Abstract
Pim kinases are involved in B-cell development and are overexpressed in B-cell chronic lymphocytic leukemia (CLL). We hypothesized that Pim kinase inhibition would affect B-cell survival. Identified from a screen of imidazo[1,2-b]pyridazine compounds, SGI-1776 inhibits Pim-1, Pim-2, and Pim-3. Treatment of CLL cells with SGI-1776 results in a concentration-dependent induction of apoptosis. To elucidate its mechanism of action, we evaluated the effect of SGI-1776 on Pim kinase function. Unlike in replicating cells, phosphorylation of traditional Pim-1 kinase targets, phospho-Bad (Ser112) and histone H3 (Ser10), and cell-cycle proteins were unaffected by SGI-1776, suggesting an alternative mechanism in CLL. Protein levels of total c-Myc as well as phospho-c-Myc(Ser62), a Pim-1 target site, were decreased after SGI-1776 treatment. Levels of antiapoptotic proteins Bcl-2, Bcl-XL, XIAP, and proapoptotic Bak and Bax were unchanged; however, a significant reduction in Mcl-1 was observed that was not caused by caspase-mediated cleavage of Mcl-1 protein. The mechanism of decline in Mcl-1 was at the RNA level and was correlated with inhibition of global RNA synthesis. Consistent with a decline in new RNA synthesis, MCL-1 transcript levels were decreased after treatment with SGI-1776. These data suggest that SGI-1776 induces apoptosis in CLL and that the mechanism involves Mcl-1 reduction.
Introduction
Pim (provirus integration site for Moloney murine leukemia virus) family proteins are highly conserved serine/threonine kinases that have been implicated in cancer progression and the development of resistance to chemotherapeutic agents (for a review, see Shah et al1 ). Three Pim kinases have been identified to date, Pim-1, -2, and -3, and elevated expression of Pim kinases have been detected in hematologic malignancies and in certain solid tumors.2-5 These kinases have similar active sites and lack regulatory domains and thus are constitutively active if expressed.6 The expression of Pim proteins is via the recruitment of the Janus kinase (JAK) after cytokine receptor activation, resulting in the induction of signal transducer and activator of transcription-driven transcription of PIM genes.7 Pim kinases have been shown to be involved in several signaling pathways, and the targets identified to date are associated with the regulation of apoptosis, cell-cycle progression, differentiation, transcription, proliferation, and tumorigenesis (reviewed by Amaravadi and Thompson6 ).
PIM1 is a coactivator of MYC, and Pim kinase phosphorylation of histone H3 at serine 10 leads to stimulation of RNA polymerase II binding, which results in increased c-Myc–driven transcription.8 It is approximated that PIM1 is required for the expression of 20% of total MYC target genes. Other target substrates include proapoptotic Bad protein, which is phosphorylated at multiple sites but predominantly at gatekeeper site Ser112 by all 3 Pim kinases.9,10 Phosphorylation of Bad leads to its sequestration from the mitochondrial surface to the cytosol by 14-3-3 and subsequent release of antiapoptotic proteins Bcl-XL and Bcl-2. Given the oncogenic nature of Pim kinases, there has been increasing interest in developing Pim kinase inhibitors for the treatment of cancer.
Pertaining to cancer biology, increased levels of Pim kinase proteins have been strongly implicated in cell survival and tumorigenesis. Elevated expression of Pim-1 has been demonstrated to induce genomic instability via disruptions in mitotic spindle checkpoints11 and also functions to protect cells from apoptosis induced by glucocorticoids,12 genotoxins,13 or cytokine withdrawal.14 Pim-1 also has been shown to interact in the p53 pathway via Mdm2.15 Pim-1 is overexpressed in lymphomas,16 acute leukemias,17 and prostate cancer,5 whereas increased expression of the human PIM2 proto-oncogene is observed in chronic lymphocytic leukemia (CLL) and non-Hodgkin lymphomas.3 Pim-2 also is required to confer rapamycin resistance in hematopoietic cells,18 and both Pim-1 and Pim-2 have been shown to be required for efficient pre–B-cell transformation by v-Abl oncogene.19 More recently, Pim-3 was reported to be aberrantly expressed in colon cancer.20 Taken together, these observations further support the rationale for the development of Pim inhibitors as therapeutic agents for hematologic malignancies.21
We hypothesized that CLL cells would be responsive to small-molecule Pim kinase inhibition and present an imidazo[1,2-b]pyridazine small molecule, SGI-1776 (Figure 1), as a Pim kinase inhibitor. CLL cells do not actively replicate DNA, and thus RNA-directed agents may be a valuable therapeutic strategy.22 Pim-1 has been shown to synergize with c-Myc in oncogenic transformation,8 and thus disruption of c-Myc activation may down-regulate c-Myc–driven oncogene transcription. We evaluated this new agent by using primary lymphocytes obtained from patients with CLL and demonstrated cytotoxicity in samples of heterogeneous patient populations. Apoptosis induction coupled with the inhibition of RNA synthesis was observed in CLL cells treated with SGI-1776. Specifically, Mcl-1 transcript and protein levels both decreased after drug treatment, whereas Bcl-2, Bcl-XL, and XIAP protein levels remained unchanged. Our results establish SGI-1776 as a potential agent for the treatment of CLL and further underscore the importance of Mcl-1 in CLL.23
Methods
Drugs and chemicals
SGI-1776 was obtained from SuperGen and was dissolved in dimethyl sulfoxide (DMSO) and stored at −20°C. All experiments, including a vehicle control, were conducted with 0.1% DMSO.
Patient samples
The present in vitro studies were conducted in primary lymphocytes obtained from patients with CLL (n = 23). For all investigations, freshly isolated leukemia lymphocytes were used. All patients signed a written informed consent to participate in this laboratory protocol, in accordance with the Declaration of Helsinki, which was approved by the institutional review board of the University of Texas M.D. Anderson Cancer Center.
Clinical laboratory end points
Determination of IgVH gene mutation status and ζ-chain–associated protein kinase-70 (ZAP-70) analysis for the patients in our study were conducted as described previously,24 and provided by the Chronic Lymphocytic Leukemia Research Consortium (University of California, San Diego, and the University of Texas M. D. Anderson Cancer Center). Fluorescent in situ hybridization analysis data were provided by the clinical cytogenetics laboratory, Department of Hematopathology at M. D. Anderson Cancer Center. The fluorescent in situ hybridization technique was used to detect the chromosome 17p deletion for p53 gene in CLL cells. The detailed methodology for the assay has been described previously.25 Chromosomal cytogenetics were determined by use of CLL lymphocytes at M. D. Anderson Cancer Center.
