Abstract
The aims of this study were to assess the feasibility of prospective pharmacogenomics research in multicenter international clinical trials of bortezomib in multiple myeloma and to develop predictive classifiers of response and survival with bortezomib. Patients with relapsed myeloma enrolled in phase 2 and phase 3 clinical trials of bortezomib and consented to genomic analyses of pretreatment tumor samples. Bone marrow aspirates were subject to a negative-selection procedure to enrich for tumor cells, and these samples were used for gene expression profiling using DNA microarrays. Data quality and correlations with trial outcomes were assessed by multiple groups. Gene expression in this dataset was consistent with data published from a single-center study of newly diagnosed multiple myeloma. Response and survival classifiers were developed and shown to be significantly associated with outcome via testing on independent data. The survival classifier improved on the risk stratification provided by the International Staging System. Predictive models and biologic correlates of response show some specificity for bortezomib rather than dexamethasone. Informative gene expression data and genomic classifiers that predict clinical outcome can be derived from prospective clinical trials of new anticancer agents.
Introduction
Multiple myeloma is an incurable malignancy that originates in the antibody-secreting bone marrow plasma cells. Median survival is approximately 3 to 4 years, but the clinical course is highly variable and difficult to predict.1,2 Therefore, there is a need to better define patient-specific treatment strategies for the use of both standard and novel therapies.
A number of clinical and laboratory features provide prognostic information, including age, performance status, tumor burden, tumor proliferative index, and hemoglobin and platelet levels, as well as serum β-2 microglobulin, albumin, creatinine, lactic dehydrogenase, and calcium.3-8 Some of these factors relate to the patient's status, whereas others reflect aspects of the tumor. A recent multivariate analysis of data from 10 000 patients identified serum albumin and β-2 microglobulin as a reliable prognostic tool, referred to as the International Staging System (ISS).2 The ISS is valid for patients of different age groups and geographies, and with respect to the 2 most common myeloma treatments of the past decade, standard-dose chemotherapy and high-dose therapy (HDT) followed by stem cell rescue.2 However, therapeutic choices for myeloma have become increasingly complex as new active agents have emerged,9,10 and their optimal use either alone or in combination with standard chemotherapy or HDT remains to be defined. As indicated in the ISS study, standard clinical prognostic factors were unable to reliably identify the highest risk patients most in need of novel therapies (defined as those with < 12 months overall survival [OS]).2
It is anticipated that genomics will help provide more precise prognostic and predictive tools.11-13 However, the practicality, utility, and challenges of prospective genomic research have only recently been explored.14 In addition, it remains unclear whether the strategies used to define prognostic genomic classifiers15 can be used to develop classifiers that predict outcome for a specific therapy.
Various molecular analyses suggest that myeloma, like other cancers, is composed of distinct subtypes that have somewhat different molecular pathologies and prognoses.11,16,17 For instance, cytogenetic studies reveal that approximately 60% to 80% of myeloma cases exhibit rearrangements of the IGH heavy chain locus, with 40% involving 5 recurrent translocations.11 Patients with the t(11;14)(q13;q32) translocation experience superior survival on treatment with HDT, whereas those with t(4;14)(p16;q32) exhibit a relatively poor survival.18-25 More specifically, although t(4;14)(p16;q32) tumors initially respond to therapy, they relapse quickly and are insensitive to alkylator salvage treatment.11,25 Deletion of chromosome 13 occurs in tumors with and without IgH translocations26 and is a significant poor prognostic factor, regardless of therapy or age.26-28
Furthermore, distinct gene expression patterns are associated with most of the molecular subtypes of myeloma,14,29-33 and these patterns are now being associated with disease prognosis.14 A recent genomic analysis of 231 myeloma cases identified 8 distinct tumor subtypes, defined via assessment of cyclin D status and other genes frequently involved in IgH translocations (referred to as TC subtypes, for Translocation and Cyclin D).33 Despite these and other molecular advances, it remains unclear how to match disease subtypes appropriately with standard myeloma therapies or the use of new agents.
To assess the technical feasibility of conducting prospective pharmacogenomics research in myeloma and, if possible, to develop and independently validate a genomic classifier of efficacy to a specific single agent, we generated gene expression data during the course of national and international phase 234,35 and phase 336 clinical trials of a novel agent, the proteasome inhibitor bortezomib (VELCADE; Millennium Pharmaceuticals, and Johnson & Johnson Pharmaceutical Research & Development). We report here the microarray results from those trials. This is the first report demonstrating the prospective development and independent validation of a genomic classifier that predicts clinical response between myeloma patients treated with a new agent (bortezomib) or an active control drug (high-dose dexamethasone; Dex).
Materials and methods
Sample collection, enrichment, data generation, and array quality control
On collection of patients' bone marrow aspirate, myeloma cells were enriched via negative selection. The RosetteSep procedure (Stem Cell Technologies, Vancouver, BC, Canada) uses a cocktail of cell-type–specific antibodies (as described in Tai et al37 ) to deplete nonplasma cells (see Document S1 for details, available on the Blood website; see the Supplemental Materials link at the top of the online article). Myeloma cells were collected and frozen. In the international studies, the first 2 samples from each site were collected and subjected to RNA isolation so that feedback on quantity and quality could be provided; ultimately, phase 2 and 3 trials provided a similar percentage of informative samples. Control samples included bone marrow–derived normal plasma cells (PCs), neutrophils, T cells, and CD71+ erythroid cells (AllCells, Berkeley, CA).
