the myeloid transcription factor PU.1 is reduced13 and its down-regulation increases APL penetrance in PML-RARα transgenic mice.17 CCAAT/enhancer binding protein α (C/EBPα) activity has also been shown to be altered in AML, as a result of mutations,18,19 down-regulation of its expression by the fusion protein AML1-ETO,14 or methylation of its promoter.20 Mutated C/EBPα proteins described in AML fail to bind to their cognate DNA target or to interact with E2F, therefore leading to a decrease of their activity. Conversely, no such mutations have been described in patients with AML associated with inv16 or t(15;17). The importance of C/EBPα in the control of normal myeloid lineage is emphasized by the fact that C/EBPα-deficient mice demonstrate a selective block in differentiation of myeloid progenitors.21 These observations underscore how a tight regulation of the myeloid genetic program by key transcriptional regulators is strictly required for normal hematopoiesis to occur. Furthermore, these observations suggest that dysregulated control of these factors plays a role in initiation of AML.
Here we investigate the phenotypic and functional nature of APL-initiating cells in a murine model. These cells, characterized as CD34+, c-kit+, FcγRIII/II+, Gr1int, and representing a promyelocytic phenotype, generate leukemia upon transplantation into recipient animals and exhibit down-regulation of C/EBPα by a methylation-dependent mechanism. As a consequence, the normal myeloid genetic program normally controlled by C/EBPα is altered, contributing to the development of APL.
Methods
Cell culture
NB4,22 HT93,23 U937PR9 and the control vector alone U937MT line,24 and U937 MT PML-RARα AHT cell lines7 were cultured in RPMI 1640 medium with 10% fetal bovine serum (HyClone). Cell viability was estimated using standard trypan blue dye exclusion. In some experiments, cells were treated with 5-aza-2′-deoxycytidine (5-Aza; 1μM or 5μM dissolved in dimethyl sulfoxide; Sigma-Aldrich) for 72 hours and/or trichostatin A (TSA; 300nM in methanol, MetOH) for 24 hours.
Luciferase assay
A total of 15 million cells were electroporated (settings: 360 mV, 960 μF) with 15 μg of a tetramer of C/EBP sites cloned from the granulocyte colony-stimulating factor (G-CSF) receptor in front of a gene coding for firefly luciferase25 and 100 ng of a vector expressing Renilla luciferase under a cytomegalovirus (CMV) promoter.26 After 24 hours, cells were lysed in passive lysis buffer (Promega), and firefly luciferase and Renilla luciferase activity was assessed (Lumat LB 9501, Berthold). Results are shown as the ratio of firefly to Renilla luciferase (relative light units [RLU]).26 Each experiment was performed in triplicate.
Bone marrow transplantation
Cells isolated from bone marrow or spleen from leukemic hMRP8 PML-RARα transgenic mice3 (FVB/N background, Ly5.1+) were resuspended in buffered saline and injected by tail vein or retro-orbital injection into 6- to 12-week-old isogeneic or congenic FVB/N mice (Ly5.2+, CD45.2+). The congenic FVB/N-5.2+ recipient line was generated by first intercrossing FVB/N mice with C57BL/Ka−Thy1.1+Ly5.2+ animals to generate FVB/N-Ly5.1/5.2+ F1 animals, and then backcrossing these heterozygous mice to FVB/N for at least 10 generations. After backcrossing, FVB/N-Ly5.1/5.2+ mice were intercrossed to generate FVB/N-Ly5.1−Ly5.2+ animals, which thereafter were maintained separately by inbreeding. Recipient mice were prepared for transplantation by cesium irradiation of 4.5 Gy. All the experiments were performed with cells isolated from leukemic founder no. 935 and leukemic founder no. 1111. Both founders displayed similar results; figures presented in this manuscript show results obtained with founder no. 935. All animal experiments were approved by the institutional review board of Harvard Medical School.
Morphologic analysis
Mice were killed by CO2 inhalation and underwent necropsy. Organs were fixed in 10% buffered formalin and processed for histopathology. Samples were embedded in paraffin; sections of 4 μm were stained with hematoxylin and eosin. Blood smears, bone marrow, and spleen cytospins were stained with Wright-Giemsa (Diff-Quik; Baxter Healthcare). Microscopy analysis was performed using a Zeiss Ax10PLAN (Zeiss MicroImaging), Canon Powershot A620 camera, run by Canon Powershot A620 software.
Flow cytometric staining and analysis
Murine bone marrow cells were analyzed and sorted on a MOFlo (MOFlo-MLS; Cytomation) or a FACSAria after exclusion of dead cells using propidium iodide staining. After lysis of erythrocytes with ACK buffer, a modified lineage depletion was performed using antibodies directed against Sca1 (clone D7; eBioscience), CD45/B220 (RM2606; Caltag), CD19 (RM7706; Calatg), CD8 (MCD0806; Caltag), and CD4 (MCD0406; Caltag) antigens, all Tricolor conjugated (omitting Gr1 antibodies from the usual depletion cocktail). Then the cells were separated using antibodies against c-kit conjugated with allophycocyanin (clone 2B8; eBioscience) and CD34 conjugated with fluorescein isothiocyanate (RAM34; eBioscience) antigens and analyzed for expression of FCγRIII/II (553143; BD Biosciences PharMingen) and Gr1 (RB6-8C5; eBioscience) antigens. For the analysis of the congenic transplantation, donor and recipient cells were distinguished with the help of Ly5.1 (A20) or Ly5.2 (104, both from eBioscience). Data were analyzed with FlowJo software (TreeStar).
Statistical analysis
Statistical differences were determined by the Student t test. Leukemia-free survival curves were measured from the date of injection of cells to the diagnosis of leukemia and analyzed using the Kaplan-Meier method (GraphPadPrism 5 software).
In vitro methylation
Plasmid DNA containing a 2.2-kb piece of the C/EBPα promoter in front of the firefly luciferase gene (pXP1 luciferase C/EBPα) was in vitro methylated by incubation with MSssI CpG methylase enzyme. A total of 15 million U937 cells were electroporated with 15 μg of methylated or unmethylated pXP1 luciferase C/EBPα together with 10 ng of CMV Renilla. Cells were lysed 6 hours after electroporation and luciferase activity was assessed.
Information on preparation of protein extracts and Western blotting, RNA isolation, and Northern blot analysis and quantitative real-time polymerase chain reaction (PCR) can be found in the supplemental data (available on the Blood website; see the Supplemental Materials link at the top of the online article).
