Abstract
Each infectious agent represents a unique combination of pathogen-associated molecular patterns that interact with specific pattern-recognition receptors expressed on immune cells. Therefore, we surmised that the blood immune cells of individuals with different infections might bear discriminative transcriptional signatures. Gene expression profiles were obtained for 131 peripheral blood samples from pediatric patients with acute infections caused by influenza A virus, Gram-negative (Escherichia coli) or Gram-positive (Staphylococcus aureus and Streptococcus pneumoniae) bacteria. Thirty-five genes were identified that best discriminate patients with influenza A virus infection from patients with either E coli or S pneumoniae infection. These genes classified with 95% accuracy (35 of 37 samples) an independent set of patients with either influenza A, E coli, or S pneumoniae infection. A different signature discriminated patients with E coli versus S aureus infections with 85% accuracy (34 of 40). Furthermore, distinctive gene expression patterns were observed in patients presenting with respiratory infections of different etiologies. Thus, microarray analyses of patient peripheral blood leukocytes might assist in the differential diagnosis of infectious diseases.
Introduction
Different classes of pathogens trigger specific pattern-recognition receptors (PRRs) differentially expressed on leukocytes.1,2 Leukocytes are components of the innate immune system (granulocytes, natural killer cells), the adaptive immune system (T and B lymphocytes), or both (monocytes and dendritic cells). Blood represents both a reservoir and a migration compartment for these cells that might have been exposed to infectious agents, allergens, tumors, transplants, or autoimmune reactions. Therefore, blood leukocytes constitute an accessible source of clinically relevant information, and a comprehensive molecular phenotype of these cells can be obtained using gene expression microarrays. This technology has already brought new perspectives in the diagnosis and prognosis of cancer,3–5 and the analysis of gene expression signatures in blood leukocytes has led to a better understanding of mechanisms of disease onset and responses to treatment.6–8
Acute infections represent a major cause of morbidity and mortality in the world,9 especially among children. Concomitantly, our ability to identify infectious agents remains inadequate, particularly if the organism is not present in the blood (or other easily accessible site). These diagnostic obstacles can delay initiation of appropriate therapy, which can result in unnecessary morbidity and even death.10 Furthermore, recent outbreaks caused by emerging pathogens9,11 and the increased risk of biologic threats foster the need for improved diagnosis of infectious diseases, especially in the acute setting.
We surmised that leukocytes isolated from the peripheral blood of patients with acute infections will carry unique transcriptional signatures that would, in turn, permit pathogen discrimination. To test this hypothesis, we analyzed gene expression patterns in blood leukocytes from patients with acute infections caused by 4 common human pathogens: (1) influenza A, an RNA virus; (2) Staphylococcus aureus and (3) Streptococcus pneumoniae, 2 Gram-positive bacteria; and (4) Escherichia coli, a Gram-negative bacterium.
Patients, materials, and methods
Patient information
Blood samples were obtained from 29 patients with E coli infections (median age, 2 months; range, 2 weeks to 16 years), 50 patients with S aureus infections (median age, 7 years; range, 1 month to 18 years), 16 with S pneumoniae (median age, 2 years; range, 2 months to 16 years), 36 with influenza A infections (median age, 1.2 years; range, 2 weeks to 36 years), and 7 healthy controls (median age, 11 months; range, 4 months to 9.5 years). Patients were divided into training and test sets according to age and antibiotic treatment (Tables 1–2). All subjects with acute infections and their controls were recruited at Children's Medical Center (CMC) in Dallas, TX. The study was approved by the Institutional Review Boards (IRBs) of the University of Texas Southwestern Medical Center and Baylor Health Care System (IRB nos. 0802-447 and 002-141) and informed consent was obtained for all patients. Microbiologic diagnosis was established by standard bacterial cultures of relevant tissue specimens or blood and by direct fluorescent antigen testing and viral cultures. All potentially eligible patients were identified on a daily basis by the investigators from both the microbiology laboratory database and inpatient admissions records. A second step was then undertaken to confirm eligibility on the basis of history, clinical findings, bacterial and viral cultures, and immunofluorescence tests. Patients with suspected (by clinical findings) or documented (by microbiologic tests) polymicrobial infections, history of immunodeficiency, chronic disease, or receiving steroids or other immunomodulatory agents were excluded. Patients were enrolled once a confirmed microbiologic diagnosis was established. Systematic testing for the presence of concomitant viral infection was initiated after the beginning of the study and respiratory viral cultures were performed in 83 of 98 (85%) patients with bacterial infections. Control samples were obtained from healthy individuals scheduled to undergo elective surgical procedures and from healthy outpatient clinic patients (Table 3).