Total RNA was isolated using a Qiagen RNAeasy isolation kit (Qiagen, Valencia, CA) and quantified by spectrophotometry; samples with at least 0.5 μg were labeled for gene expression profiling in 2 batches (Document S1), using the Affymetrix GeneChip microarray system (Affymetrix, Santa Clara, CA). A standard T7-based amplification protocol (Affymetrix) was used to convert 2.0 μg RNA (if available) to biotinylated cRNA. cRNA for each sample was hybridized to the U133A/B arrays in triplicate; operators, chip lots, clinical sites, and scanners (GeneArray 3000; Affymetrix) were controlled throughout. Data processing used Affymetrix MAS5.0. Quality control metrics determined by Affymetrix and Millennium Pharmaceuticals included the percentage present (> 25%), scale factor (< 14), β-actin 3′/5′ ratio (< 15), and background (< 120) (Table S1). Samples falling outside these metrics were excluded from subsequent analysis.
The myeloma purity score examines expression of genes described as highly expressed in myeloma cells and their normal plasma precursor cells (205692_s_at CD38 antigen [P45]; 201286_at syndecan-1 [SDC1]; 201891_s_at β-2 microglobulin [B2M]; 211528_x_at B2M) compared with genes expressed highly in erythroid cells (37986_at erythropoietin receptor [EPOR]; 209962_at EPOR; 205838_at glycophorin A [GYPA]), neutrophils (203948_s_at myeloperoxidase [MPO]; 203591_s_at colony-stimulating factor 3 receptor [CSFR3] [granulocyte]; 204039_at CCAAT/enhancer binding protein α [CEBPA]; 214 523_at CCAAT/enhancer binding protein ϵ [CEBPE]), or T cells (209603_at GATA binding protein 3 [GATA3]; 209604_s_at GATA binding protein 4 [GATA4]; 205456_at CD3E antigen, ϵ polypeptide). Myeloma score = expression of myeloma markers / expression of (erythroid + neutrophil + T cell) markers. The data set is available at Gene Expression Omnibus (http://www.ncbi.gov/geo/).
Clinical studies and efficacy
The APEX phase 3 trial (039) was conducted at 93 centers in the United States, Canada, Europe, and Israel from June 2002 to October 2003.36 A total of 669 patients with myeloma who had relapsed following 1 to 3 prior therapies were randomly assigned to treatment with bortezomib 1.3 mg/m2 or high-dose Dex; Dex patients who experienced progressive disease (PD) were permitted to crossover to receive bortezomib in a companion study (040). The 040 study also directly enrolled 263 patients who had more than 3 prior therapies; these “non-crossover” patients were also eligible for pharmacogenomics research. The SUMMIT phase 2 trial (025) enrolled 202 patients with relapsed and refractory myeloma at 14 centers in the United States.35 Patients received bortezomib 1.3 mg/m2 for no more than 8 cycles. The CREST phase 2 trial (024) had a similar design, except the 54 enrolled patients had either relapsed or refractory disease, and they received bortezomib 1.0 or 1.3 mg/m2.34 Phase 2 investigators had the option to add Dex 20 mg if patients had suboptimal response; however, clinical and genomic studies report activity of single-agent bortezomib by censoring outcome data at the time of adding Dex.
Review boards at all participating institutions approved the studies; all patients provided written informed consent. Additional consent was provided for pharmacogenomics analysis. The studies were conducted in accordance with the Declaration of Helsinki and International Conference on Harmonisation Good Clinical Practice guidelines.
Outcome definitions
Clinical response was treated as a categorical variable, whereas OS was treated as a censored continuous-time variable. OS (days) was assessed from the date patients received their first dose of study drug, without regard to other subsequent therapies. Patients were classified as achieving complete response (CR), partial response (PR), minimal response (MR), no change (NC), or PD, using European Group for Bone Marrow Transplantation criteria.38 In brief, PD requires 25% increase in paraprotein, whereas MR, PR, and CR require at least 25%, 50%, and 100% decreases, respectively. NC is the absence of response or progression but, in this study, required at least 2 measures of stable disease. The efficacy data of the genomics subset were manually reviewed to reconfirm classifications (Document S1).
Data analysis
Only the 9200 probe sets with strongest between-sample variance relative to their in-sample replicate variance were retained for further analysis (B.B., E.K., G.M., manuscript in preparation). Repeated expression measurements on a given sample were summarized by the log of their median value.
Gene set enrichment analysis (GSEA).
Analysis used GSEA software (version 1.0; Broad Institute, http://www.broad.mit.edu/gsea/) and C2 curated functional gene sets from the Molecular Signature Database (MSigDB)39 (Document S1). Analysis was performed on the full set of bortezomib samples from all trials, as well as on samples from individual trials, including 039 bortezomib and 039 Dex separately. Gene sets satisfying the default multiple hypothesis testing threshold (FDR q value < .25) and having nominal P values no more than .025 were identified (56 associated with response [CR/PR/MR; R] and 16 associated with PD). These were classified as to whether they also had a less stringent (nominal P ≤ .05), but consistent, association with the phenotype in analysis of samples from at least 2 individual trials. Gene sets were then ranked, first by consistent association with phenotype, then by FDR q value.
Analysis of clinical response.