Results
Identification and characterization of stage-specific myeloid cells in murine wild-type bone marrow
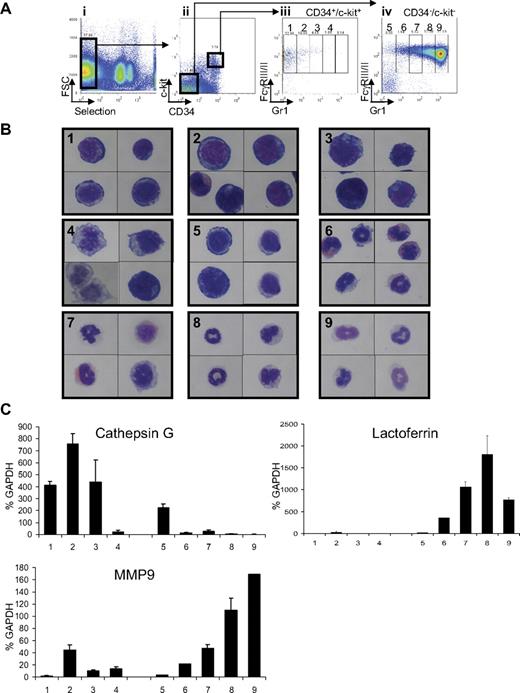
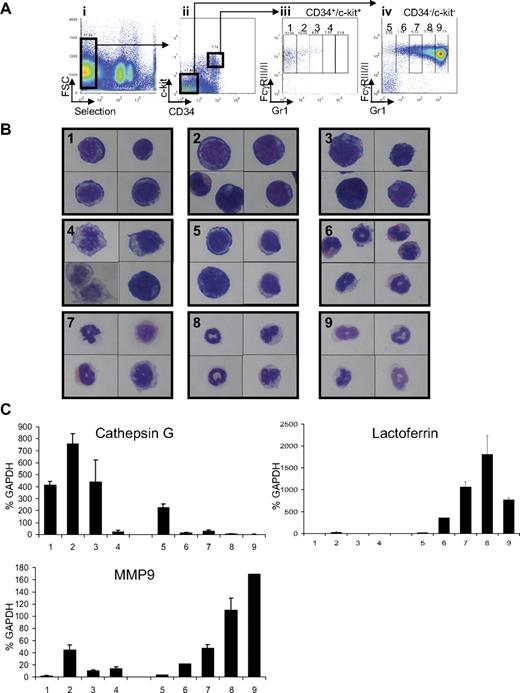
Although a number of protocols have been developed to isolate murine hematopoietic stem cells27 and multipotential progenitors,28,29 the prospective isolation of more differentiated progenitor populations, including promyelocytes, has not been well described. To characterize the nature of the leukemic progenitors in APL, murine wild-type bone marrow cells were first fractionated into distinct stages of differentiation along the myeloid lineage using flow cytometric analysis. Total wild-type bone marrow cells were depleted of B cells, T cells, and Sca1+ cells (used as a marker for hematopoietic stem cells; Figure 1Ai). Depleted cells were then separated using c-kit and CD34 markers (both expressed by hematopoietic progenitor cells; Figure 1Aii). Both double-positive (c-kit+/CD34+) and double-negative populations (c-kit−/CD34−) were further analyzed for expression of the myeloid markers FcγRIII/III and Gr1. Cells positive for FcγRIII/III expression were then divided into 5 subpopulations depending on their levels of Gr1 expression (FcγRIII/III+ from Gr1− to Gr1+; Figure 1Aiii-iv). Subpopulation 1, defined as c-kit+/CD34+, FcγRIII/II+, and Gr1−, corresponds to the granulocytic monocytic-restricted progenitor population previously defined by Akashi et al.28 Subpopulations 2 to 4 and 5 to 9 are characterized by a gradual increase in Gr1 expression. Subpopulations 5 to 9 exhibit loss of c-kit and CD34 expression, suggesting commitment toward myeloid differentiation (Figure 1Aiii-iv).
Definition of the myeloid subsets in bone marrow of wild-type FVB/N mice. (Ai) Cells expressing Sca1, CD45/B220, CD19, CD8, and CD4 were depleted with antibodies. Expression of CD34 and c-kit defined 2 populations (Aii). Each of these 2 populations (CD34+/c-kit+ and CD34−/c-kit−) was sorted according to expression of Gr1 and FcγRIII/II and divided into 4 subpopulations (1-4; Aiii) for cells isolated from the CD34+/c-kit+ fraction and 5 subpopulations (5-9) for cells isolated from the CD34−/c-kit− fraction (Aiv). (B) Sorted cells from CD34+/ckit+ exhibit the morphology of myeloblasts to promyelocytes (subpopulation 1-4), whereas sorted cells from CD34−/c-kit− exhibit the morphology of increasingly mature myeloid cells from metamyelocytes to mature “band cells.” Sorted cells were subjected to Wright-Giemsa staining, and pictures were acquired at an original magnification ×100 with oil. Numbers in each panel represent the 9 fractions shown in Aiii and iv. (C) Expression of cathepsin G (a primary granule gene), lactoferrin (a secondary granule gene), and MMP9 (a tertiary granule gene) in CD34+/ckit+ and CD34−/c-kit− sorted cells demonstrate a pattern of granule gene expression correlating with their morphology. Gene expression was assessed by quantitative real-time PCR and normalized to GAPDH levels. Numbers on the x-axis represent the 9 fractions shown in panels Aiii and iv.
Definition of the myeloid subsets in bone marrow of wild-type FVB/N mice. (Ai) Cells expressing Sca1, CD45/B220, CD19, CD8, and CD4 were depleted with antibodies. Expression of CD34 and c-kit defined 2 populations (Aii). Each of these 2 populations (CD34+/c-kit+ and CD34−/c-kit−) was sorted according to expression of Gr1 and FcγRIII/II and divided into 4 subpopulations (1-4; Aiii) for cells isolated from the CD34+/c-kit+ fraction and 5 subpopulations (5-9) for cells isolated from the CD34−/c-kit− fraction (Aiv). (B) Sorted cells from CD34+/ckit+ exhibit the morphology of myeloblasts to promyelocytes (subpopulation 1-4), whereas sorted cells from CD34−/c-kit− exhibit the morphology of increasingly mature myeloid cells from metamyelocytes to mature “band cells.” Sorted cells were subjected to Wright-Giemsa staining, and pictures were acquired at an original magnification ×100 with oil. Numbers in each panel represent the 9 fractions shown in Aiii and iv. (C) Expression of cathepsin G (a primary granule gene), lactoferrin (a secondary granule gene), and MMP9 (a tertiary granule gene) in CD34+/ckit+ and CD34−/c-kit− sorted cells demonstrate a pattern of granule gene expression correlating with their morphology. Gene expression was assessed by quantitative real-time PCR and normalized to GAPDH levels. Numbers on the x-axis represent the 9 fractions shown in panels Aiii and iv.
To precisely characterize each subpopulation, cells were sorted and analyzed by morphology (Figure 1B). Cells isolated from CD34+/c-kit+ (subpopulations 1-4) are morphologically immature and have a large nucleus and scant cytoplasm. Conversely, cells isolated from the CD34−/c-kit− fraction (subpopulations 5-9) have a granulocytic appearance, the cells are smaller, and their nuclei demonstrate the segmented shape characteristic of granulocytic cells. Fractions 6 and 7 included eosinophils characterized by the presence of red granules in their cytoplasm. The identity of the sorted cells was confirmed by quantitative real-time PCR analysis (Figure 1C; supplemental Figure 1) for granule genes used as markers of myeloid differentiation. Granule genes are sequentially expressed during myeloid differentiation, with cathepsin G, myeloperoxidase, elastase, and proteinase3 expressed in primary granules; lactoferrin in secondary granules; and matrix metallopeptidase 9 (MMP9) in tertiary granules.30,31 More immature cells (isolated from CD34+/c-kit+) express high levels of primary granule genes (cathepsin G, myeloperoxidase, elastase 2, and proteinase3) and low levels of lactoferrin and MMP9. More differentiated cells (subpopulations 6-9, isolated from CD34−/c-kit−) express lactoferrin, with a peak in subpopulation 8 (appearing to be bands) and do not express cathepsin G. More differentiated cells (subpopulations 8 and 9) express the highest levels of MMP9 (Figure 1C). In addition, cells isolated from the CD34+/c-kit+ fraction display a strong expression of the early myeloid marker Fms-like tyrosine kinase 3, whereas its expression decreases in mature cells isolated from the CD34−/c-kit− fraction (supplemental Figure 1). In summary, morphologic analysis corroborated by gene expression studies demonstrated that this novel fractionation method can isolate myeloid cells at specific stages of differentiation.
Expansion of the promyelocytic compartment in a murine model of APL
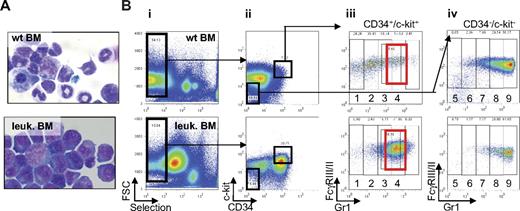
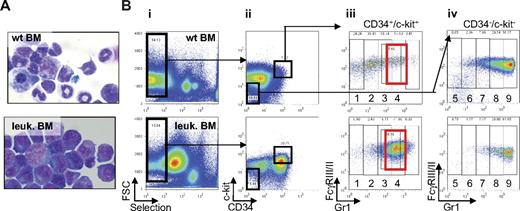
To study the pathogenesis of APL, we took advantage of the migration inhibitory factor related protein-8 (MRP8) hPML-RARα murine model.3 In this model, the human PML-RARα gene is expressed using regulatory elements of the myeloid-specific MRP8 promoter in transgenic mice.3 Because of significant mortality in these mice as a result of epidermal papillomatosis, the APL model uses a transplantation approach. Cells isolated from the spleen of leukemic transgenic mice are used to generate a similar leukemia after transplantation into isogenic recipients. This leukemia mimics human APL disease, with an arrest of neutrophilic differentiation at the promyelocytic stage, responsiveness to ATRA, and arsenic treatment and therefore represents a relevant model to study APL. We isolated cells from MRP8 hPML-RARα leukemic spleen and injected them into sublethally irradiated isogenic FVB/N recipients. All mice developed leukemia within 30 to 40 days after transplantation, as previously described.3 Leukemic mice presented splenomegaly (supplemental Figure 2) and numerous blast cells in the bone marrow and spleen (Figure 2A).