Processing of blood samples
All blood samples were collected in acid-citrate-dextrose tubes (BD Vacutainer, Becton Dickinson, Franklin Lakes, NJ) at the CMC and immediately delivered at room temperature to the Baylor Institute for Immunology Research (Dallas, TX) for processing. Peripheral blood mononuclear cells (PBMCs) from 3 to 4 mL blood were isolated via Ficoll gradient and immediately lysed in RLT reagent (Qiagen, Valencia, CA) with β-mercaptoethanol (BME) and stored at −80°C (within 4-6 hours from the time of blood draw) in the same laboratory by the same team to standardize the quality and handling of RNA samples.
Microarray assay
Total RNA was isolated using the RNeasy kit (Qiagen) according to the manufacturer's instructions, and RNA integrity was assessed by using an Agilent 2100 Bioanalyzer (Agilent, Palo Alto, CA).
Affymetrix GeneChips.
Double-stranded cDNA was generated from 2 to 5 μg total RNA, followed by single-round in vitro transcription with biotin-labeled nucleotides, using the Affymetrix RNA transcript labeling kits (Affymetrix, Santa Clara, CA). Biotinylated cRNA targets were purified using the Sample Cleanup Module (Affymetrix), and subsequently hybridized, according to the manufacturer's standard protocols, to Affymetrix HGU133A GeneChips (which contain 22 283 probe sets). Arrays were scanned using an Affymetrix confocal laser scanner. Expression results of a set of genes were confirmed by real-time polymerase chain reaction (PCR). Details are shown in Document S1 (available on the Blood website; see the Supplemental Materials link at the top of the online article).
Illumina BeadChips.
These microarrays consist of 50mer oligonucleotide probes attached to 3-μm beads, which are lodged into microwells at the surface of a glass slide. Samples were processed and data acquired by Illumina (San Diego, CA). Targets were prepared using the Illumina RNA amplification kit (Ambion, Austin, TX). cRNA targets were hybridized to Sentrix Hu6 BeadChips (> 46 000 probes), which were scanned on an Illumina BeadStation 500. The Illumina Beadstudio software was used to assess fluorescent hybridization signals.
Raw data obtained for all 144 microrarray analyses are deposited in the public gene expression database GEO (www.ncbi.nlm.nih.gov/geo/) (accession no. GSE6269).
Microarray data analysis
Microarray Suite, version 5.0 (MAS 5.0; Affymetrix) software was used to assess fluorescent hybridization signals, to normalize signals, and to evaluate signal detection calls. Raw signal intensity values for each probe set were analyzed by algorithms in MAS 5.0. A maximum of 8 samples was assigned randomly for hybridization and staining each run day to minimize technical variability.