Isolation of lymphocytes
Whole blood was collected in heparinized tubes and diluted with phosphate-buffered saline (PBS) and layered onto Ficoll-Hypaque (specific gravity, 1.086; Life Technologies). Mononuclear cells were isolated using standard Ficoll-Hypaque density gradient centrifugation.26 The lymphocytes then were washed twice with cold PBS and resuspended in 10 mL of RPMI 1640 supplemented with 10% autologous plasma. The cell number and mean cell volume were determined by use of a Coulter channelyzer (Coulter Electronics). The lymphocytes were then maintained at a concentration of 107 cells/mL for all experiments.
Kinase assays
Kinase inhibition was measured by the use of radiometric assays performed by KinaseProfiler service (Millipore Corporate Headquarters). SGI-1776 was screened against more than 200 human kinases available commercially. Similar to other small molecule kinase inhibitors, SGI-1776 is an ATP competitive compound. To test selectivity, many protein kinase families were tested for inhibition with 1 μmol/L concentration of inhibitor. The “gold standard” direct radiometric assay format was used as described in detail by the manufacturer.27 In short, assays contained a peptide substrate, known purified recombinant human kinases, gamma-labeled ATP, magnesium ion, and a fixed concentration (1 μmol/L) of SGI-1776. Radioactive phosphorylated product was quantitated with and without inhibitor.
Once the selectivity of the inhibitor for kinase was defined, assays were performed to determine the inhibitory constant for Pim kinases species. Pim kinase inhibition by SGI-1776 was determined by the use of IC50 Profiler Express from Millipore. In a final reaction volume of 25 μL, 5 to 10 mU of Pim-1 (human)/Pim-2 (human)/Pim-3 (human) is incubated with 8 mmol/L of MOPS, pH 7.0; 0.2 mmol/L ethylene diamine tetraacetic acid; 100 μmol/L KKRNRTLTV;10 mmol/L MgAcetate; and [γ-32P-ATP] (specific activity approximately 500 cpm/pmol, concentration as required). The reaction was initiated by the addition of the MgATP mix. After incubation for 40 minutes at room temperature, the reaction was stopped by the addition of 5 μL of a 3% phosphoric acid solution. Then, 10 μL of the reaction was spotted onto a P30 filtermat and washed 3 times for 5 minutes in 75 mmol/L phosphoric acid and once in methanol before it was dried and measured via a scintillation counter.
Apoptosis assay
CLL cells were treated with DMSO alone or with 0.3, 1, 3, or 10 μmol/L SGI-1776 for 24 hours. Cells (106) were washed, resuspended in 100 μL of annexin binding buffer (Roche), and mixed with 5 μL of annexin–fluorescein isothiocyanate (FITC) solution (BD Pharmingen) and 5 μL of propidium iodide (PI; Sigma-Aldrich) solution with 2.5 μg/mL DNase-free RNase A (Roche). At least 10 000 cells were measured per sample by the use of a Becton Dickinson FACSCalibur flow cytometer. Caspase inhibitor N-benzyloxycarbonyl-valyl-alanyl-aspartyl-fluoromethylketone (ZVAD) was obtained from Alexis Biochemicals, and cells were treated with DMSO alone or 25 μmol/L ZVAD with or without 10 μmol/L SGI-1776 treatment for 24, 48, and 72 hours and then analyzed by flow cytometry.
Immunoblot analysis
CLL cells were treated with DMSO alone or with 0.3, 1, 3, or 10 μmol/L SGI-1776 for 4, 8, or 24 hours. The cell pellets were washed with ice-cold PBS and lysed at 4°C in radioimmunoprecipitation assay buffer supplemented with 1 mini Complete Protease Inhibitor (Roche) tablet per 10 mL of buffer, and lysate protein content was measured by the use of a DC protein assay kit (Bio-Rad) according to the manufacturer's instructions. Aliquots (30-50 μg) of total protein was loaded onto either 12% or 4% to 12% sodium dodecyl sulfate polyacrylamide gels and transferred to nitrocellulose membranes (GE Osmonics Labstore) as previously described.22 The membranes were blocked at room temperature for 1 hour in Odyssey blocking buffer (LI-COR Inc), then incubated with primary antibodies overnight at 4°C against the following: Bad (Cell Signaling Technologies), phospho-Bad (Ser112; Cell Signaling), Bak (Millipore), Bax (BD Pharmingen), Bcl-2 (Dako), Bcl-XL (BD Transduction Laboratories), c-Myc (clone C33; Santa Cruz Biotechnology Inc), c-Myc (Ser62; Abcam), caspase-3 (clone 8G10; Cell Signaling Technologies), glyceraldehyde 3-phosphate dehydrogenase (GAPDH; Cell Signaling), total histone H3 (Cell Signaling), histone H3(Ser10; Millipore), Mcl-1 (Santa Cruz), total p53 (Oncogene Research Products or Cell Signaling), PARP (BD Pharmingen), Pim-1 (clone 12H8; Santa Cruz), Pim-2 (Sigma), Pim-3 (Abgent), and XIAP (BD Transduction). After washing, membranes were incubated with infrared-labeled secondary antibodies (LI-COR Inc) for 1 hour and then visualized with the use of an LI-COR Odyssey Infrared Imager.
Uridine incorporation
CLL cells were incubated with SGI-1776 for 24 hours. One hour before the removal of each aliquot, 10 μCi/mL [3H]uridine was added to the cell cultures, and then the cells were analyzed by use of a multiscreen assay system (Millipore) as previously described.22 Radioactivity was measured in SGI-1776–treated and DMSO-treated (control) cells and expressed as percent of control.
Real-Time reverse-transcription polymerase chain reaction (RT-PCR)
RNA was isolated with the RNeasy Mini kit (QIAGEN, Inc), and the gene expression levels were measured on an ABI Prism 7900 Sequence Detection System (Applied Biosystems) by the use of one-step real-time TaqMan RT-PCR. Taqman primers and probes for MCL-1 and 18S were purchased from Applied Biosystems. Each RNA sample was assayed in triplicate, and the relative gene expression levels were normalized with 18S.