Differential expression of genes with respect to clinical response was assessed by 2-sided t test with unequal variances. Predictive models were built using a linear predictor score40 on the top 100 differentially expressed genes. To assess the accuracy of the predictive modeling method on the entire dataset, a standard bootstrap procedure was used,41 in which the data were repeatedly divided into separate training and test sets. Each time, genes were selected, and a predictive model was built on the training set; the model was then applied to the test set to assess accuracy, sensitivity (Sn), and specificity (Sp). To determine whether predictive accuracy differed significantly from what might be expected at random, outcome values were repeatedly randomly permuted among samples, and the bootstrap procedure was reapplied. Empirical distributions of accuracies for true and permuted outcomes were then compared.
Analysis of OS.
We used the Cox proportional hazards model to assess strength of association of individual probe sets with OS. Predictive models were built using the method of Bair and Tibshirani,42 as follows. The 100 probe sets most strongly associated with outcome were selected for the model. We computed the top 2 principal components of these genes' expression on the training samples. Test data were mapped onto the space defined by the principal component vectors, and Cox modeling was used to assess strength of association of the transformed test data with outcome. For visualization of the models using Kaplan-Meier curves, the linear predictor score from the Cox model was used to divide test samples in equally sized high- and low-risk groups.
Results
Sample collection and genomic data generation in multicenter clinical trials
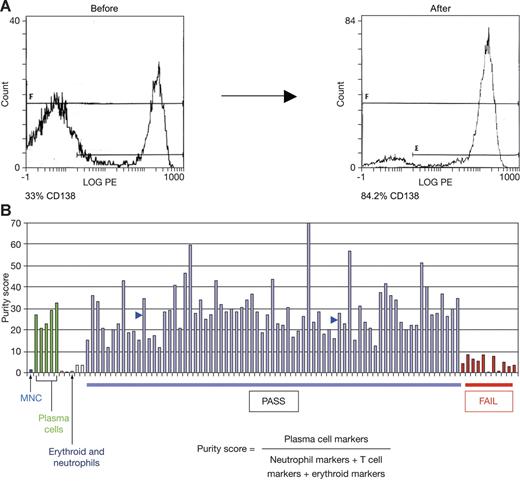
The phase 2 and phase 3 clinical trials of bortezomib for the treatment of myeloma included a research component to investigate the feasibility of pharmacogenomics in a prospective setting; tumor samples were provided from 89 centers in 12 different countries. A pretreatment bone marrow aspirate was collected during routine screening procedures. The percentage of tumor cells in aspirates varied from approximately 5% to greater than 75%. All samples were therefore subject to an enrichment procedure in an effort to increase tumor content to at least 60% to 80%, a level consistent with prior genomic studies of cancer biology and outcome.14,43,44 Fluorescence cell sorting analysis (FACS) of samples before and after enrichment demonstrated that enrichment could yield samples of 80% to 90% tumor cells (Figure 1A). FACS analyses were not practical at all participating centers. Therefore, we assessed sample purity via analysis of a myeloma purity score derived from microarray data. Samples with low tumor cell purity were excluded from further analyses (Figure 1B).
Bone marrow aspirate enrichment procedure effectively depletes nontumor cells. (A) Bone marrow aspirate samples before and after enrichment were subject to CD138 staining and FACS analysis. (B) Myeloma purity score is elevated in control plasma cell samples (> 90% pure) relative to bone marrow mononuclear cells (MNCs), neutrophils, and erythroid cells. Two enriched patient samples of 84% and 91% tumor purity by FACS analysis had scores of 35 and 28, respectively (blue arrows). A score of at least 10 (at least 3-fold elevated relative to the score for nonplasma cell types) was set as a threshold for further analysis.
Bone marrow aspirate enrichment procedure effectively depletes nontumor cells. (A) Bone marrow aspirate samples before and after enrichment were subject to CD138 staining and FACS analysis. (B) Myeloma purity score is elevated in control plasma cell samples (> 90% pure) relative to bone marrow mononuclear cells (MNCs), neutrophils, and erythroid cells. Two enriched patient samples of 84% and 91% tumor purity by FACS analysis had scores of 35 and 28, respectively (blue arrows). A score of at least 10 (at least 3-fold elevated relative to the score for nonplasma cell types) was set as a threshold for further analysis.
Sample attrition was observed at each step in the process of generating gene expression data (Table 1). Approximately 60% of samples exhibited RNA quantity and quality adequate for hybridization. Of these samples, about 85% generated high-quality microarray data, and 85% passed the assessment of tumor-cell enrichment with the myeloma purity score. Results were generally consistent between trials (Table 1). The bortezomib dataset consists of 169 patients evaluable for response and 188 evaluable for OS, whereas the Dex dataset has 70 and 76 evaluable for response and OS, respectively. The details for each trial are provided in Table S2.
For each trial we examined a series of clinical and prognostic variables to ensure that the subsets of patients with genomic data were representative of the general trial populations (Table 2). No bias was observed with regard to age, sex, number of prior therapies, or myeloma isotype. For some trials the response rate, time to progression (TTP), or survival values of the genomics subset were indicative of a worse outcome. Although serum albumin and serum β-2 microglobulin were elevated in the genomics subset of trial 025, this was not observed in other trial data. However, the genomics subset of each trial did exhibit a higher baseline tumor burden in the bone marrow aspirate (Table 2), indicating that successful sampling is partly related to extent of marrow disease. The data suggest that genomic subsets are reasonable representations of study populations as a whole, except for an overrepresentation of patients with greater tumor burden.
Because of differences in entry criteria, there are differences between the trial populations in terms of median number of prior therapies, time from diagnosis, and response rate (Table 1; Table S3). For example, trial 025 enrolled relapsed patients who were refractory to their last prior therapy (median number of prior lines of therapy, 6), whereas trial 039 specified 1 to 3 prior lines of therapy. Accordingly, the median number of prior lines of therapy in genomics subsets of trials 025 and 039 are 6 and 2, respectively.