Normalization of signal values per chip was achieved using the MAS 5.0 global method of scaling to the target intensity value of 500 per GeneChip. Analysis was restricted to probe sets for which a present (P) call was obtained in at least 75% of GeneChips in at least one patient class evaluated (quality control probes). A gene expression analysis software program, GeneSpring, version 7.1 (Agilent), was used to perform statistical analysis, hierarchical clustering, and classification of samples. Nonparametric univariate tests (Mann-Whitney U or Fisher exact test) were used to rank genes on the basis of their ability to discriminate between predefined groups of patients. The ability of the top ranked (ie, classifier) genes to discriminate the predefined class of pathogen was determined by the K-Nearest Neighbors (kNN) method (Document S2 presents details).12
Results
Patient characteristics
We analyzed PBMCs from 29 patients with E coli infections, 50 patients with S aureus infections, 16 patients with S pneumoniae infections, and 36 patients with influenza A infections. We chose young patients because of fewer concomitant diseases and therapies than possibly present in older adults. Patients with underlying immunosuppression, receiving immunomodulatory therapy including corticosteroids, or with significant chronic medical problems were excluded. The median duration of hospitalization at the time of blood draw was 3 days (range, 0-9 days) and the median duration of symptoms was 7 days (range, 1-22 days). The clinical diagnoses included acute respiratory infections, bacteremia, localized abscesses, bone and joint infections, urinary tract infections, and meningitis (Tables 1–2). Patients were treated according to standard hospital protocols and, as such, antimicrobial therapy was promptly initiated in the emergency department.
Step-wise data analysis strategy
To determine whether blood leukocytes isolated from patients with acute infections carry gene expression signatures that allow discrimination between pathogen type, a step-wise analysis was conducted. (1) Statistical group comparison: differentially expressed genes were identified in pair-wise comparisons using the nonparametric Mann-Whitney test. Hierarchical clustering ordered the genes according to their expression levels, revealing reciprocal patterns of expression between the 2 groups. (2) Sample classification: genes capable of discriminating 2 groups of patients, that is, classifiers, were identified through comparison of patient groups of comparable age range and treated with similar classes of antimicrobials (training set). These genes were then evaluated within the same set of patients in a leave-one-out cross-validation scheme. (3) Independent validation of classifier genes: the same genes were tested for their ability to classify an independent group of patients (test set). The patients included in the training sets used for the identification of the classifier genes were selected very carefully to avoid potential confounding factors. After that careful selection, the classifier genes (also described as transcriptional markers) were then evaluated in a new group of patients that was heterogeneous and therefore more representative of a realistic clinical setting (test set). (4) Independent validation across microarray platforms and chips: the results were then further validated in another set of patients (40 new patients and 6 used in previous analyses) using a different microarray platform (Illumina BeadChip or Affymetrix U133plus2 chips).
Transcriptional signatures discriminate patients with influenza A infection from those with bacterial infections
To identify genes differentially expressed between samples from patients with either influenza or bacterial infections, 11 patients with influenza A infections and 12 patients with E coli or S pneumoniae infections were selected as a training set on the basis of similar age groups and antibiotic class treatment. There were no significant differences between the influenza A and the bacterial infection training groups in median age (11 months [range, 1-20 months] versus 4 months [range, 2-23 months]; P = .22) or days of hospitalization prior to sample collection (2 days [range, 1-2 days] versus 2.5 days [range, 2-5 days], P = .06). All 11 patients with influenza A infections were receiving β-lactam antibiotics, as compared with 10 of 12 in the bacterial infection group (P = .16). There were no statistically significant differences in the relative proportions of neutrophils, lymphocytes, and monocytes in PBMCs from the 2 groups (Table S1).
Statistical group comparisons of patients with influenza A and those with bacterial infections yielded 854 differentially expressed genes (P < .01; Table S2), of which 394 were relatively overexpressed in influenza A infections and 460 were overexpressed in bacterial infections. Patients with influenza A displayed a prominent type I interferon (IFN) signature (Figure 1A), including genes coding for antiviral molecules such as myxovirus resistance genes (MX1, MX2); 2′-5′-oligoadenylate synthetases (OAS1, OAS2); guanylate-binding protein 1 (GPB-1); and CIG5 (viperin, virus inhibitory protein, endoplasmic reticulum-associated, IFN-inducible). Genes regulating transcription and translation represent up to 25% of the 460 probe sets expressed at higher levels in the bacterial infection group.