Results
Identification of SGI-1776 as a lead compound
The X-ray structure of Pim-1 kinase in complex with AMP-PNP (pdb entry: 1XR1) was used for reference protein coordinates in the creation of a model for virtual screening. A large number of external and internal databases of virtual compounds were prepared for screening. Predictive calculations of pharmaceutical properties were used to select compounds. Virtual “hits” from this screen were evaluated and refined to a selection of approximately 250 structures. These were further evaluated and analyzed by the use of criteria for solubility, permeability, molecular weight, and Lipinski-like criteria that narrowed the “hits” to approximately 100. Two chemical scaffolds were selected for evaluation in in vitro assays: imidazo[1,2-b]pyridazines was one of the scaffolds. Comparative evaluation of these 2 pharmacophores suggested the imidazo[1,2-b]pyridazines series exhibited more favorable substitution capabilities and better drug-like properties. Further optimization of this scaffold led to the identification of SGI-1776 as the candidate for further development.
Pim kinase inhibition by SGI-1776
SGI-1776 at a concentration of 1 μmol/L was screened against a panel of kinases by the use of radiolabeled biochemical assays and was found to be highly selective for Pim kinases without any effects on cell cycle kinases, including CDKs and Aurora kinases. Other kinases, such as Chk1, IκB kinase (IKK), c-Jun N-terminal kinase (JNK), Abl, Raf, and mitogen-activated protein kinases, also were not affected. Protein kinase A and B and phosphoinositide-3 kinase (PI3K) activity ranged between 90% and 100% (Figure 1B). In addition to Pim kinases, there was a 40% decrease in c-Kit kinase activity. Flt-3 and TrkA were the 2 other kinases that were inhibited by SGI-1776 at this concentration. Additional assays, using wide range of SGI-1776 concentrations, were performed to determine inhibitory constants for 3 Pim kinase species. Specifically, the half-maximal inhibitory concentration (IC50) values for Pim-1, Pim-2, and Pim-3 were 7 nmol/L, 363 nmol/L, and 69 nmol/L, respectively (Figure 1C). The other 2 enzymes affected at nanomolar concentrations of SGI-1776 were Flt-3 (44 nmol/L) and haspin (34 nmol/L).
Structure and functional activity of imidazo[1,2-b]pyridazine compound SGI-1776. (A) Chemical structure of SGI-1776. (B) Kinase inhibition by SGI-1776. Several human kinases were tested for selective inhibition by 1 μmol/L SGI-1776 using Millipore kinase profiler assay as described in “Kinase assays.” (C) In vitro kinase assays of Pim-1, -2, and -3 with varying concentrations of SGI-1776 to determine IC50 values for each Pim kinase species. The IC50 Profiler Express Assay is described in “Kinase assays.”
Structure and functional activity of imidazo[1,2-b]pyridazine compound SGI-1776. (A) Chemical structure of SGI-1776. (B) Kinase inhibition by SGI-1776. Several human kinases were tested for selective inhibition by 1 μmol/L SGI-1776 using Millipore kinase profiler assay as described in “Kinase assays.” (C) In vitro kinase assays of Pim-1, -2, and -3 with varying concentrations of SGI-1776 to determine IC50 values for each Pim kinase species. The IC50 Profiler Express Assay is described in “Kinase assays.”
Expression of Pim kinases in CLL lymphocytes
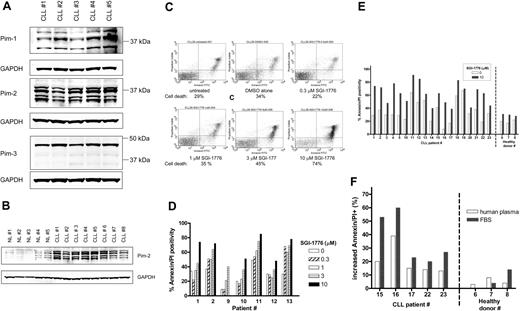
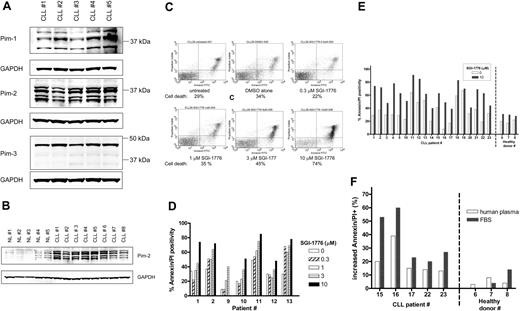
Expression of the target protein, Pim kinase, in CLL cells was determined by immunoblot analysis (Figure 2A), and various isoforms for both Pim-1 and Pim-2 were detected, whereas Pim-3 protein was detected largely as a single band. Because PIM2 gene expression levels were previously reported to be greater in CLL,3,28 we evaluated the relative Pim-2 protein levels in CLL cells compared with normal lymphocytes from healthy donors (Figure 2B). Significantly greater levels of Pim-2 protein were detected in untreated CLL cells (n = 8) compared with untreated normal lymphocytes (n = 5).
Expression of Pim kinase and the effect of SGI-1776 on apoptosis induction in CLL primary cells by SGI-1776. (A) Protein expression of Pim-1, -2, and -3 in untreated CLL primary cells. CLL lymphocytes were lysed and analyzed by immunoblot for Pim kinases with GAPDH as a loading control. (B) Elevated Pim-2 protein levels in CLL primary cells compared with normal lymphocytes. CLL lymphocytes and normal lymphocytes obtained from healthy donors were analyzed by immunoblot for Pim-2 protein as described in panel A. The IgVH mutation status for the CLL patients were as follows: unmutated (nos. 1, 4, 5, and 6), mutated (nos. 3, 7, 8), and undetermined (no. 2). (C) Flow cytometry analysis of annexin-FITC/PI staining of CLL primary cells (patient no. 1) that were either untreated; treated with 0.1% DMSO vehicle alone; or with 0.3, 1, 3, or 10 μmol/L SGI-1776 for 24 hours. (D) Graph of cell death in CLL samples (n = 7) from treatment with either 0, 0.3, 1, 3, or 10 μmol/L SGI-1776 for 24 hours. (E) Graph of cell death in untreated samples or samples treated with 10 μmol/L SGI-1776 for 24 hours in CLL (n = 19) and lymphocytes from healthy donors (n = 3). (F) Graph of the increase in apoptosis in CLL cells treated with 10 μmol/L SGI-1776 for 24 hours cultured in either media supplemented with autologous human plasma or FBS. Annexin V binding assay was used for apoptosis assay as described in “Apoptosis assay.”