Identification of the population of cells accumulating in leukemic bone marrow of MRP8 hPML-RARα mice. (A) Cytospins of single-cell suspensions of total bone marrow from wild-type (top) and leukemic mice (bottom) after ACK treatment stained with Wright-Giemsa (original magnification ×63). The leukemia developed after transplantation with 2.5 × 105 to 5 × 106 cells from total leukemic spleen of a MRP8 hPML-RARα mouse. Mice were killed and analyzed when moribund. (B) Phenotypic analysis of leukemic cells (bottom panel) isolated from bone marrow display an increase of immature cells (CD34+/c-kit+ fraction) and accumulation of cells in subpopulations 3 and 4 that reflects the block at promyelocytic stage of differentiation (red rectangle) together with a decrease of more mature cells (subpopulations 5-9) compared with wild-type bone marrow (top panel). Negatively selected cells (Bi) from bone marrow were separated into CD34+/c-kit+ and CD34−/c-kit− fractions (Bii). CD34/c-kit double-positive cells were further fractionated into 4 fractions (subpopulations 1-4) according to Gr1 levels (Biii), and CD34/c-kit double-negative cells were further fractionated into 5 fractions according to Gr1 levels (subpopulations 5-9; Biv).
Identification of the population of cells accumulating in leukemic bone marrow of MRP8 hPML-RARα mice. (A) Cytospins of single-cell suspensions of total bone marrow from wild-type (top) and leukemic mice (bottom) after ACK treatment stained with Wright-Giemsa (original magnification ×63). The leukemia developed after transplantation with 2.5 × 105 to 5 × 106 cells from total leukemic spleen of a MRP8 hPML-RARα mouse. Mice were killed and analyzed when moribund. (B) Phenotypic analysis of leukemic cells (bottom panel) isolated from bone marrow display an increase of immature cells (CD34+/c-kit+ fraction) and accumulation of cells in subpopulations 3 and 4 that reflects the block at promyelocytic stage of differentiation (red rectangle) together with a decrease of more mature cells (subpopulations 5-9) compared with wild-type bone marrow (top panel). Negatively selected cells (Bi) from bone marrow were separated into CD34+/c-kit+ and CD34−/c-kit− fractions (Bii). CD34/c-kit double-positive cells were further fractionated into 4 fractions (subpopulations 1-4) according to Gr1 levels (Biii), and CD34/c-kit double-negative cells were further fractionated into 5 fractions according to Gr1 levels (subpopulations 5-9; Biv).
Next, the novel sorting strategy we developed was used to compare the relative composition of myeloid cells in bone marrow and spleen of leukemic versus wild-type animals. Flow cytometry revealed a strong increase in the number of c-kit+/CD34+ cells in leukemic mice compared with wild-type mice (5-fold increase in the bone marrow and 11-fold in the spleen, Figure 2Bii; supplemental Figure 3). In leukemic mice, bone marrow and spleen cells show accumulation of subpopulations 3 and 4 (72-fold increase in the bone marrow and 19-fold in the spleen; Figure 2Biii, red rectangle, and supplemental Figure 3), which present promyelocytic characteristics (CD34+, c-kit+, FcγRIII/II+, Gr1int). Expression of the human PML-RARα fusion mRNA (data not shown) and protein (see Figure 4B) confirmed that these cells are derived from the transplanted MRP8 hPML-RARα leukemic splenic cells. In addition, a drastic reduction in the more mature subpopulations (subpopulations 6-9) was observed in leukemic bone marrow and spleen (Figure 2Biv; supplemental Figure 3). These mature cells partially originated from recipient myelopoiesis as observed after transplantation in FVB/N congenic mice (supplemental Figure 4).
APL progenitors are leukemia-initiating cells
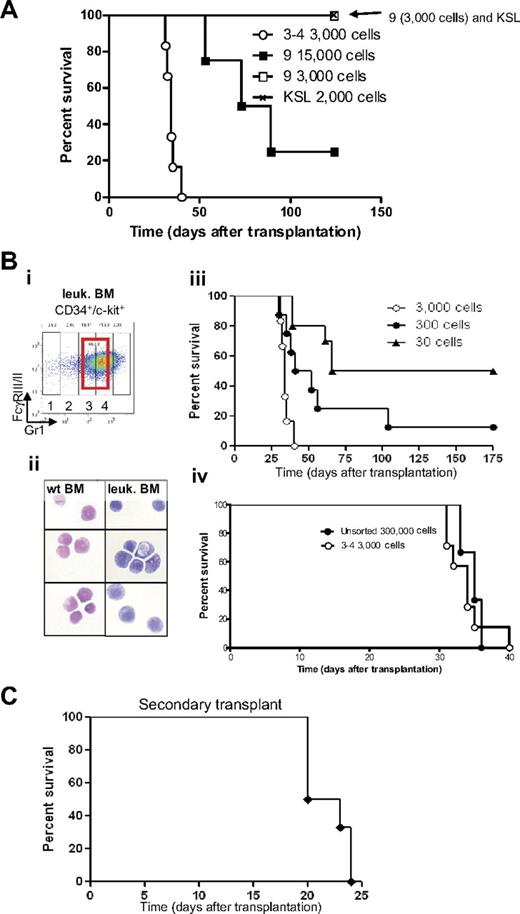
We assessed whether different cell populations isolated from bone marrow of leukemic mice were able to generate leukemia by sorting specific subpopulations and transplanting them into sublethally irradiated mice (Figure 3A). Injection of 2000 c-Kit+, Sca-1+, lin− cells (KSL, Figure 3A crosshatched line), which include the hematopoietic stem cell in normal mice,27 or injection of 3000 late myeloid differentiated cells (subpopulation 9, CD34− c-kit− FcγRIII/II+ Gr1high cells; Figure 3A open square), did not lead to development of disease. In contrast, injection of 3000 “promyelocytic-like cells” accumulating in leukemia (CD34+, c-kit+, FcγRIII/II+, Gr1int cells, subpopulations 3-4; Figure 3A open circle) was able to generate leukemia with a very short latency (median survival [MS] of 34.5 days). Cells characterized by CD34− c-kit− FcγRIII/II+ Gr1high cells (subpopulation 9) were capable to raise leukemia only at low efficiency and with long latency when transplanted at a high dose (Figure 3A, 15 000 cells [filled square]).