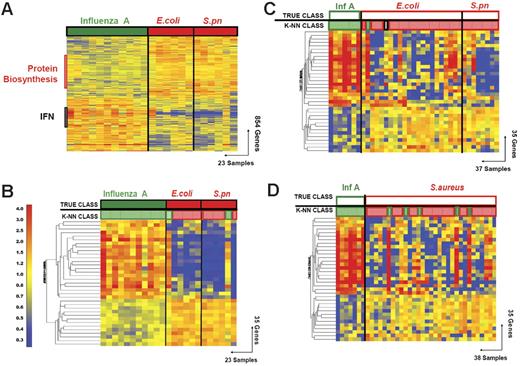
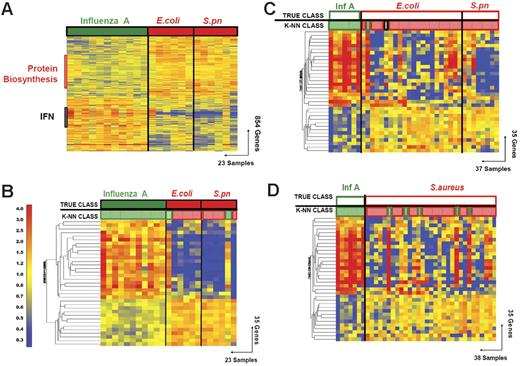
Discriminating patients with influenza A virus infection from patients with bacterial infections. (A) Hierarchical clustering of 854 genes obtained from Mann-Whitney rank test comparison (P < .01) between 2 groups: influenza A (Inf A, 11 samples, green rectangle) and bacterial infections (red rectangle) with E coli (E.coli, 6 samples) or S pneumoniae (S.pn, 6 samples). Transformed expression levels are indicated by color scale, with red representing relatively high expression and blue indicating relatively low expression compared to the median expression for each gene across all donors. The black bar indicates IFN-inducible genes (IFN), and the red bar indicates genes involved in protein biosynthesis. Genes are listed in Table S2. (B) A supervised learning algorithm was used to identify 35 genes presenting the highest capacity to discriminate the 2 classes (Tables 1–2 and S3). Leave-one-out cross-validation of the training set with 35 genes classified the samples with 91% accuracy. The predicted class is indicated by light-colored solid rectangles (green for influenza A and red for bacteria). Two patients with bacterial infections were misclassified. (C) The 35 classifier genes thus identified were tested on an independent set of patients (open rectangles), including 7 new patients with influenza A (green), 23 with E coli, and 7 with S pneumoniae (red) infections. The 37 samples in this test set were classified with 95% accuracy (predicted class is indicated by light-colored rectangles). One patient was misclassified and one patient was indeterminate in class prediction (gray box). (D) The 35 classifier genes identified in panel B were tested on an independent set of patients (open squares), including 7 new patients with influenza A (Inf A), and 31 with S aureus infections. The 38 samples were classified with 87% accuracy.
Discriminating patients with influenza A virus infection from patients with bacterial infections. (A) Hierarchical clustering of 854 genes obtained from Mann-Whitney rank test comparison (P < .01) between 2 groups: influenza A (Inf A, 11 samples, green rectangle) and bacterial infections (red rectangle) with E coli (E.coli, 6 samples) or S pneumoniae (S.pn, 6 samples). Transformed expression levels are indicated by color scale, with red representing relatively high expression and blue indicating relatively low expression compared to the median expression for each gene across all donors. The black bar indicates IFN-inducible genes (IFN), and the red bar indicates genes involved in protein biosynthesis. Genes are listed in Table S2. (B) A supervised learning algorithm was used to identify 35 genes presenting the highest capacity to discriminate the 2 classes (Tables 1–2 and S3). Leave-one-out cross-validation of the training set with 35 genes classified the samples with 91% accuracy. The predicted class is indicated by light-colored solid rectangles (green for influenza A and red for bacteria). Two patients with bacterial infections were misclassified. (C) The 35 classifier genes thus identified were tested on an independent set of patients (open rectangles), including 7 new patients with influenza A (green), 23 with E coli, and 7 with S pneumoniae (red) infections. The 37 samples in this test set were classified with 95% accuracy (predicted class is indicated by light-colored rectangles). One patient was misclassified and one patient was indeterminate in class prediction (gray box). (D) The 35 classifier genes identified in panel B were tested on an independent set of patients (open squares), including 7 new patients with influenza A (Inf A), and 31 with S aureus infections. The 38 samples were classified with 87% accuracy.