Tumor samples were collected between 2001 and 2004; microarray hybridizations were performed in 2 batches separated by 9 months. Replicate hybridizations allowed us to assess within-patient reproducibility and between-patient variations prior to selecting the approximately 9200 most differentially expressed probe sets for further analysis.
Comparison of dataset with published myeloma biology
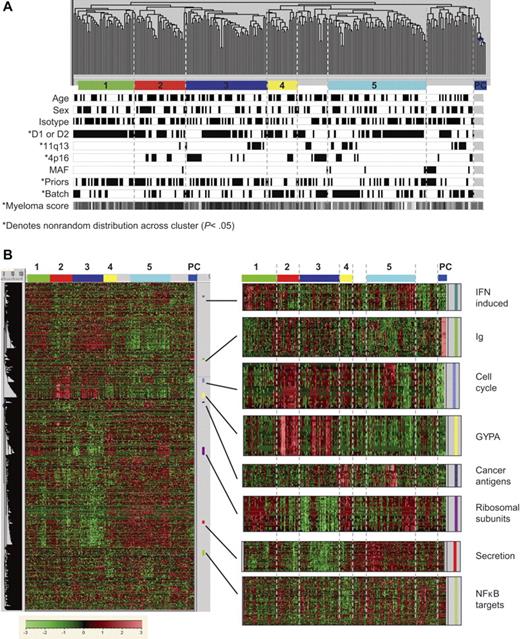
Our genomics approach differs from that of prior myeloma studies14,29,30,45 in that samples were collected at multiple sites and subjected to a negative-selection procedure to enrich for tumor cells. Therefore, we closely examined how the data might have been influenced by demographic, clinical, and technical parameters, using unsupervised hierarchical clustering. Figure 2A shows a dendrogram of 264 myeloma patient samples and 6 normal plasma cell control samples. Patients with different age, sex, and myeloma isotype were randomly distributed (Figure 2A) across these groups. Further, there was no significant clustering of samples that originated at the same clinical center.
Sample relationships are influenced by clinical and gene-expression characteristics. Two hundred sixty-four myeloma patient samples and 6 normal plasma cell control samples were subject to unsupervised hierarchical clustering based on 9174 differentially expressed probe sets. Highly related branches (labeled groups 1-5) were identified by setting a fixed similarity metric (GeneMaths software; Applied Maths, Austin, TX) and requiring at least 12 samples for membership; unlabeled samples comprise various smaller groups. (A) Patient attributes are encoded below the sample dendrogram. Attributes with nonrandom distribution (P < .05) are indicated by asterisks. Black is associated with age older than 60 years, female sex, IgG isotype, 1 or 2 prior therapies, hybridization batch 1 (trials 024, 025, and 040), and low purity score. White is associated with age 60 years and younger, male sex, other isotypes, 3 or more prior therapies, hybridization batch 2 (trial 039), and high purity score. (B) An overview of the 9174 differentially expressed probe sets, with an expansion of specific functional groups.
Sample relationships are influenced by clinical and gene-expression characteristics. Two hundred sixty-four myeloma patient samples and 6 normal plasma cell control samples were subject to unsupervised hierarchical clustering based on 9174 differentially expressed probe sets. Highly related branches (labeled groups 1-5) were identified by setting a fixed similarity metric (GeneMaths software; Applied Maths, Austin, TX) and requiring at least 12 samples for membership; unlabeled samples comprise various smaller groups. (A) Patient attributes are encoded below the sample dendrogram. Attributes with nonrandom distribution (P < .05) are indicated by asterisks. Black is associated with age older than 60 years, female sex, IgG isotype, 1 or 2 prior therapies, hybridization batch 1 (trials 024, 025, and 040), and low purity score. White is associated with age 60 years and younger, male sex, other isotypes, 3 or more prior therapies, hybridization batch 2 (trial 039), and high purity score. (B) An overview of the 9174 differentially expressed probe sets, with an expansion of specific functional groups.
However, a nonrandom distribution was observed for clinical study, number of prior therapies, array hybridization batch, myeloma purity score, and, consistent with a recent report,14 myeloma TC subtype. Because several of these factors are interrelated (eg, patients from trial 039 had fewer prior therapies and their samples were hybridized in one batch), it was difficult to discern which factors influence the clustering. We investigated the influence of prior therapies by examining the distribution of samples from trial 025, which have a varied number of prior therapies and were hybridized in a single batch. In fact, patients from trial 025 in groups 1 to 3 had fewer lines of prior therapy (mean = 3.7) than those in groups 4 to 5 (mean = 5.1) (P = .053), suggesting that distribution of these samples is at least in part influenced by the extent of prior therapy.
Analysis of gene expression patterns within this dataset revealed several features in common with previously reported studies of myeloma (Figure 2B). These include a reduced expression of genes associated with immune function (IGH, IGL)29 and heterogeneous overexpression of cancer antigens, interferon-induced genes, and genes involved in protein synthesis and proliferative pathways.29,30,46,47 We also noted differential expression of various genes related to protein secretion and endoplasmic reticulum stress, as well as NF-κB transcription targets (Figure 2B).