Identification of the APL-initiating cells. (A) Kaplan-Meier survival curve of sublethally irradiated mice transplanted with different sorted cell populations isolated from leukemic bone marrow. Injections of sorted cells from subpopulations 3 and 4 (3000 cells) and subpopulation 9 (3000 and 15 000 cells) and 2000 KSL (2000 cells, c-kit+, Sca1+, lineage−) isolated from leukemic bone marrow were injected into sublethally irradiated mice. Cells isolated from subpopulations 3 and 4 were able to generate leukemia with a latency of 38 days (○, n = 8). Mice injected with a high concentration of subpopulation 9 (15 000 cells, ■, n = 4) developed leukemia with incomplete penetrance, whereas none of the mice injected with 3000 cells from subpopulation 9 (□, n = 6) or KSL (crosshatched line, n = 6) developed leukemia. (B) Cells accumulating in leukemic bone marrow and their counterparts isolated from wild-type bone marrow were sorted (subpopulations 3 and 4; Bi; red bold rectangle in CD34+/c-kit+ fraction) and stained (Bii) with Wright-Giemsa (original magnification ×100). (Biii) As few as 30 cells accumulating from populations 3 and 4 initiate leukemia in sublethally irradiated recipient mice. Decreasing amounts of cells were injected into sublethally irradiated recipient mice (3000 cells, ○, n = 6; 300 cells, ●, n = 8: and 30 cells, ▲, n = 10). (Biv) Injection of cells accumulating in leukemic bone marrow (subpopulations 3 and 4) results in a 100-fold enrichment of LICs compared with injection of unsorted cells (○, injection of 3000 sorted cells, n = 6, compare with ●, injection of 300 000 unsorted leukemic bone marrow cells, n = 3). (C) Kaplan-Meier survival curve of mice after secondary transplantation. Thirty sorted LICs from subpopulations 3 and 4 (CD34+, c-kit+, FcγRIII/II+, Gr1int) were used in the primary transplantation. Subsequently, 2.5 × 105 to 5 × 106 unfractionated leukemic spleen cells were isolated from a moribund primary leukemic mouse and transplanted into sublethally irradiated secondary recipients (n = 6), which subsequently developed a secondary leukemia.
Identification of the APL-initiating cells. (A) Kaplan-Meier survival curve of sublethally irradiated mice transplanted with different sorted cell populations isolated from leukemic bone marrow. Injections of sorted cells from subpopulations 3 and 4 (3000 cells) and subpopulation 9 (3000 and 15 000 cells) and 2000 KSL (2000 cells, c-kit+, Sca1+, lineage−) isolated from leukemic bone marrow were injected into sublethally irradiated mice. Cells isolated from subpopulations 3 and 4 were able to generate leukemia with a latency of 38 days (○, n = 8). Mice injected with a high concentration of subpopulation 9 (15 000 cells, ■, n = 4) developed leukemia with incomplete penetrance, whereas none of the mice injected with 3000 cells from subpopulation 9 (□, n = 6) or KSL (crosshatched line, n = 6) developed leukemia. (B) Cells accumulating in leukemic bone marrow and their counterparts isolated from wild-type bone marrow were sorted (subpopulations 3 and 4; Bi; red bold rectangle in CD34+/c-kit+ fraction) and stained (Bii) with Wright-Giemsa (original magnification ×100). (Biii) As few as 30 cells accumulating from populations 3 and 4 initiate leukemia in sublethally irradiated recipient mice. Decreasing amounts of cells were injected into sublethally irradiated recipient mice (3000 cells, ○, n = 6; 300 cells, ●, n = 8: and 30 cells, ▲, n = 10). (Biv) Injection of cells accumulating in leukemic bone marrow (subpopulations 3 and 4) results in a 100-fold enrichment of LICs compared with injection of unsorted cells (○, injection of 3000 sorted cells, n = 6, compare with ●, injection of 300 000 unsorted leukemic bone marrow cells, n = 3). (C) Kaplan-Meier survival curve of mice after secondary transplantation. Thirty sorted LICs from subpopulations 3 and 4 (CD34+, c-kit+, FcγRIII/II+, Gr1int) were used in the primary transplantation. Subsequently, 2.5 × 105 to 5 × 106 unfractionated leukemic spleen cells were isolated from a moribund primary leukemic mouse and transplanted into sublethally irradiated secondary recipients (n = 6), which subsequently developed a secondary leukemia.
To assess whether the CD34+, c-kit+, FcγRIII/II+, Gr1int cells (subpopulations 3 and 4) that accumulate in the bone marrow of leukemic mice were enriched in leukemia-initiating cells (LICs), sorted cells were transplanted into sublethally irradiated isogeneic recipients (Figure 3Bi-ii). Recipient mice were transplanted with 3000, 300, or 30 sorted leukemic cells (subpopulations 3 and 4). All recipient mice transplanted with 3000 cells developed fatal leukemia with a latency of less than 40 days (Figure 3Biii open circle). The onset of leukemia was “dose-dependent” because injection of decreasing numbers of cells (Figure 3Biii, 300 cells [filled circle], and 30 cells [filled triangle]) led to development of a fatal leukemia with a significant longer latency (MS = 34 days for 3000 cells, 46.5 days for 300 cells, and 120.5 days for 30 cells; supplemental Table 1 contains details of the calculation of statistical significance). Furthermore, transplantation of 3000 (open circle) CD34+, c-kit+, FcγRIII/II+, Gr1int cells (populations 3 and 4) gave rise to leukemia with latency similar to 300 000 unfractionated leukemic bone marrow cells (Figure 3Biv filled circle; MS = 34 and 35 days, respectively), indicating a 100-fold enrichment in LICs in populations 3 and 4.
Mice transplanted with LICs that developed leukemia had a phenotype similar to mice transplanted with unfractionated leukemic spleen cells, that is, enlarged spleen (supplemental Figure 2), pale bone with cytopenia, infiltration of myeloid cells in the liver, enlarged lymph nodes (supplemental Figure 5), and the abnormal presence of c-kit+ cells in the peripheral blood (data not shown). Moreover, spleen cells from moribund leukemic mice previously injected with 30 sorted cells (Figure 3Bi subpopulations 3-4) generated leukemia in secondary transplanted animals with complete penetrance and an MS of 21.5 days (Figure 3C). Bone marrow and spleen of these leukemic secondary transplanted mice demonstrated fractions similar to the primary leukemias as determined by flow cytometry (supplemental Figure 3).
In summary, our data suggest that subpopulations 3 and 4 (CD34+, c-kit+, FcγRIII/II+, Gr1int), expanded in bone marrow and spleen of leukemic mice, have the characteristic of LICs, which are arrested in myeloid differentiation at a stage corresponding to promyelocytes and self-renew in recipient animals, therefore generating leukemia in vivo.
C/EBPα is down-regulated in APL-initiating cells, and its decreased expression after reduced gene dosage increases APL penetrance
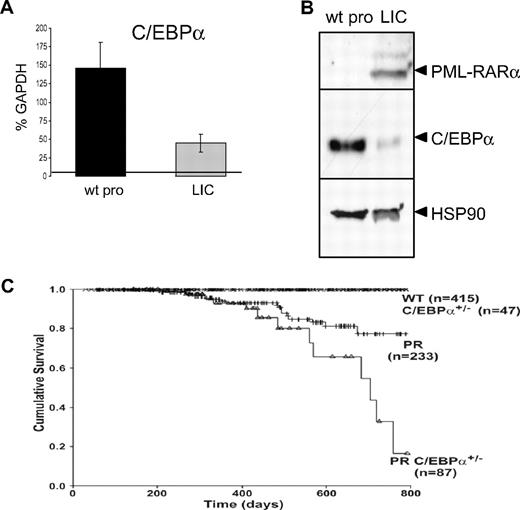
The APL-initiating cells induce a disease characterized not only by altered self-renewal but also by a block in the myeloid differentiation program. As C/EBPα is essential for myeloid differentiation in steady-state hematopoiesis,21,32,33 we reasoned that its down-regulation would contribute to the pathogenesis of APL cancer cells. To test this hypothesis, C/EBPα expression was measured in APL-initiating cells (LICs) and compared with its expression in promyelocytic phenotypic counterpart cells isolated from bone marrow of wild-type mice. Both C/EBPα RNA (Figure 4A) and protein (Figure 4B) were markedly reduced in APL-initiating cells compared with promyelocytes isolated from wild-type littermate controls, suggesting that down-regulation of C/EBPα is involved in the pathogenesis of APL in this model.