Expression of Pim kinase and the effect of SGI-1776 on apoptosis induction in CLL primary cells by SGI-1776. (A) Protein expression of Pim-1, -2, and -3 in untreated CLL primary cells. CLL lymphocytes were lysed and analyzed by immunoblot for Pim kinases with GAPDH as a loading control. (B) Elevated Pim-2 protein levels in CLL primary cells compared with normal lymphocytes. CLL lymphocytes and normal lymphocytes obtained from healthy donors were analyzed by immunoblot for Pim-2 protein as described in panel A. The IgVH mutation status for the CLL patients were as follows: unmutated (nos. 1, 4, 5, and 6), mutated (nos. 3, 7, 8), and undetermined (no. 2). (C) Flow cytometry analysis of annexin-FITC/PI staining of CLL primary cells (patient no. 1) that were either untreated; treated with 0.1% DMSO vehicle alone; or with 0.3, 1, 3, or 10 μmol/L SGI-1776 for 24 hours. (D) Graph of cell death in CLL samples (n = 7) from treatment with either 0, 0.3, 1, 3, or 10 μmol/L SGI-1776 for 24 hours. (E) Graph of cell death in untreated samples or samples treated with 10 μmol/L SGI-1776 for 24 hours in CLL (n = 19) and lymphocytes from healthy donors (n = 3). (F) Graph of the increase in apoptosis in CLL cells treated with 10 μmol/L SGI-1776 for 24 hours cultured in either media supplemented with autologous human plasma or FBS. Annexin V binding assay was used for apoptosis assay as described in “Apoptosis assay.”
Apoptosis induction by SGI-1776 in CLL lymphocytes
CLL lymphocytes were treated with varying concentrations of SGI-1776, then stained with annexin V-FITC/PI and analyzed by flow cytometry to measure apoptosis induction (Figure 2C). In vitro incubation of primary CLL cells (n = 7), with 1, 3, and 10 μmol/L SGI-1776 for 24 hours resulted in an average increase in apoptosis of 10%, 22%, and 38%, respectively, compared with untreated cells (Figure 2D). Incubation of CLL cells with SGI-1776 for 48 or 72 hours further increased the percentage of apoptotic cells (data not shown).
To compare the cytotoxicity of SGI-1776 in CLL cells and normal lymphocytes, additional CLL samples (n = 19) were evaluated, and normal lymphocytes from the peripheral blood of healthy donors was obtained (n = 3). Both types of cells were treated with 10 μmol/L SGI-1776 for 24 hours, and the levels of apoptosis were measured by annexin V-FITC/PI staining (Figure 2E). Because more than 95% of the SGI-1776 is bound to human plasma protein, at this concentration (10 μmol/L), the free level of the drug is less than 500 nmol/L. Excluding samples that were majority spontaneously apoptotic, the average increase in apoptosis in treated CLL cells was 32%. In contrast, little-to-no cytotoxicity was observed in the lymphocytes from healthy donors, where the increase in apoptosis averaged only 5%. These experiments were conducted with cells cultured in media supplemented with autologous plasma, and similar results were obtained when cells were cultured with fetal bovine serum (FBS)–supplemented media, with even greater cytotoxicity toward CLL cells (Figure 2F). These data further elucidate that SGI-1776 binds to human plasma much more than to the FBS protein.
To determine whether caspase activation is associated with SGI-1776–induced apoptosis in CLL lymphocytes, ZVAD was used as a pan-caspase inhibitor. CLL cells were treated with vehicle alone, 10 μmol/L SGI-1776, or 10 μmol/L SGI-1776 in combination with 25 μmol/L ZVAD, then stained for annexin V-FITC/PI and analyzed by flow cytometry after 24 and 48 hours (data not shown). The inclusion of ZVAD did not decrease the levels of apoptotic cells; thus, caspase activation does not appear to be critical for the mechanism of action SGI-1776 in CLL. In separate experiments, ZVAD treatment blocked fludarabine-induced apoptosis (data not shown). SGI-1776–induced apoptosis was observed in heterogeneous patient populations, and there was disparity in the expression levels of traditional CLL prognostic markers, including ZAP-70, β2-microgobulin, IgVH mutation status, Rai stage, and number of previous treatments (supplemental Table 1, available on the Blood website; see the Supplemental Materials link at the top of the online article).
Effects of SGI-1776 on Pim kinase targets
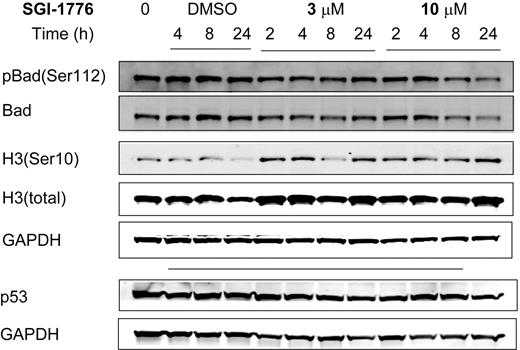
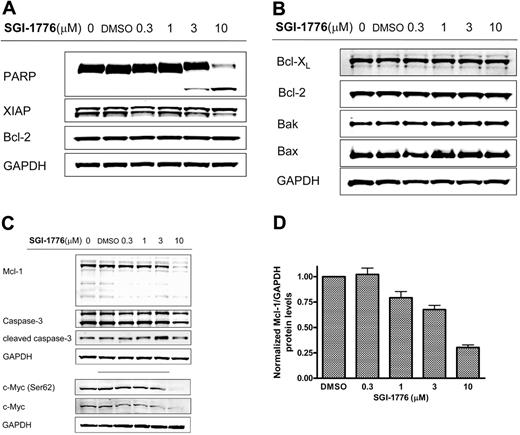
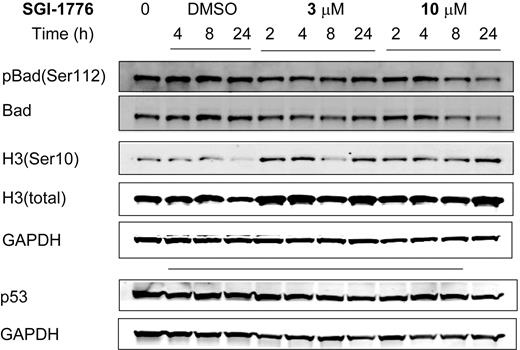
To elucidate its mechanism of action, we evaluated the effect of SGI-1776 on Pim kinase function. CLL cells were treated with vehicle DMSO alone or 3 or 10 μmol/L SGI-1776; harvested after 2, 4, 8, and 24 hours; and compared with untreated CLL cells. The total and phosphorylated protein levels of traditional Pim-1 kinase targets Bad and histone H3 then were analyzed by immunoblot analysis. Phosphorylation of Bad at Ser112 and histone H3 at Ser10 residues was not decreased by SGI-1776 treatment in CLL, unlike in replicating cell types, suggesting an alternative mechanism in CLL (Figure 3). Stabilization of p53 was not observed in SGI-1776–treated CLL cells as detected by immunoblot using a p53 antibody (Oncogene Research Products). To further confirm this reactivity, 3 additional samples were tested with different p53 antibodies (Cell Signaling). Pim-1 protein levels also were unaffected by SGI-1776 treatment in CLL cells (supplemental Figure 1). Apoptosis induction was further confirmed by immunoblot of PARP cleavage (Figure 4A); however, there was no significant change observed in the expression level of antiapoptotic proteins Bcl-2, XIAP (Figure 4A), or Bcl-XL (Figure 4B) by immunoblot. There were also no observed changes in the protein levels for proapoptotic Bak or Bax (Figure 4B).