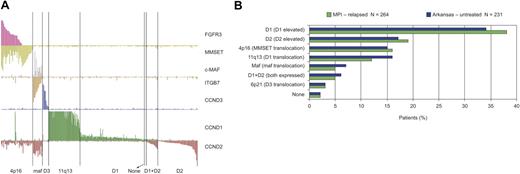
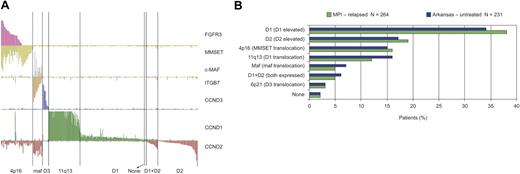
Recently, a study highlighted the overexpression of D-type cyclins and other common IgH translocation targets and suggested that newly diagnosed myeloma comprises 8 distinct TC subtypes.33 These TC subtypes were also observed in this dataset of relapsed/refractory patients collected from multiple clinical centers (Figure 3A). Notably, the frequencies of each TC subtype were very similar in the 2 different datasets (Figure 3B). Figure 3A highlights additional hallmark features of myeloma gene expression, including loss of FGFR3 expression in a subset of t(4;14)(p16;q32)–positive samples48 and correlation between overexpression of c-MAF transcription factor and the c-MAF target gene cyclin D2.45 Together these observations indicate that the current genomic dataset, derived from national and international clinical trials, is consistent with previously described data.
All samples assigned to TC subtypes based on expression of D cyclins and translocation target genes (n = 264). (A) The TC subtypes of 264 relapsed myeloma samples are shown. The y-axis shows normalized expression level of each gene; subtypes were determined as in Bergsagel et al.33 (B) A comparison of the TC subtype frequency for relapsed patients in Millennium Pharmaceuticals (MPI) studies (green) and for newly diagnosed patients (blue) as defined at the University of Arkansas.33
All samples assigned to TC subtypes based on expression of D cyclins and translocation target genes (n = 264). (A) The TC subtypes of 264 relapsed myeloma samples are shown. The y-axis shows normalized expression level of each gene; subtypes were determined as in Bergsagel et al.33 (B) A comparison of the TC subtype frequency for relapsed patients in Millennium Pharmaceuticals (MPI) studies (green) and for newly diagnosed patients (blue) as defined at the University of Arkansas.33
We noted that a subset of samples express genes generally detected in erythroid or myeloid lineages, including GYPA and CD14 (Figure 2B). The origin of this expression pattern remains unclear. However, such expression has been noted in both normal plasma cells and myeloma cells after positive selection,14 and in such studies this expression was associated with better prognosis on treatment with HDT.14
Can pretreatment gene expression predict response?
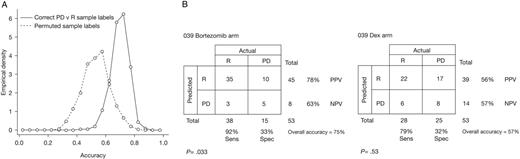
Response rates in the various bortezomib trials are shown in Table 2. To investigate whether information in pretreatment tumor samples could predict whether patients would respond to bortezomib, we first used a bootstrap approach, in which samples were repeatedly split into random train and test sets. The mixing of patients from different trials minimized the influence of known and unknown confounding variables. To best distinguish any predictive signal and interpret subsequent biology we initially focused on patients who had either PD or R. A linear predictor classifier40 to distinguish PD and R was developed in each training set and evaluated on the held-out test data. As shown in Figure 4A, the median test-set accuracy was 70.2% (mean = 69.8%); this accuracy exceeded the accuracy obtained when sample labels in the training set were permuted (mean = 53.6%, 95th percentile = 69.4%).
Prediction scores. (A) Data from all bortezomib-treated patients analyzed in bootstrap; the empirical distributions of prediction accuracies for all test sets are shown. Note that the median value of the accuracies for the correctly labeled samples (70.2%) is higher than 95% of the accuracies for the permuted sample labels (95th percentile = 69.4%). Thus, the 2 distributions are significantly different. (B) A classifier for trials 025 and 040 was used to predict the response of patients receiving bortezomib and patients receiving Dex in trial 039. Accuracy of response prediction for bortezomib-treated patients is significant (P < .033; 75% overall accuracy) but not significant (P = .53; 57% overall accuracy) for patients treated with Dex. No significant accuracy is observed when all test samples are simply predicted as the most popular response category (P > .999).
Prediction scores. (A) Data from all bortezomib-treated patients analyzed in bootstrap; the empirical distributions of prediction accuracies for all test sets are shown. Note that the median value of the accuracies for the correctly labeled samples (70.2%) is higher than 95% of the accuracies for the permuted sample labels (95th percentile = 69.4%). Thus, the 2 distributions are significantly different. (B) A classifier for trials 025 and 040 was used to predict the response of patients receiving bortezomib and patients receiving Dex in trial 039. Accuracy of response prediction for bortezomib-treated patients is significant (P < .033; 75% overall accuracy) but not significant (P = .53; 57% overall accuracy) for patients treated with Dex. No significant accuracy is observed when all test samples are simply predicted as the most popular response category (P > .999).
Because data came from several multisite studies with different patient populations, we next assessed whether a predictor developed with data from one study could be validated on another. Using samples from the earliest trial (025), a classifier was developed to distinguish PD and R, and bootstrap validation within trial 025 suggested the classifier should have significant accuracy on other similar data (73% average accuracy, 95% of test sets showing > 55% accuracy). However, this classifier exhibited an overall accuracy of 55% (Sn = 58%, Sp = 47%; P = .77) on testing in the bortezomib arm of trial 039, and 57% (Sn = 64%, Sp = 48%; P = .41) in the Dex arm. Lack of significance with the samples from trial 039 as an independent test set may relate to differences in patient populations enrolled in these distinct trials (notably, the higher response rate to bortezomib in trial 039), the relatively small sample size of the training set, disease heterogeneity, or a combination of these factors.