C/EBPα is down-regulated in APL-initiating cells, and its haploinsufficiency increases APL penetrance. (A) Comparison of C/EBPα RNA expression in APL-initiating cells (LICs) and their phenotypic counterpart (wild-type promyelocytes, wt pro) isolated from a wild-type bone marrow (fractions 3 and 4, Figure 2Biii) by quantitative real-time PCR normalized to GAPDH levels (representative of 3 independent experiments, P < .003, a 3-fold decrease for C/EBPα RNA levels). (B) Western blot analysis of C/EBPα protein (middle panel) in CD34+, c-kit+, FcγRIII/II+, Gr1int cells isolated from wild-type (wt) and APL-initiating cells (LICs; proteins extracted from 20 000 cells were loaded on each lane). Cells isolated from leukemic mice expressed the fusion protein PML-RARα (top panel). Loading was assessed with an antibody against HSP90 (bottom panel). (C) Kaplan-Meier analysis for leukemia-free survival in wild-type (crossed line), C/EBPα+/− (crossed line), hCathepsinG PML-RARα (PR, hatched line), and C/EBPα+/− x hCathepsinG PML-RARα mice (PR C/EBPα+/−, ▵). The cumulative survival was plotted with respect to time in days. The cumulative probability of death resulting from APL was significantly higher in hCathepsinG PML-RARα C/EBPα+/− compared with hCathepsinG PML-RARα animals (P = .02).
C/EBPα is down-regulated in APL-initiating cells, and its haploinsufficiency increases APL penetrance. (A) Comparison of C/EBPα RNA expression in APL-initiating cells (LICs) and their phenotypic counterpart (wild-type promyelocytes, wt pro) isolated from a wild-type bone marrow (fractions 3 and 4, Figure 2Biii) by quantitative real-time PCR normalized to GAPDH levels (representative of 3 independent experiments, P < .003, a 3-fold decrease for C/EBPα RNA levels). (B) Western blot analysis of C/EBPα protein (middle panel) in CD34+, c-kit+, FcγRIII/II+, Gr1int cells isolated from wild-type (wt) and APL-initiating cells (LICs; proteins extracted from 20 000 cells were loaded on each lane). Cells isolated from leukemic mice expressed the fusion protein PML-RARα (top panel). Loading was assessed with an antibody against HSP90 (bottom panel). (C) Kaplan-Meier analysis for leukemia-free survival in wild-type (crossed line), C/EBPα+/− (crossed line), hCathepsinG PML-RARα (PR, hatched line), and C/EBPα+/− x hCathepsinG PML-RARα mice (PR C/EBPα+/−, ▵). The cumulative survival was plotted with respect to time in days. The cumulative probability of death resulting from APL was significantly higher in hCathepsinG PML-RARα C/EBPα+/− compared with hCathepsinG PML-RARα animals (P = .02).
To further confirm that C/EBPα down-regulation is involved in APL, we used a second murine model.5 This model is distinct from that used in the previous studies, in that it is a transgenic rather than a transplantation model. In this case, the human cathepsin G (hCathepsin G) regulatory elements were used to express PML-RARα in transgenic mice. These animals demonstrate a low incidence of APL with a long latency period.5 We hypothesized that, if PML-RARα contributed to down-regulation of C/EBPα in these mice, then further reduction of C/EBPα might result in increased penetrance and/or decreased latency of the disease. We tested this hypothesis by breeding hCathepsin G PML-RARα mice to C/EBPα+/− mice. C/EBPα+/− mice express approximately 50% of wild-type protein in bone marrow (Y. Yamamoto and D.G.T., unpublished data, November 2003). The latency was decreased and penetrance of development of APL and subsequent mortality were significantly higher in hCathepsin G PML-RARα C/EBPα+/− mice (PR [hCathepsin G PML-RARα] C/EBPα+/−, △) compared with hCathepsin G PML-RARα mice (PR, crossed line; Figure 4C). These results again strongly implicate down-regulation of C/EBPα in the development of APL.
Expression of PML-RARα induces down-regulation of C/EBPα expression and activity in human APL cell lines
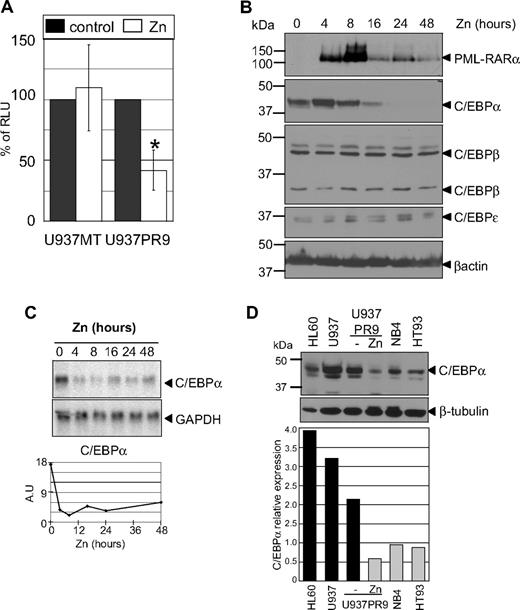
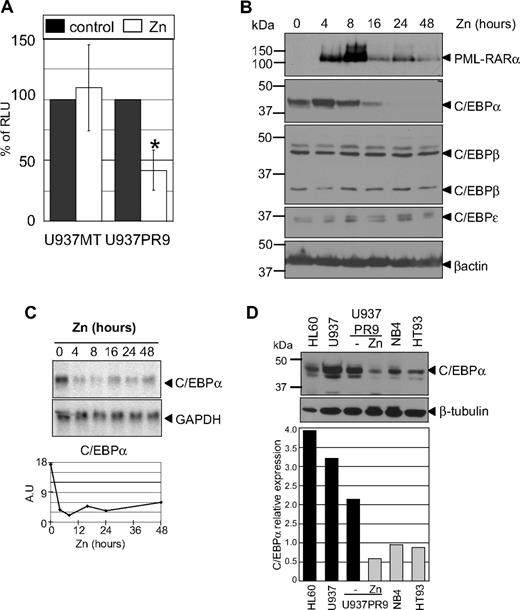
To further investigate whether the effect of PML-RARα was specific to C/EBPα and/or other C/EBP family members, we used human APL cell lines. U937PR9 cells can be induced to express PML-RARα after treatment with zinc sulfate (ZnSO4, zinc) and were used to assess C/EBP transcriptional activity. Cells were transfected with a luciferase reporter construct containing a tetramer of C/EBP binding sites.25 Expression of PML-RARα decreased the activity of the C/EBP reporter by 60% (P < .001). This effect was not the result of a nonspecific effect of zinc, as activity of the C/EBP reporter in U937 MT control cells did not change after treatment with zinc (Figure 5A).
PML-RARα induces a decrease of C/EBP activity as a result of decreased RNA and protein expression. (A) C/EBPα activity in myeloid cells with and without expression of PML-RARα. U937PR9 cells and U937 cells stably transfected with the empty vector (U937MT) or a vector expressing PML-RARα (U937PR9) were pretreated 16 hours with 100μM Zn2+SO4 (Zn) to induce PML-RARα expression. Then,15 million cells were electroporated with a vector driving expression of luciferase cloned in front of 4 C/EBP sites from the G-CSF receptor promoter25 together with a CMV-Renilla vector driving expression of Renilla luciferase.26 Cells were lysed 7 hours after electroporation. Firefly luciferase activity was normalized to Renilla luciferase activity (RLU). *P < .001. (B) Western blot analysis demonstrates a selective decrease in C/EBPα protein after induction of PML-RARα. U937PR9 cells were treated with 100μM Zn2+SO4 (Zn) for the indicated time. Cells were lysed at the indicated time in radioimmunoprecipitation assay buffer and separated onto 10% acrylamide-bis acrylamide gel. After transfer onto a PVDF membrane, the immmunoblot was incubated with an anti-RARα antibody and anti-C/EBPα, β, and ϵ antibodies. Loading control was assessed with an antibody against β-actin. (C) C/EBPα RNA decreases over several days after induction of PML-RARα. Northern blot of U937PR9 cell treated with 100μM Zn2+SO4 (Zn). Cells were collected at the indicated times. A total of 8 μg of RNA were electrophoresed and transferred onto a nylon filter. The filter was hybridized with a probe staining C/EBPα mRNA (top panel). GAPDH (bottom panel) was used to assess the equal loading of each sample. Signals were quantified using densitometric analysis of PhosphorImager data. The amount of C/EBPα mRNA was normalized to GAPDH expression and plotted over time according to arbitrary units (A.U.; ratio C/EBPα to GAPDH RNA). (D) C/EBPα protein expression is decreased in human myeloid cells expressing PML-RARα. C/EBPα protein was detected by Western blot analysis (top panel) in myeloid cell lines, which did not express PML-RARα (HL60, U937, and U937PR9 without zinc) and do express PML-RARα (U937PR9 + zinc, NB4, and HT93). Loading was assessed with an antibody against β-tubulin (bottom panel). The expression of C/EBPα was quantified using the ImageJ program (http://rsb.info.nih.gov/ij/) and normalized to the expression of β-tubulin, and plotted on the bar graph below the blot. Relative expression of C/EBPα (C/EBPα expression divided by β-tubulin expression) is represented with black bars for the cell lines that do not express PML-RARα (HL60, U937, and U937PR9 without zinc) and gray bars for the cell lines that do express PML-RARα (U937PR9 + zinc, NB4, and HT93).