The kNN algorithm identified 35 genes that discriminated patients with acute influenza infection from acute bacterial infections (Figure 2; Tables 4 and S3). Leave-one-out cross-validation of this training set correctly classified 21 of the 23 samples (91% accuracy) to either the influenza A or the bacterial infection groups (Figure 1B).
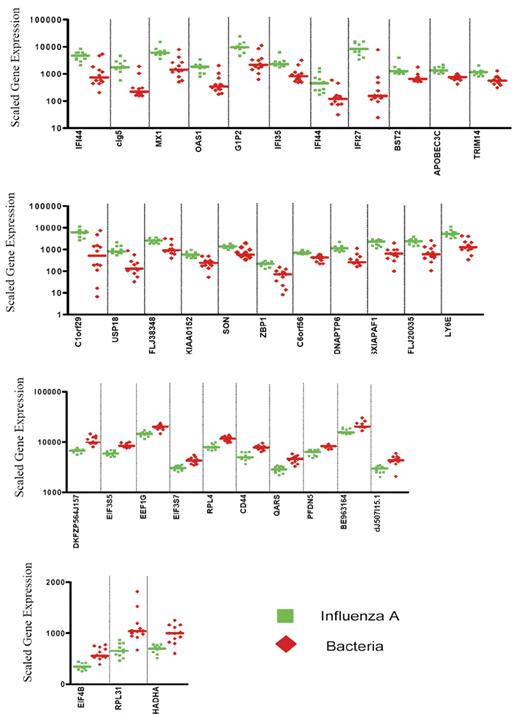
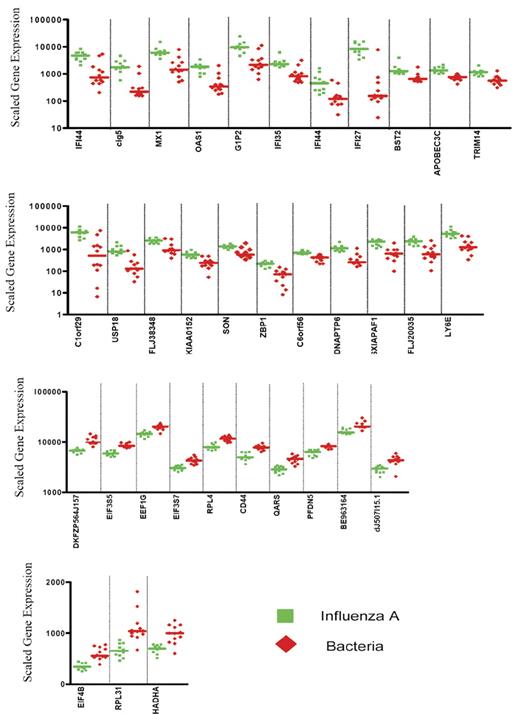
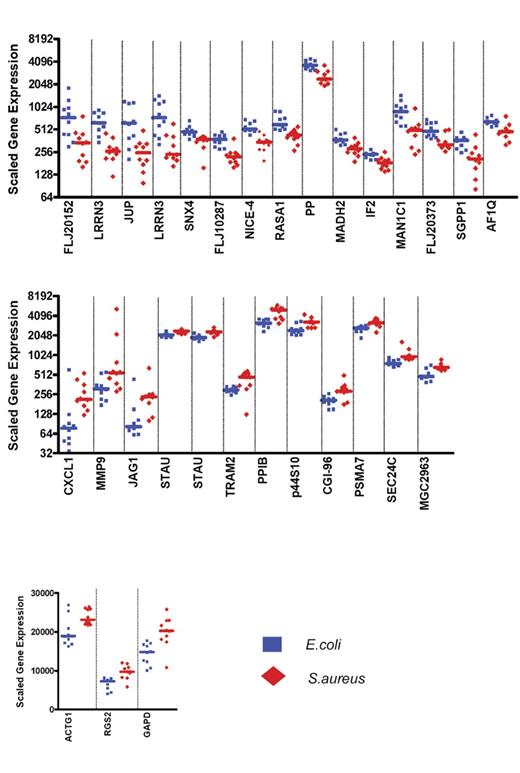
Expression levels of the 35 classifier genes discriminating patients with influenza A infection from patients with bacterial infections. Scaled gene expression values (average difference intensity) are plotted for the 35 classifier genes represented in Figure 1B that discriminate between samples from patients with influenza A (11 samples, green squares) and bacterial infections (6 samples with E coli and 6 samples with S pneumoniae, red diamonds). Each plot represents one sample, lines represent median expression.