We next built a response classifier using data from both the 025 and 040 trials (67 samples from patients with R or PD) and tested it on data from trial 039. As shown in Figure 4B, response in the bortezomib arm was predicted with an overall accuracy of 75%, (Sn = 92%, Sp = 33%; P = .033). However, response prediction for the Dex arm was 57% (Sn = 79%, Sp = 32%; P = .53), suggesting that the classifier has some specificity for bortezomib. The 100 probe sets comprising this classifier are listed in Table S4. Finally, we obtained similar results when these predictive analyses included patients with NC, who were grouped with PD patients to form a nonresponse (NR) category. Although an 025 trial NR versus R classifier was unable to significantly predict outcome of the test set from trial 039, an 025 + 040 trials NR versus R classifier exhibited 63% (P = .03) and 54% (P = .3) overall accuracy when tested in the bortezomib and Dex arms of trial 039, respectively (Table S5). Median bootstrap accuracy of the NR versus R predictor was also 63% (mean = 63.1%); this accuracy exceeded that obtained when the sample labels were permuted (median = 49.2%, mean = 49.4%, 95th percentile = 62.7%; Figure S1). Similar accuracy was noted when the number of probe sets used in the classifier was varied from 50 to 500 (data not shown).
Although we have used the permutation approach and the Fisher exact test to establish that our predictions are significant, it is also common to compare prediction accuracies with that of the best constant model (which is 72% for trial 039 bortezomib arm, and 68% for the bootstrap of all bortezomib-treated patients). Our PD versus R models do not significantly outperform the best constant model. However, this appears to be due to the response classes being unbalanced: a subsampling analysis (Figure S2) shows that our prediction method significantly outperforms the best constant model in the case of equal numbers of PD and R patients. The more-balanced NR versus R prediction also shows significant accuracy (63%) compared with the best constant model (46%; see Document S1 for details).
In summary, we observed a statistically significant prediction of response when data were combined across all the studies or from 2 studies, but not with the 025 study alone.
Genes and pathways associated with response
A number of the probe sets in the 025 + 040 trials classifier (Table S4) represent genes of known function. Among those overexpressed in PD are ribosomal (RPS7, RPS13), mitochondrial (COX7C, UQCRH), ER stress (SERP1), DNA repair (APEX1, REC14), and cancer-associated (NRAS, NPM1) genes. Those overexpressed in patients achieving R include components of the PI3 kinase pathway (PIK3R1, DAPP1) and other signaling molecules (TYROBP, RRAGC, LYK5).
We further examined the biology of bortezomib sensitivity by applying GSEA,49 an algorithm that correlates all approximately 18 000 genes represented on the arrays with a phenotype (R or PD) and highlights known or experimentally annotated sets of genes that are enriched in these phenotypes. This analysis included bortezomib data from trials 024 and 039 as well as that of the samples from 025 + 040 trials. The most significant gene sets relatively highly expressed in samples from responsive patients are shown in Table 3. These include adhesion, cytokines, NF-κB activity, and hypoxia gene sets. Gene sets elevated in samples from patients classified as PD (Table 3) include protein synthesis, mitochondrial function and RNA transcription/splicing. Among the NF-κB targets correlated with R were IL8, IL15, CXCL5, CFLAR, ICAM, and NFKB2, suggesting that expression of a subset of NF-κB targets characterizes myeloma cells more sensitive to bortezomib. This is consistent with various preclinical studies of bortezomib's mechanism of efficacy, showing inhibition of NF-κB signaling and subsequent apoptosis of myeloma50,51 and other cells52,53 on treatment with bortezomib.
Several gene sets elevated in samples from patients achieving R encode adhesion molecules, indicating that more adhesive myelomas may be sensitive to bortezomib. This interpretation is also supported by preclinical experiments showing that fibronectin adhesion increases sensitivity of myeloma cells to bortezomib54 while reducing sensitivity to melphalan and doxorubicin.55 Interestingly, on analysis of the smaller datasets from the 039 trial, several gene sets highlighted in Table 3 are strongly correlated with R or PD (P < .05) in the bortezomib arm but not the Dex arm; these include brentani cell adhesion and cytokine pathway (Table 3, associated with R), and translation factors and ribosomal proteins (Table 3, associated with PD). Such results imply that these pathway observations are specific to bortezomib.
Can pretreatment gene expression predict survival?