PML-RARα induces a decrease of C/EBP activity as a result of decreased RNA and protein expression. (A) C/EBPα activity in myeloid cells with and without expression of PML-RARα. U937PR9 cells and U937 cells stably transfected with the empty vector (U937MT) or a vector expressing PML-RARα (U937PR9) were pretreated 16 hours with 100μM Zn2+SO4 (Zn) to induce PML-RARα expression. Then,15 million cells were electroporated with a vector driving expression of luciferase cloned in front of 4 C/EBP sites from the G-CSF receptor promoter25 together with a CMV-Renilla vector driving expression of Renilla luciferase.26 Cells were lysed 7 hours after electroporation. Firefly luciferase activity was normalized to Renilla luciferase activity (RLU). *P < .001. (B) Western blot analysis demonstrates a selective decrease in C/EBPα protein after induction of PML-RARα. U937PR9 cells were treated with 100μM Zn2+SO4 (Zn) for the indicated time. Cells were lysed at the indicated time in radioimmunoprecipitation assay buffer and separated onto 10% acrylamide-bis acrylamide gel. After transfer onto a PVDF membrane, the immmunoblot was incubated with an anti-RARα antibody and anti-C/EBPα, β, and ϵ antibodies. Loading control was assessed with an antibody against β-actin. (C) C/EBPα RNA decreases over several days after induction of PML-RARα. Northern blot of U937PR9 cell treated with 100μM Zn2+SO4 (Zn). Cells were collected at the indicated times. A total of 8 μg of RNA were electrophoresed and transferred onto a nylon filter. The filter was hybridized with a probe staining C/EBPα mRNA (top panel). GAPDH (bottom panel) was used to assess the equal loading of each sample. Signals were quantified using densitometric analysis of PhosphorImager data. The amount of C/EBPα mRNA was normalized to GAPDH expression and plotted over time according to arbitrary units (A.U.; ratio C/EBPα to GAPDH RNA). (D) C/EBPα protein expression is decreased in human myeloid cells expressing PML-RARα. C/EBPα protein was detected by Western blot analysis (top panel) in myeloid cell lines, which did not express PML-RARα (HL60, U937, and U937PR9 without zinc) and do express PML-RARα (U937PR9 + zinc, NB4, and HT93). Loading was assessed with an antibody against β-tubulin (bottom panel). The expression of C/EBPα was quantified using the ImageJ program (http://rsb.info.nih.gov/ij/) and normalized to the expression of β-tubulin, and plotted on the bar graph below the blot. Relative expression of C/EBPα (C/EBPα expression divided by β-tubulin expression) is represented with black bars for the cell lines that do not express PML-RARα (HL60, U937, and U937PR9 without zinc) and gray bars for the cell lines that do express PML-RARα (U937PR9 + zinc, NB4, and HT93).
To determine whether down-regulation of C/EBPα was responsible for the observed decrease of total C/EBP activity, expression levels of C/EBP family member proteins were analyzed by Western blot in U937PR9 cells. Both C/EBPβ and C/EBPϵ expression remained constant after induction of PML-RARα (Figure 5B), whereas C/EBPα protein was markedly reduced (3.2-fold and 19-fold after 16 and 24 hours, respectively, of zinc treatment; Figure 5B). Similarly, C/EBPα RNA levels were down-regulated (3.5-fold and 5-fold after 16 and 24 hours, respectively, of zinc treatment; Figure 5C), but no change was detected in the expression of C/EBPβ and C/EBPϵ RNA (data not shown). Similar treatment was performed in the U937 control cell line, and the levels of the C/EBP family members did not show any changes (supplemental Figure 6). The decrease in C/EBPα expression led to reduction of RNA expression of its target genes, including the G-CSF receptor25 as well as an increase in c-myc, an indirect target of C/EBPα in myeloid cells34 (data not shown). To demonstrate that the reduction in C/EBPα caused by PML-RARα was not restricted to U937PR9 cells, C/EBPα endogenous levels were compared in different human myeloid cell lines. Cells that do not express PML-RARα (ie, HL60, U937, and U937PR9 not treated with zinc) demonstrated significantly higher endogenous levels of C/EBPα protein than cell lines harboring PML-RARα (P < .014 between cells without and cells with PML-RARα; ie, NB4,22 HT93,23 and zinc-treated U937PR9), suggesting that PML-RARα expression leads to decreased levels of C/EBPα RNA and protein (Figure 5D). In conclusion, these experiments demonstrate that PML-RARα specifically down-regulates C/EBPα, and not other C/EBPs, and therefore that inhibition of C/EBPα expression results in the decrease of C/EBP activity.
C/EBPα down-regulation is the result of PML-RARα–mediated methylation
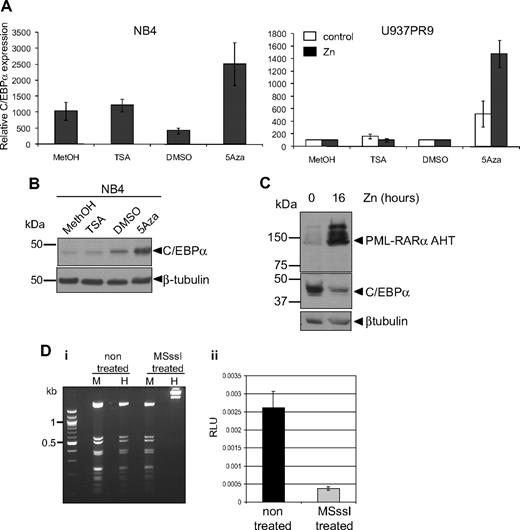
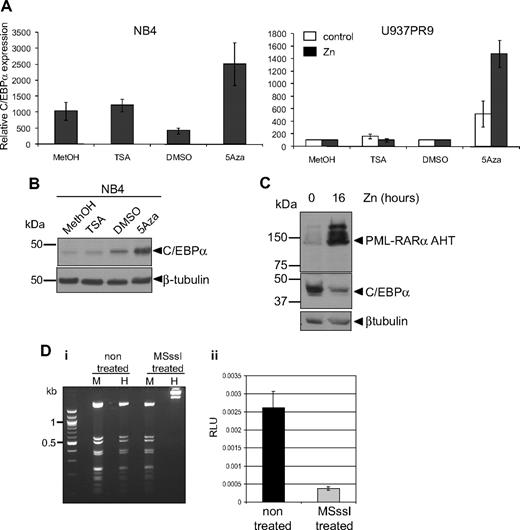
We further investigated the mechanisms leading to down-regulation of C/EBPα by PML-RARα. It has been shown that PML-RARα recruits a corepressor complex harboring histone deacetylase7 and DNA methylase activity.6 To determine whether C/EBPα repression was mediated by one of these pathways, APL cell lines (NB4 and HT93) were treated with either the DNA methylase inhibitor 5-Aza or the histone deacetylase inhibitor TSA. Treatment with 5-Aza resulted in a significant increase in C/EBPα RNA, whereas treatment with TSA had no significant effect (Figure 6A left graph). Induction of C/EBPα expression after treatment with 5-Aza could also be observed at the protein level (Figure 6B). Similarly, induction of PML-RARα in U937PR9 after zinc treatment (Figure 6A right graph, black bars) showed up-regulation of C/EBPα RNA in the presence of 5-Aza (Figure 6A right graph). In contrast, C/EBPα RNA was not affected by treatment with TSA with or without PML-RARα in U937PR9 cells (Figure 6A right graph). To further exclude that C/EBPα expression is repressed via a histone deacetylase pathway, we used a PML-RARα mutant (PML-RARα AHT), which is unable to recruit the histone deacetylase complex through its N-Cor domain.7,35 This mutant protein was expressed in U937 cells,7 and C/EBPα protein levels were assessed. In the presence of PML-RARα AHT, C/EBPα expression was repressed (Figure 6C), consistent with the results obtained with TSA treatment. We therefore conclude that PML-RARα–mediated repression of C/EBPα is driven through a DNA methylation pathway.