Immunoblot analysis of potential target proteins in CLL primary cells treated with SGI-1776. CLL primary cells (patient no. 5) were either untreated, treated with 0.1% vehicle DMSO alone, or treated with 3 or 10 μmol/L SGI-1776, and cells were harvested after 2, 4, 8, and 24 hours and lysed. The protein levels of pBad(Ser112), total Bad, histone H3(Ser10), total histone H3, and p53 were analyzed by the use of immunoblot and normalized with GAPDH as a loading control.
Immunoblot analysis of potential target proteins in CLL primary cells treated with SGI-1776. CLL primary cells (patient no. 5) were either untreated, treated with 0.1% vehicle DMSO alone, or treated with 3 or 10 μmol/L SGI-1776, and cells were harvested after 2, 4, 8, and 24 hours and lysed. The protein levels of pBad(Ser112), total Bad, histone H3(Ser10), total histone H3, and p53 were analyzed by the use of immunoblot and normalized with GAPDH as a loading control.
Immunoblot analysis of anti- and proapoptotic proteins and cell death in CLL cells treated with SGI-1776. CLL primary cells were treated with 0.1% vehicle DMSO alone or 0.3, 1, 3, or 10 μmol/L SGI-1776 for 24 hours, then harvested and lysed. The protein levels of (A) PARP, XIAP, and Bcl-2 were immunoblotted from lysates (patient no. 2); (B) Bcl-XL, Bcl-2, Bak, and Bax (patient no. 4); and (C) Mcl-1, caspase-3, and cleaved caspase-3, total c-Myc, and phospho c-Myc(Ser62) were analyzed by immunoblot and normalized by the use of GAPDH as a loading control. (D) Quantitation of Mcl-1 protein levels normalized to GAPDH levels in CLL cells (patients nos. 1-4) treated with SGI-1776 as detected by immunoblot. The results represent the mean ± SEM from 4 patient samples.
Immunoblot analysis of anti- and proapoptotic proteins and cell death in CLL cells treated with SGI-1776. CLL primary cells were treated with 0.1% vehicle DMSO alone or 0.3, 1, 3, or 10 μmol/L SGI-1776 for 24 hours, then harvested and lysed. The protein levels of (A) PARP, XIAP, and Bcl-2 were immunoblotted from lysates (patient no. 2); (B) Bcl-XL, Bcl-2, Bak, and Bax (patient no. 4); and (C) Mcl-1, caspase-3, and cleaved caspase-3, total c-Myc, and phospho c-Myc(Ser62) were analyzed by immunoblot and normalized by the use of GAPDH as a loading control. (D) Quantitation of Mcl-1 protein levels normalized to GAPDH levels in CLL cells (patients nos. 1-4) treated with SGI-1776 as detected by immunoblot. The results represent the mean ± SEM from 4 patient samples.
However, in contrast, Mcl-1 protein levels were markedly reduced after SGI-1776 treatment, in a dose-dependent manner. After 24 hours, there was a decrease in Mcl-1 protein levels at 1, 3, and 10 μmol/L SGI-1776 (Figure 4C). The reduction in Mcl-1 protein does not appear to be as a result of caspase-mediated Mcl-1 cleavage because there was no detectable increase in the cleaved Mcl-1. Consistent with this observation, there was no increase in caspase-3 activation by cleavage of procaspase-3. At 1 and 3 μmol/L SGI-1776, the Mcl-1 protein levels were approximately 80% and 70% of vehicle control, and at 10 μmol/L SGI-1776, the Mcl-1 protein was approximately only 30% of vehicle control cells (Figure 4D). Because MCL-1 is one of the c-Myc–driven genes, we evaluated the effect of SGI-1776 on phosphorylation of Ser62 of c-Myc, one of the target phosphorylation sites for Pim kinases. Further immunoblot analysis revealed a decrease in both c-Myc(Ser62) and total c-Myc protein levels after SGI-1776 treatment (Figure 4C).
Inhibition of RNA synthesis by SGI-1776 in CLL lymphocytes
Because the reduction in Mcl-1 protein did not appear to be a result of Mcl-1 cleavage, we examined the potential transcription inhibition by SGI-1776 and evaluated the potential inhibition of c-Myc–driven transcription by measuring total RNA synthesis. After treatment with 0.3, 1, 3, or 10 μmol/L SGI-1776, there was a decrease in total RNA synthesis to approximately 50% of control at 10 μmol/L, which was measured with a radioactive uridine incorporation assay (n = 3; Figure 5A). Interestingly, in one patient sample, there was no observed inhibition in total RNA synthesis (CLL #14). However, when specific transcripts were measured by the use of real-time RT-PCR and normalized to 18S transcripts, in all samples, there was a decrease in Mcl-1 transcript levels in a time-dependent (Figure 5B) and dose-dependent (Figure 5C) manner. At 10 μmol/L SGI-1776, a decrease in Mcl-1 transcript levels can be detected as early as 4 hours after treatment, and after 8 hours, the transcript levels decrease to less than 50% of control (Figure 5B). A dose–response also was observed, and after 24 hours, Mcl-1 transcript levels were approximately 80% of control at 1 and 3 μmol/L SGI-1776, which was further reduced to approximately 35% of control at 10 μmol/L SGI-1776 (Figure 5C).