The 039 randomized trial demonstrated superior OS with bortezomib versus Dex (30 versus 24 months; P = .027; 22-month median follow-up, 44% events occurred).56 A significant TTP and survival advantage was also observed at a preplanned interim analysis, at which time all patients were permitted to receive bortezomib and 62% of the patients in the Dex arm subsequently received single-agent bortezomib.36
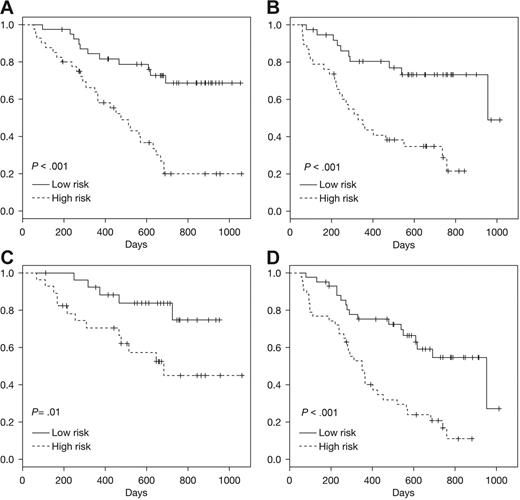
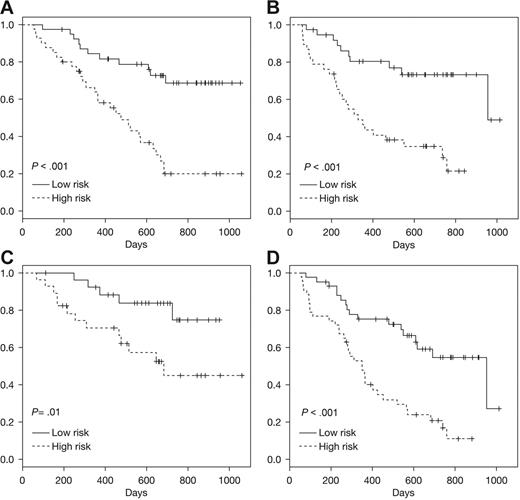
We used gene expression data from patients in 025 + 040 trials to develop a survival classifier42 that was then tested with data from the 039 trial. As shown in Figure 5A, this gene expression classifier stratified the patients in trial 039 receiving bortezomib into high- and low-risk groups that were significantly associated with risk of death (P < .001). The classifier also effectively stratified the patients enrolled in the Dex arm of trial 039 (P < .001; Figure 5B). It is possible this survival classifier and the underlying probe sets may be prognostic of survival independent of the specific therapy administered. However, there may be some specificity for bortezomib (as observed with the response classifier) that is masked by the subsequent use of bortezomib in the majority of patients enrolled in the Dex arm. Additional analyses and comparisons with other myeloma pharmacogenomics datasets will be required to address these possibilities.
Prediction of survival using Super PC. A survival classifier based on 025 + 040 trials was used to identify high- and low-risk groups within an independent test dataset derived from 039 patients. Kaplan-Meier analyses of the actual survival of these predicted high-/low-risk patient groups is shown for test set (A) trial 039 bortezomib, (B) trial 039 Dex, (C) ISS = 1 for patients from 039 trial (bortezomib or Dex), (D) ISS = 2 to 3 for patients from 039 trial (bortezomib or Dex).
Prediction of survival using Super PC. A survival classifier based on 025 + 040 trials was used to identify high- and low-risk groups within an independent test dataset derived from 039 patients. Kaplan-Meier analyses of the actual survival of these predicted high-/low-risk patient groups is shown for test set (A) trial 039 bortezomib, (B) trial 039 Dex, (C) ISS = 1 for patients from 039 trial (bortezomib or Dex), (D) ISS = 2 to 3 for patients from 039 trial (bortezomib or Dex).
To determine whether the pretreatment gene expression provides data not already captured by prognostic clinical variables, we assessed the survival of patients predicted to be high- or low-risk by ISS.2 These risk groups are relevant for various myeloma therapies2 and also discern high/low risk in the 039 trial patients (data not shown). As shown in Figure 5C-D, the gene expression classifier enables further, significant stratification in patients identified as low risk (ISS = 1; Figure 5C) and high risk (ISS = 2-3; Figure 5D) respectively, indicating that clinical staging and genomic information are not redundant but are likely to be complementary.
The probe sets comprising this survival classifier (Table S6) do not overlap with the response classifier. This is not surprising, because the survival and response end points are only partially related. Overexpression of adhesion-related genes (CDH1, CD36) are correlated with longer survival, suggesting there may be biologic consistencies, but a more detailed examination of response and survival pathways will be required.
Discussion
Clinical genomics offers great promise to improve cancer diagnosis, prognosis, and treatment selection. However, this type of research requires large datasets derived from well-characterized, uniformly treated patients with appropriate end point data.12,42,57 This study describes a myeloma gene expression dataset derived from large prospective clinical trials, and the lessons from this research highlight both the challenges and advantages of the implementation of similar research in the future.
The first challenge was sample attrition (Table 1). Inadequate RNA, because of insufficient tumor sampling or RNA degradation, precluded use of approximately 50% of collected samples. Across these myeloma trials, patient consent, sample acquisition, and data generation/quality control produced only limited losses; however, even small losses at each stage compounded the attrition issue.
Second, the necessary analysis of data from multiple clinical trials and comparisons between trials were made more difficult because of differences between trials. Patients in the phase 2 trials had experienced more prior therapy and were less responsive than patients enrolled in the phase 3 trial (Table 1; Table S3).34-36 It will be interesting to compare further these data from relapsed patients with data from newly diagnosed myeloma. The near identical frequency of TC subtypes in both relapsed and newly diagnosed myeloma (Figure 3B) indicates the TC categories do not define any subgroup of patients that is rapidly lost after first-line therapy; other ways of comparing these datasets may reveal such high-risk patient types. A final caveat to future studies is the time required for prospective research. In this example, despite bortezomib's rapid advance to phase 3 trials in myeloma, more than 4 years elapsed between the initial sample collection in phase 2 trials and the genomic analysis of the updated survival data from the phase 3 trial.
Despite such issues and differences in purity methodologies, these clinical trials yield a myeloma dataset consistent with a previous single-center study (Figure 3).33 The data are primarily derived from patients who were subsequently treated with bortezomib but includes a subset of control patients whose treatment was Dex.