Expression levels of the 35 classifier genes discriminating patients with influenza A infection from patients with bacterial infections. Scaled gene expression values (average difference intensity) are plotted for the 35 classifier genes represented in Figure 1B that discriminate between samples from patients with influenza A (11 samples, green squares) and bacterial infections (6 samples with E coli and 6 samples with S pneumoniae, red diamonds). Each plot represents one sample, lines represent median expression.
The ability of the identified classifier genes to discriminate influenza A from the bacterial infections was then validated with independent sets of samples (test sets). The first test set of patients included 7 new patients with influenza A and 30 patients with bacterial infections (7 with S pneumoniae and 23 with E coli infections). Patients were included in the test set without regard to age or type of antibiotic treatment (age: influenza A, 4 years [range, 3 weeks to 36 years]; E coli, 2 months [range, 2 weeks to 16 years]). Predictor genes correctly classified 35 of the 37 samples (95% accuracy; Figure 1C). One sample (INF48) was misclassified and one sample was of indeterminate classification (INF120).
The 35 classifier genes were then evaluated in a second test set, consisting of 7 patients with influenza A infection and 31 patients with S aureus infection, yielding 87% accuracy in discrimination (Figure 1D). Test sets were again selected without regard to age or type of antibiotic treatment (age: influenza A, 4 years [range, 3 weeks to 36 years]; S aureus, 7 years [range, 3 months to 15 years]). Five S aureus samples were misclassified (INF62, INF70, INF89, INF221, and INF242).
About one third of the patients with bacterial infection displayed elevated expression levels of IFN-related genes. This signature, however, had limited effects on classification outcomes because samples obtained from patients with bacterial infections lacked the reciprocal expression signature characteristic of influenza infection (underexpressed genes in influenza compared to bacterial infection) and also in part because expression levels of IFN-inducible genes were lower in samples from patients with bacterial infections (Figure 1C). It is yet unclear as to whether elevated levels of expression of IFN-inducible genes can be attributed to a response to the documented bacterial infection itself,13 or an undiagnosed or preceding viral infection.
Thus, transcriptional signatures of host response to influenza infection and bacterial infection can be identified. These signatures permit the discrimination between these causative agents.
Transcriptional signatures discriminate patients with E coli infections from those with S aureus infections
To identify genes differentially expressed between patients with E coli and S aureus infections, 10 patients per group were selected as training set. There were no significant differences between the E coli and the S aureus infection training groups in median age (2 months [range, 3.5 months to 16 years] versus 12 months [range, 4 months to 10 years]; P = .06). Each group included 6 patients treated with β-lactam antibiotics and 4 with other antibiotic classes. Total peripheral leukocyte counts and the relative proportions of the peripheral blood cell types between the 2 groups were not significantly different (Table S1). The median number of days of hospitalization prior to sample collection was 2 days for the E coli group and 4 days for the S aureus group (P = .01), a difference that may be accounted for by the time interval typically required for definitive microbiologic diagnosis.