(A) Inhibition of RNA synthesis in CLL primary cells treated with SGI-1776. CLL primary cells (patients 3, 4, and 14) were incubated with 0.1% DMSO alone or with 0.3, 1, 3, or 10 μmol/L SGI-1776 for 24 hours. Then, 1 hour before the cells were harvested, [3H]uridine was added to the cell culture as described in “Uridine incorporation.” The results represent an average of triplicate experiments ± SEM. (B) Time-dependent reduction of MCL-1 transcript levels in CLL cells treated with SGI-1776. CLL primary cells (patients nos. 15, 16, and 17) were treated with 0.1% vehicle DMSO alone or 10 μmol/L SGI-1776 for 0, 2, 4, 8, and 24 hours, and then the RNA was isolated. MCL-1 transcript levels were measured by the use of real-time RT-PCR and normalized with 18S transcripts. Each RNA sample was assayed in triplicate, and the results are expressed as a percentage of the MCL-1 expression level in cells treated with vehicle DMSO alone. The results represent an average ± SEM of triplicate experiments. (C) Dose-dependent reduction of MCL-1 transcript levels in CLL cells treated with SGI-1776. CLL primary cells (patients nos. 2, 4, 12, and 14) were treated with 0.1% vehicle DMSO alone or with 0.3, 1, 3, or 10 μmol/L SGI-1776 for 24 hours, and transcript levels were measured in triplicate using real-time RT-PCR, and normalized with 18S transcript.
(A) Inhibition of RNA synthesis in CLL primary cells treated with SGI-1776. CLL primary cells (patients 3, 4, and 14) were incubated with 0.1% DMSO alone or with 0.3, 1, 3, or 10 μmol/L SGI-1776 for 24 hours. Then, 1 hour before the cells were harvested, [3H]uridine was added to the cell culture as described in “Uridine incorporation.” The results represent an average of triplicate experiments ± SEM. (B) Time-dependent reduction of MCL-1 transcript levels in CLL cells treated with SGI-1776. CLL primary cells (patients nos. 15, 16, and 17) were treated with 0.1% vehicle DMSO alone or 10 μmol/L SGI-1776 for 0, 2, 4, 8, and 24 hours, and then the RNA was isolated. MCL-1 transcript levels were measured by the use of real-time RT-PCR and normalized with 18S transcripts. Each RNA sample was assayed in triplicate, and the results are expressed as a percentage of the MCL-1 expression level in cells treated with vehicle DMSO alone. The results represent an average ± SEM of triplicate experiments. (C) Dose-dependent reduction of MCL-1 transcript levels in CLL cells treated with SGI-1776. CLL primary cells (patients nos. 2, 4, 12, and 14) were treated with 0.1% vehicle DMSO alone or with 0.3, 1, 3, or 10 μmol/L SGI-1776 for 24 hours, and transcript levels were measured in triplicate using real-time RT-PCR, and normalized with 18S transcript.
Discussion
Pim kinase inhibitors are emerging as a new class of cancer therapeutics, and the prevalence of increased Pim kinase expression across different cancer types suggests that Pim inhibitors may be a treatment modality for a variety of cancers. A recent report29 describes the development of a Pim-1 mAb as a cancer therapeutic. The crystal structure for the active site of Pim-1 has been reported,30 and an interesting feature of the enzyme is the presence of a proline residue in the location typically occupied by a methionine in the hinge region of other kinases, which alters the substrate specificity of the active site. This results in the absence of a canonical hydrogen bond donor in the hinge region that is a determining element for inhibitor binding in numerous kinases.31 Initially, several small molecules with common core scaffolds, including bisindolylmaleimides and flavonoids, were identified to be inhibitory toward Pim kinases.32 More recently, structural analysis identified imidazo[1,2-b]pyridazines as Pim kinase inhibitors with in vitro antileukemic activity,33 and SGI-1776 (Figure 1A) was identified as a lead compound.
Previously, overexpression of Pim kinase gene was reported in CLL.3 A direct comparison of the Pim 2 proteins in CLL versus normal lymphocytes elucidated over expression of Pim 2 protein (Figure 2B). Pim-1 has been reported to exist as 3 forms, as a 44-kDa form (Pim-1L), a 33-kDa form (Pim-1S), and a third 37-kDa form that is believed to be a posttranslationally modified variant. Both Pim-1L and Pim-1S exhibit kinase activity,34,35 and in the CLL lymphocytes analyzed in this report, all 3 forms were detected. Pim-2 has been reported as 3 isoforms and Pim-3 as 1 isoform, and our results are consistent.10,36
SGI-1776 at nanomolar concentrations inhibited all 3 Pim kinases (Figure 1C). In contrast, during whole-cell studies in which primary CLL lymphocytes were used, 3 and 10 μmol/L drug levels were needed for a biologic effect. This difference in concentration may be attributable to greater than 95% binding of the SGI-1776 to protein in human plasma. Consistent with this postulate, CLL cells incubated with 10% FBS rather than 10% autologous serum had an increased apoptosis with SGI-1776.
In vivo animal model system data clearly demonstrated that the required biologic dose of SGI-1776 was achieved in plasma. When experiments were conducted in mice, at the 300 mg/kg oral dosing, the plasma concentrations of the drug reached to approximately 3 μmol/L with a half life of 6 hours, and the oral bioavailability was 40%. This is the free (unbound to plasma protein) drug in plasma and, interestingly, the concentration of the drug is much greater in tumor cells compared with plasma levels. For example, at 140 mg/kg dose, the concentration of the drug in tumor at 4 and 12 hours was 10-fold and greater than 25-fold higher, respectively.