C/EBPα expression is repressed by methylation rather than by acetylation. (A) NB4 (left panel) or U937PR9 (right panel) treated (■) or not (□) every 24 hours with 100μM zinc (Zn) were treated for 72 hours with 1μM (NB4) or 5μM, respectively, 5-Aza or for 24 hours with 300nM TSA. C/EBPα mRNA expression was detected by quantitative real-time PCR and normalized to GAPDH expression. This graph is representative of 3 independent experiments. Methanol (MetOH) and dimethyl sulfoxide are vehicle controls for TSA and 5-Aza, respectively. (B) C/EBPα protein was assessed in NB4 cells treated with TSA or 5-Aza by Western blot. Equal loading of the protein was assessed with β-tubulin. (C) Western blot of C/EBPα protein in U937MT PML-RARα AHT mutant cells either untreated or treated for 16 hours with 100μM Zn. Equal loading was assessed with an antibody against β-tubulin. (D) In vitro methylation represses C/EBPα promoter activity. (Di) A vector expressing luciferase driving by C/EBPα promoter sequence was treated in vitro with the CpG methyltransferase MSssI. Efficiency of the methylation was assessed by digesting the non–MSssI-treated and the MSssI-treated vector with a methylation-insensitive enzyme MspI (M) and a methylation-sensitive enzyme, HpaII (H). (Dii) U937 cells were electroporated with the methylated (MSssI) or untreated vector (nontreated) driving luciferase activity. Shown are relative luciferase units (RLU) representing the ratio between the firefly activity driven by the C/EBPα promoter before and after methylase treatment normalized to the CMV Renilla activity driven by a control vector.26 When the C/EBPα promoter was methylated, relative luciferase activity was decreased by 85% compared with the activity obtained with the unmethylated vector (P < .001).
C/EBPα expression is repressed by methylation rather than by acetylation. (A) NB4 (left panel) or U937PR9 (right panel) treated (■) or not (□) every 24 hours with 100μM zinc (Zn) were treated for 72 hours with 1μM (NB4) or 5μM, respectively, 5-Aza or for 24 hours with 300nM TSA. C/EBPα mRNA expression was detected by quantitative real-time PCR and normalized to GAPDH expression. This graph is representative of 3 independent experiments. Methanol (MetOH) and dimethyl sulfoxide are vehicle controls for TSA and 5-Aza, respectively. (B) C/EBPα protein was assessed in NB4 cells treated with TSA or 5-Aza by Western blot. Equal loading of the protein was assessed with β-tubulin. (C) Western blot of C/EBPα protein in U937MT PML-RARα AHT mutant cells either untreated or treated for 16 hours with 100μM Zn. Equal loading was assessed with an antibody against β-tubulin. (D) In vitro methylation represses C/EBPα promoter activity. (Di) A vector expressing luciferase driving by C/EBPα promoter sequence was treated in vitro with the CpG methyltransferase MSssI. Efficiency of the methylation was assessed by digesting the non–MSssI-treated and the MSssI-treated vector with a methylation-insensitive enzyme MspI (M) and a methylation-sensitive enzyme, HpaII (H). (Dii) U937 cells were electroporated with the methylated (MSssI) or untreated vector (nontreated) driving luciferase activity. Shown are relative luciferase units (RLU) representing the ratio between the firefly activity driven by the C/EBPα promoter before and after methylase treatment normalized to the CMV Renilla activity driven by a control vector.26 When the C/EBPα promoter was methylated, relative luciferase activity was decreased by 85% compared with the activity obtained with the unmethylated vector (P < .001).
To further demonstrate that DNA methylation could down-regulate C/EBPα RNA, we investigated the effect of DNA methylation on C/EBPα promoter activity in vitro. A vector containing 2.2 kb of the human C/EBPα proximal promoter was cloned in front of a luciferase reporter gene, subjected to in vitro methylation by treatment with CpG methylase MSssI, and subsequently electroporated into U937 cells. Comparison of luciferase activity of the methylated versus the unmethylated vector revealed that the methylated C/EBPα promoter exhibited 86% reduction of its activity (P < .001; Figure 6Dii). Altogether, these results demonstrate that PML-RARα down-regulates C/EBPα through a process involving DNA methylation.
Discussion
The work presented here aimed at isolating and characterizing the APL-initiating cell population. Isolation of these cells will result in better understanding the mechanisms leading to the development and relapse of the disease. Using an approach combining cell-surface marker analysis and the property of myeloid cells to sequentially activate expression of granule genes, we developed a technique to isolate cells at specific stages along the myeloid differentiation pathway, including promyelocytes, in normal murine bone marrow. This novel flow cytometric strategy appears to be a very powerful tool to identify and isolate cells at specific stages of differentiation along the myeloid lineage. The same promyelocytic phenotype (CD34+, c-kit+, FcγRIII/II+, Gr1int) characterized a population of myeloid committed progenitors that accumulates in an APL murine model. This finding was consistent with previous observations of abnormal expression of CD34+ and Gr1+ in cells isolated from leukemic spleen and bone marrow of different APL murine models.36-38 In agreement with the human APL phenotype, these CD34+, c-kit+, FcγRIII/II+, Gr1int promyelocyte-like cells (high expression of primary granules and low or no detectable expression of secondary and tertiary granules) accumulate in hematopoietic sites in this model, with a 72-fold increase in bone marrow and a 19-fold increase in the spleen of leukemic mice. To determine whether accumulation of these cells in leukemic mice represented the cause or a consequence of the leukemia, we transplanted this and other subpopulations into recipient mice to analyze whether they were capable of initiating leukemia. Transplantation of KSL cells isolated from leukemic mice bone marrow failed to generate leukemia in recipient animals. Moreover, transplantation of late myeloid cells (subpopulation 9, CD34−, c-kit−, FcγRIII/II+, Gr1high cells) generated leukemia in recipient mice inefficiently and with long latency. By transplanting dilutions of subpopulations 3 and 4, we demonstrated that as few as 30 sorted cells were sufficient to generate leukemia in recipient animals with short latency, demonstrating that subpopulations 3 and 4 display properties of an APL-initiating cell. Moreover, by comparing the latencies of the number of transplanted subpopulation 3 and 4 cells versus the number of transplanted total bone marrow cells, we determined a 100-fold enrichment in this LIC compartment. This leukemia was further transplantable into secondary recipients. Leukemias initiated after injection of subpopulations 3 and 4 were similar to the ones generated by injection of total leukemic spleen cells, indicating that these cells have the ability to recapitulate the leukemic phenotype by themselves. Although our results suggest that the APL-initiating cell is the promyelocyte, we have not excluded other populations, such as KSL. KSL are not the targets of MRP8 hPML-RARα transgene expression in this model, as the MRP8 regulatory elements direct expression in myeloid cells rather than stem cells,39 and we could not detect PML-RARα RNA in KSL cells in this model by real-time PCR (data not shown). However, the hypothesis that APL-initiating cells are solely found in the committed myeloid promyelocyte-like population is consistent with previous findings in human APL.11 This result, along with our findings reported here, suggests that APL cancer stem cells have a more mature phenotype compared with those of other AMLs.40
APL is characterized by expression of the PML-RARα fusion protein and the ability of the cells to differentiate in the presence of ATRA. Genes reactivated during ATRA treatment and the mechanisms underlying their reactivation are well characterized.13,41,42 However, comprehension of the initiation of APL is poorly understood. Expression of PML-RARα is required to trigger the initiation of the disease, but the other mechanisms necessary for APL to fully develop are not entirely characterized.38,43-45 Additional gene deregulations or mutations have been shown to enhance APL development: loss of PU.1,13,17 coexpression of Fms-like tyrosine kinase 3-ITD,46 and expression of activated Kras.47 We found that, in APL-initiating cells, C/EBPα expression is reduced compared with their wild-type promyelocytic counterparts. The importance of C/EBPα dysregulation in APL development is highlighted by the fact that, when crossed with C/EBPα+/− mice, hCathG PML-RARα mice exhibit decreased latency and increased penetrance of APL. This result is supported by the observations that expression of C/EBPα in combination with ATRA treatment has a synergic effect on survival of leukemic MRP8 hPML-RARα mice compared with treatment with ATRA alone, and this effect is more marked for C/EBPα than other C/EBP family members, such as C/EBPϵ.48,49 Furthermore, loss of C/EBPα may contribute to induction of the differentiation block in APL through its role in regulation of the transcription factor PU.1.13 Down-regulation of C/EBPα expression and activity were detected in APL cell lines, and deregulation of C/EBPα has frequently been observed in AML, highlighting its importance in the maintenance of normal myelopoiesis.