We identified a pretreatment gene expression pattern and predictive classifier that is significantly associated with subsequent response to bortezomib but not Dex. Although the association with response appears to be subtle, the significance is supported by bootstrap analyses as well as testing of independent data.57,58 This comparison of predictive accuracy for bortezomib and Dex is not complicated by the previously mentioned confounding variables (extent of prior therapies and prognostic features) because the independent test data derives from patients enrolled in a randomized study that controlled for the number of prior therapies and β-2M levels in each study arm.36 The apparent specificity suggests that there are distinct subsets of patients sensitive to bortezomib or Dex and that these subsets can be distinguished by pretreatment tumor gene expression. A distinct genomic classifier based on OS also showed statistical significance when tested with independent data. However, at this time, we cannot determine whether this association is specific for bortezomib treatment, because the majority of patients in the Dex arm were subsequently treated with bortezomib.
An overview of the gene sets significantly associated with response to bortezomib (Table 3) highlighted pathways, such as NF-κB activity and cell adhesion, whose functions were already clearly implicated as relevant to bortezomib activity in vitro.50-54 This overlap between genomic analyses of clinical specimens and preclinical model systems is encouraging and suggests that some preclinical systems may provide relevant information regarding the drug sensitivity of patients. Many of the pathways associated with PD regulate protein biosynthesis and mitochondrial function, which could relate to protein load in secretory myeloma cells59,60 or the status of mitochondrial apoptotic pathways.61 Consistent with the response prediction, some of these pathways (eg, adhesion- and cytokine-related gene sets) appeared to be bortezomib specific when data for bortezomib versus Dex were compared (Table 3). It will be important to induce and/or inhibit these pathways in model systems to test whether their activity confers sensitivity or resistance to bortezomib, Dex, and/or other anticancer agents.
We note that the survival classifier described here captures outcome-related information that is distinct from clinical prognostic variables (eg, serum albumin and β-2M) as demonstrated by the significant capacity to discern risk groups within the high- as well as the low-risk ISS groups (Figure 5). Studies in lymphoma have drawn similar conclusions.62 Multivariate analyses to integrate the genomic and clinical variables are being investigated; it is hoped that merging these complementary data will enable a better understanding of both clinical trial populations and individual patients.
The predictive accuracy required of a clinical diagnostic for myeloma treatment has not yet been defined. Requirements may vary according to disease stage, therapeutic options (single-agent versus combination regimens), and whether therapy is likely to achieve disease control or cure. Although the classifier described here is promising, further refinement is necessary before it can be considered for clinical use in predicting patient response to single-agent bortezomib in the relapsed setting. The 75% overall accuracy (92% Sn, 33% Sp) might be improved with more patient samples, or it may be that there is not adequate information in the RNA levels of pretreatment, purified myeloma samples to make a significantly more accurate prediction. Additional research is needed to assess the relevance of these genomic predictors in newly diagnosed myeloma and in the context of multiagent therapy that is fundamental to more-effective treatment of myeloma. Key data for such analyses will emerge from genomic research in other large clinical trials, including Total Therapy 2 and 3,14,29 as well as the ongoing HOVON cooperative trial comparing vincristine, doxorubicin, and Dex with bortezomib, doxorubicin, and Dex as induction therapy in newly diagnosed patients. These analyses will help to rapidly highlight the patient groups that benefit from drug combinations as well as those still in need of novel therapies.
Authorship
Contribution: G.M., P.R., P.S., D.S., and K.C.A. designed the research; E.K., P.R., P.S., and K.C.A. performed the research; G.M., B.B., S.R., E.K., A.F., W.L.T., D.S., and D.-L.E. contributed vital new reagents or analytical tools; G.M., B.B., E.K., A.F., and K.C.A. collected the data; G.M., C.M., B.B., F.Z., W.J.C., S.R., E.K., Y.H., P.R., W.L.T., A. Broyl, P.S., J.D.S., P.L.B., D.-L.E., A. Boral, and K.C.A. analyzed the data; and G.M., B.B., P.R., W.L.T., P.S., J.D.S., P.L.B., A. Boral, and K.C.A. wrote the paper.
Conflict-of-interest disclosure: Several of the authors (G.M., B.B., F.Z., S.R., E.K., Y.H., W.L.T., D.S., D.-L.E., and A. Boral) have declared a financial interest in Millennium Pharmaceuticals, Inc, whose product was studied in the present work. Several of the authors (G.M., B.B., S.R., E.K., A.F., W.L.T., D.-L.E., and A. Boral) are currently employed by Millennium Pharmaceuticals, Inc. D.S. is currently employed by Genentech. The remaining authors declare no competing financial interests.
A list of the members of the APEX study group appears as Document S2.
Correspondence: George Mulligan, Clinical Research/Translational Medicine, Millennium Pharmaceuticals, Inc, 40 Landsdowne St, Cambridge, MA 02139; e-mail: george.mulligan@mpi.com.
Presented in poster form at the 47th annual meeting of the American Society of Hematology, Atlanta, GA, December 12, 2005.63
The online version of this article contains a data supplement.
An Inside Blood analysis of this article appears at the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
We thank all patients participating in these clinical trials. We thank MPMx, J. Brown, A. Bolt, S. Kim, A. Damokosh, G. Tucker-Kellogg, M. Lane, M. Morrissey, J. Larsen, T. Hideshima, M. Kauffman, and J. Adams. We also thank R. Roubenoff, H. Danaee, T. Myers, and Y. Meng for critical review of the manuscript.
This work was supported in part by Millennium Pharmaceuticals and Johnson & Johnson Pharmaceutical Research and Development.