Because SGI-1776 targets, Pim kinases, were expressed in CLL cells, we explored the mechanism of action of SGI-1776 in these leukemic lymphocytes. Although the phosphorylation of Pim targets such as Bad, histone H3, p21, and p27 were not affected by SGI-1776, apoptosis induction was observed in CLL lymphocytes. Given the quiescent nature of CLL lymphocytes once exited from the bone marrow, it is consistent that traditional Pim targets were unaffected by SGI-1776.
A possibility exists that other kinases inhibited by SGI-1776 (Figure 1B) may play significant roles in CLL. However, Flt-3 is largely associated with acute myeloid leukemia (AML),37 and thus SGI-1776 also may be a viable strategy for the treatment of AML but not a target in CLL. Similarly, TrkA is also associated with AML and neurogenesis,38,39 without any association to CLL identified to date. Likewise, haspin is a mitotic histone kinase involved with metaphase and chromosome alignment and thus is unlikely to be a critical component in replicationally quiescent CLL signaling.40,41
Recent studies have demonstrated that Pim-1 and Pim-2 stabilize c-Myc protein in vivo,42 specifically through phosphorylation at Ser62 of c-Myc. Unlike other proteins associated with proliferation, gene expression of MYC is independent of cell cycle.43 Despite relatively inactive cell cycling, CLL lymphocytes actively synthesize RNA for maintenance that is driven by proto-oncogenes such as MYC, and hence we determined the effect of SGI-1776 on c-Myc phosphorylation. Treatment of CLL cells with SGI-1776 resulted in reduction in phospho-Myc (Ser62), correlating with a decrease in total c-Myc protein levels. In agreement, there was a dose-dependent inhibition of RNA in SGI-1776–treated CLL lymphocytes. Thus, the SGI-1776–mediated reduction in RNA synthesis may result from the combination of disrupted Pim-associated c-Myc–driven transcription and destabilization of c-Myc protein.
It is approximated that MYC binds up to 15% of all human genes,44 and MCL-1 is one of the c-Myc target genes.45 BAX and BCL2 are also c-Myc targets; however, both protein half lives are greater than 10 hours,46,47 whereas the half-life for Mcl-1 is less than 2 hours.48 In agreement, Bax and Bcl-2 proteins levels remain unaffected, whereas significant reduction in both the Mcl-1 transcript (Figure 5B-C) and protein levels (Figure 4C-D) were observed after SGI-1776 treatment in CLL lymphocytes. Consistent with our observations, inhibition of c-Myc expression with antisense oligonucleotides correlates with loss of the antiapoptotic protein Mcl-1.49
Although there is no specific genetic marker for CLL, Mcl-1 protein has been demonstrated to be of particular importance for CLL cell survival, and depletion of Mcl-1 protein via transcription and/or translation inhibitors results in cell death.23 Of clinical significance, high levels of Mcl-1 mRNA and protein have been inversely correlated with in vitro response to chemotherapeutic agents and with the failure of response to fludarabine in CLL patients.50 Several factors from the microenvironment provide considerably increased Mcl-1 protein levels in CLL cells51 and occur via increased transcription of the MCL-1 gene. Disruption of c-Myc–driven transcription is advantageous for targeting Mcl-1 protein as a result of the prevalence of AU-rich elements in its transcripts, leading to rapid turnover of MCL-1 mRNA. Hence it will be important to evaluate SGI-1776 when cells are cocultured with stromal cells to simulate the microenvironment.
Our results suggest that SGI-1776 may be a new therapeutic for the treatment of CLL. A recent report52 revealed that Pim-1 kinase is regulated by heat shock proteins and that Hsp90 inhibitor geldanamycin prevents the heat-shock stabilization of Pim-1. As such, we are currently investigating SGI-1776 treatment with Hsp90 inhibitors as a combination strategy in CLL. To understand the implications of microenvironment factors on Pim kinases, we are also evaluating the effect of stromal cell support on Pim kinase expression and sensitivity to SGI-1776. Taken together, Pim kinase inhibition may be a valuable approach toward cancer therapy, and further investigation is warranted.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Yuling Chen for obtaining blood samples and Susan Lerner for providing information on patient characteristics and clinical laboratory observations. V.G. and W.G.W. are members of the CLL Research Consortium.
This work is supported in part by grant CA57629 from the National Cancer Institute, Department of Health and Human Services.
National Institutes of Health
Authorship
Contribution: L.S.C. designed research, performed experiments, and analyzed results; V.G. supervised research and analyzed data; W.G.W. identified patients and reviewed the manuscript; D.B. and S.R. provided input on the compound and its primary and secondary pharmacodynamic activities; and L.S.C and V.G. wrote the paper.
Conflict-of-interest disclosure: S.R. and D.B. are employees of SuperGen Inc. The remaining authors declare no competing financial interests.
Correspondence: Varsha Gandhi, Department of Experimental Therapeutics and the Department of Leukemia, University of Texas M. D. Anderson Cancer Center, 1515 Holcombe Blvd, Houston, TX 77030; e-mail: vgandhi@mdanderson.org.