Different mechanisms have been described to explain C/EBPα loss of function in cancers: mutations14,19,50 and transcriptional repression by aberrant DNA methylation in AML, including APL,12,51 as well as in solid tumors.52,53 In this study, the decrease in C/EBPα activity is not the result of expression of a mutated form, but rather a reduction of its expression. We detected a decrease of C/EBPα protein and RNA expression in the presence of PML-RARα, possibly resulting from a methylation-mediated mechanism, as observed in human cell lines. In accordance with this finding, a recent study in human APL samples described increased C/EBPα promoter methylation,51 consistent with the ability of PML-RARα to recruit corepressor complexes. However, the mechanisms underlying C/EBPα down-regulation are not fully identified yet. Careful in silico analysis of the C/EBPα proximal promoter sequence did not reveal any obvious consensus or degenerate retinoic acid response element binding sites. Moreover, the PML-RARα effect on C/EBPα repression does not seem to be mediated via direct binding to its proximal promoter, as we could not detect this activity by electrophoretic mobility shift assay performed on the 1-kb C/EBPα proximal promoter (data not shown). However, we cannot exclude the possible binding of PML-RARα to a distal regulatory element. We recently demonstrated that methylation of a distal regulatory region resulting in down-regulation of the transcription factor PU.1 in multiple myeloma.54 Preliminary studies of methylation status of murine C/EBPα promoter regions did not lead to the identification of a methylation pattern in leukemic cells (data not shown). The distal elements regulating C/EBPα remain unknown but could be targets of PML-RARα–mediated methylation leading to C/EBPα repression. Aberrant methylation of APL patients samples has recently been detected in a region just upstream of the C/EBPα promoter,51 approximately 1 kb upstream of the transcription start site, but we could not detect differential methylation of this region in our studies. PML-RARα recruitment to C/EBPα regulatory elements could also occur through an indirect mechanism. For instance, PML-RARα has been described to interact with Sp155 and to repress target genes without a canonical retinoic acid response element within their promoter.56 The C/EBPα proximal promoter contains 2 sites that bind to Sp1 by electrophoretic mobility shift assay (data not shown) that could enable recruitment of PML-RARα, and 2 potential Sp1 binding sites in the previously described methylated region upstream of the promoter.51 Identification and definition of the distal regulatory elements mediating C/EBPα expression will be necessary to test whether C/EBPα, like PU.1,54 can be down-regulated by DNA methylation of distal elements in specific diseases.
In conclusion, identification of APL cancer–initiating cells will lead to a greater understanding of the mechanisms underlying the initiation of APL. It remains to be demonstrated whether such understanding will lead to design of novel treatments targeting the residual cancer-initiating cells postulated to be responsible for relapse after effective treatment.10 In a more general perspective, this novel approach will be very useful for studying other AML subtypes and identification of their cancer-initiating cells. Similar sorting strategies will allow further characterization of the specific block along the differentiation program that occurs in different AML subtypes. In particular, a direct quantification of the subpopulations that aberrantly accumulate in leukemic bone marrows can be performed and compared with their normal counterpart subpopulations. Furthermore, this strategy offers the advantage of analyzing normal and malignant cells blocked at identical stages of differentiation, instead of comparing unfractionated cells isolated from healthy and leukemic bone marrow.50 Several attempts to characterize stage-specific myeloid cells by flow cytometry have been undertaken in healthy human samples.30 These studies have demonstrated that it is important to isolate specific subpopulations of AML cells to accurately assess dysregulation of critical gene targets.57,58 Therefore, it will be essential to use this strategy in the analysis of cancer-initiating cells in leukemia and other tumors to identify new mechanisms of leukemogenesis, particularly when dysregulation of gene expression is involved.
In conclusion, we demonstrate that a committed myeloid cell with a promyelocytic phenotype (CD34+, c-kit+, FcγRIII/II+, Gr1int) has the property of APL-initiating cells, whose transformation is caused by expression of the fusion protein PML-RARα associated with deregulation of the myeloid program resulting from down-regulation of C/EBPα.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Note added in proof:
Similar down-regulation of C/EBPα has been noted in human APL patient blasts. (Sukhai M, Thomas M, Goswami R, Xuan Y, Pintor dos Reis P, Kamel-Reid P. Deregulation of transcription factors GATA-1, GATA-2, and C/EBPα in acute promyelocytic leukemia. Blood (ASH Annual Meeting Abstracts). 2008;112:781a. Abstract 2241.)
Acknowledgments
The authors thank Pier-Giuseppe Pellicci and Francesco Grigani for the kind gift of U937AHT cells, Scott Kogan for the kind gift of leukemic spleens from leukemic founders no. 935 and no. 1111, all members of the Tenen laboratory for helpful discussions, Junyan Zhang for excellent assistance with mouse husbandry, Christopher Hetherington for quantitative real-time RT-PCR analysis, David Gonzalez for assistance in molecular experiments, and the Harvard Stem Cell and Dana-Farber Cancer Institute flow cytometry core facilities for expert assistance with multicolor flow cytometry and high-speed cell sorting.
This work was supported by the National Institutes of Health (NIH/NCI POICA66996) and the Harvard Stem Cell Institute (D.G.T.), the Fondation Recherche Médicale (F.C.G.), the European Hematology Association (M.A.-J.), Kakenhi (19591126, 20592125, and 21591246) and the Shimizu Foundation (Research Grant for 2006; H.H.), the Flight Attendant Medical Research Institute (E.L.), the Italian Foundation for Cancer Research (award Leonino Fontana e Maria Lionello; A.D.R.), and Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP; grants 01/07974-3 and 01/08693-8; B.A.S.-L.). This work was supported by the Burroughs Wellcome Fund (Career Award; A.J.W.).
National Institutes of Health
Authorship
Contribution: F.C.G. and M.A.-J. designed and performed research, analyzed data, and wrote the paper; H.H. designed and performed research and analyzed data; A.E., E.L., P.Z., B.A.S.-L., and A.D.R. performed research, analyzed data, and wrote the paper; D.N. analyzed data; A.J.W. contributed vital reagents; and E.M.R. and D.G.T. designed research, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The present address of H.H. is Department of Transfusion Medicine and Cell Therapy, Kyoto University, Kyoto, Japan.
Correspondence: Daniel G. Tenen, Center for Life Sciences, Rm 437, 3 Blackfan Cir, Boston, MA 02115; e-mail: dtenen@bidmc.harvard.edu.
References
Author notes
F.C.G. and M.A.-J. are dual first authors of this work.