![Figure 1. Structure and functional activity of imidazo[1,2-b]pyridazine compound SGI-1776. (A) Chemical structure of SGI-1776. (B) Kinase inhibition by SGI-1776. Several human kinases were tested for selective inhibition by 1 μmol/L SGI-1776 using Millipore kinase profiler assay as described in “Kinase assays.” (C) In vitro kinase assays of Pim-1, -2, and -3 with varying concentrations of SGI-1776 to determine IC50 values for each Pim kinase species. The IC50 Profiler Express Assay is described in “Kinase assays.”](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/114/19/10.1182_blood-2009-03-212852/4/m_zh89990944160001.jpeg?Expires=1767813768&Signature=rOIta89fZIt0wGZFHK3L2nwQ~I29lL9Jiea1JOJlwsg~CEPwCX37B-aH00AI8leBBQ5jqScTv1a2GRReje5vMB2E5nC8i3zb-0N~SBNrfJ2wr3Jfv6KEkt1FpPZdtC~qmLcPE6IRcXdPqDEchdUSWHy5qz8PRsioOcQOECADGOwgdfBq2tbhL9YwMaYWS6Me1Oy5vGx-TjOtoq1N6toffNc8MjRMIOJCg~STgQIvSTSb6ffUYb4CM8XrKkt-S3XBLB-VMDxhnmnZ~3i4YtGo27c4St24jeIDgFM2TtoH6S~ccwGtvwqwhjqNXKOB9j4F-7PdFcVc0DFEFM7JVTzL4Q__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 5. (A) Inhibition of RNA synthesis in CLL primary cells treated with SGI-1776. CLL primary cells (patients 3, 4, and 14) were incubated with 0.1% DMSO alone or with 0.3, 1, 3, or 10 μmol/L SGI-1776 for 24 hours. Then, 1 hour before the cells were harvested, [3H]uridine was added to the cell culture as described in “Uridine incorporation.” The results represent an average of triplicate experiments ± SEM. (B) Time-dependent reduction of MCL-1 transcript levels in CLL cells treated with SGI-1776. CLL primary cells (patients nos. 15, 16, and 17) were treated with 0.1% vehicle DMSO alone or 10 μmol/L SGI-1776 for 0, 2, 4, 8, and 24 hours, and then the RNA was isolated. MCL-1 transcript levels were measured by the use of real-time RT-PCR and normalized with 18S transcripts. Each RNA sample was assayed in triplicate, and the results are expressed as a percentage of the MCL-1 expression level in cells treated with vehicle DMSO alone. The results represent an average ± SEM of triplicate experiments. (C) Dose-dependent reduction of MCL-1 transcript levels in CLL cells treated with SGI-1776. CLL primary cells (patients nos. 2, 4, 12, and 14) were treated with 0.1% vehicle DMSO alone or with 0.3, 1, 3, or 10 μmol/L SGI-1776 for 24 hours, and transcript levels were measured in triplicate using real-time RT-PCR, and normalized with 18S transcript.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/114/19/10.1182_blood-2009-03-212852/4/m_zh89990944160005.jpeg?Expires=1767813768&Signature=VT4PuW6Qb3uCs7IKVh5La26bvvFqDporxOb1G6F7Kh3g~JWd-vi0uswnGtWwWRlOC--l4roF~C9xhSju~lWyYmzqiMWHUtVFSmbk6yDo3a3fYXryICXl2-7~~2tusCttDmdLRIQqIC3QOoJYrVWeHVTk2GWPCGZYnt5vvYdArqoKn~BGFBHfQMSLPf8iatyg5KA-ik7wXcTIH7GWbBs1HzEQiTJNofe7JqbMeHrxZ5b9PXTtCt6TCCgXrUQqiis5pJVTTAH1uTFKsPTEkcaBTzzK1bW1kmM62-L~LBN8cSnPuxfosezBPMvLRNM8ZRnSkcBY3LQdZiukhBMTCKh9lA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

![Figure 1. Structure and functional activity of imidazo[1,2-b]pyridazine compound SGI-1776. (A) Chemical structure of SGI-1776. (B) Kinase inhibition by SGI-1776. Several human kinases were tested for selective inhibition by 1 μmol/L SGI-1776 using Millipore kinase profiler assay as described in “Kinase assays.” (C) In vitro kinase assays of Pim-1, -2, and -3 with varying concentrations of SGI-1776 to determine IC50 values for each Pim kinase species. The IC50 Profiler Express Assay is described in “Kinase assays.”](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/114/19/10.1182_blood-2009-03-212852/4/m_zh89990944160001.jpeg?Expires=1767915826&Signature=Izc78XS7DlJOYPrTQqdQxt1L5zX8yJ7q-UZtknDVY1kn156s4sKjg23Q-nA3qU4M1IEFKY1ab0B2zy0lrAcIR8tFjRqwbvJCPFyQVpNxuxCFu2Z1YgnsyEbqjCA39Tp2YMaZYnOghu89v4U2-JCliejA5edKgSObjmYpGRrsX88fIhQowWaIcHt94hGnVddKrU9KF-Mjz7WvxcTkwiWI7HU9kVfcyBnlOhyciaX5jcXMRS5s4Gxgn-51Gdq4M5YD1wGqN4qd37zOlYX7o-KwZP0aQ2gntmVGSWUZbI6qQBr9rRoAg8Kh1fYFvQPWBz8BUywnK~RubUtxEyJ0uM9PUw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

![Figure 5. (A) Inhibition of RNA synthesis in CLL primary cells treated with SGI-1776. CLL primary cells (patients 3, 4, and 14) were incubated with 0.1% DMSO alone or with 0.3, 1, 3, or 10 μmol/L SGI-1776 for 24 hours. Then, 1 hour before the cells were harvested, [3H]uridine was added to the cell culture as described in “Uridine incorporation.” The results represent an average of triplicate experiments ± SEM. (B) Time-dependent reduction of MCL-1 transcript levels in CLL cells treated with SGI-1776. CLL primary cells (patients nos. 15, 16, and 17) were treated with 0.1% vehicle DMSO alone or 10 μmol/L SGI-1776 for 0, 2, 4, 8, and 24 hours, and then the RNA was isolated. MCL-1 transcript levels were measured by the use of real-time RT-PCR and normalized with 18S transcripts. Each RNA sample was assayed in triplicate, and the results are expressed as a percentage of the MCL-1 expression level in cells treated with vehicle DMSO alone. The results represent an average ± SEM of triplicate experiments. (C) Dose-dependent reduction of MCL-1 transcript levels in CLL cells treated with SGI-1776. CLL primary cells (patients nos. 2, 4, 12, and 14) were treated with 0.1% vehicle DMSO alone or with 0.3, 1, 3, or 10 μmol/L SGI-1776 for 24 hours, and transcript levels were measured in triplicate using real-time RT-PCR, and normalized with 18S transcript.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/114/19/10.1182_blood-2009-03-212852/4/m_zh89990944160005.jpeg?Expires=1767915826&Signature=jogqJSzgPBxZ2qwVa7KXom~l3OvM7kZTrd8dfANv9trYlhvsTij3V-O491~Tu~TVSlaMaM0T8tbj3F-WA5l5clNCrj5nf17pjCplqFM35D66JxfUjyL1~xzw5fZC3khw8ePPak3YQVzgCGZ2JBTlQniXHdO8tAbLZmR5gM0aSvGYE6mEVvG2YAERMumsqXZj36ZNO~K9a3XuuOqh4CVZvtyFA6owBopYgavZy10lwzKD3xJj9bL0nGShjrLQODSx2IlsQsd~bX8zsVL9CDO5uacrLUzNltAJNCW1A5FlzKPEStlWhiv9SRrU18BgeQu~F24BLZokqBMA-Kq-JCa2TA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)