Statistical group comparisons yielded 211 genes with significantly different expression levels (P < .01; Table S4; Figure 3A). Expression levels of a selection of genes were independently confirmed by real-time PCR (Figure S1; Method S1). A number of genes overexpressed in S aureus compared to E coli are associated with neutrophil activity, including chemoattractant molecules such as CXCL1 (CXC chemokine ligand 1, GRO-1) and PPIB (cyclophilin B).14,15 Furthermore, the matrix metalloproteinase 9 (MMP9) plays an important role in neutrophil extravasation and migration16 ; secretory granule proteoglycan 1 (PRG1) participates in packaging of granule proteins in human neutrophils17 ; and ALOX5AP activates arachidonate 5-lipoxygenase and prolongs the capacity of neutrophils to synthesize leukotrienes.18 Finally, neutrophils have recently been identified as the main source of S100A8 and S100A9 (Calgranulin A and B, alias MRP 8 and 14) in a S aureus infection model.19 These results suggest that “neutrophil activity” may, in part, explain differences in levels of gene expression between samples obtained from patients with E coli and S aureus infections. Previous studies in patients with systemic lupus erythematosus (SLE) demonstrated a “granulopoiesis signature” that was associated with the presence of low-density neutrophils that copurified with mononuclear cells during density gradient centrifugation.6 In line with these findings, low-density cells were found in PBMCs isolated from 11 randomly selected patients with acute S aureus infection (data not shown).
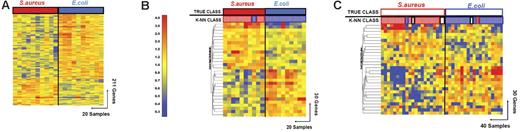
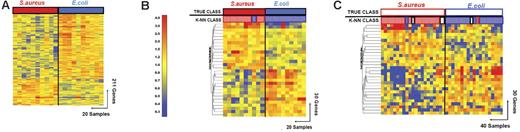
Discriminating patients with S aureus infections from patients with E coli infections. (A) Hierarchical clustering of 211 genes obtained from Mann-Whitney rank test comparison (P < .01) between 2 groups: Staphylococcus aureus (S aureus, 10 samples, red rectangle) and Escherichia coli (E coli, 10 samples, blue rectangle) infections. Transformed expression levels are indicated by color scale, with red representing relative high expression and blue indicating relative low expression compared to the median expression for each gene across all donors. Genes are listed in Table S4. (B) A supervised learning algorithm was used to identify 30 genes presenting the highest capacity to discriminate the 2 classes (Table S6). Leave-one-out cross-validation of the training set with 30 classifier genes grouped the samples with 95% accuracy. (C) The 30 classifier genes thus identified were tested on an independent set of patients (open rectangles), including 21 new patients with S aureus and 19 with E coli infections. The 40 samples in this test set were predicted with 85% accuracy (predicted class is indicated by light-colored rectangles). Of these 40 samples, only 2 were misclassified, whereas the class of 4 other samples could not be determined (open rectangles).
Discriminating patients with S aureus infections from patients with E coli infections. (A) Hierarchical clustering of 211 genes obtained from Mann-Whitney rank test comparison (P < .01) between 2 groups: Staphylococcus aureus (S aureus, 10 samples, red rectangle) and Escherichia coli (E coli, 10 samples, blue rectangle) infections. Transformed expression levels are indicated by color scale, with red representing relative high expression and blue indicating relative low expression compared to the median expression for each gene across all donors. Genes are listed in Table S4. (B) A supervised learning algorithm was used to identify 30 genes presenting the highest capacity to discriminate the 2 classes (Table S6). Leave-one-out cross-validation of the training set with 30 classifier genes grouped the samples with 95% accuracy. (C) The 30 classifier genes thus identified were tested on an independent set of patients (open rectangles), including 21 new patients with S aureus and 19 with E coli infections. The 40 samples in this test set were predicted with 85% accuracy (predicted class is indicated by light-colored rectangles). Of these 40 samples, only 2 were misclassified, whereas the class of 4 other samples could not be determined (open rectangles).
Thirty classifier genes that discriminate between the training set of patients with E coli and S aureus infections were identified (Figure 4; Tables 5 and S6). In leave-one-out cross-validation 19 of 20 samples were classified correctly (95% accuracy; Figure 3B). One patient with a S aureus infection (INF89) was misclassified. The classifier genes were validated with an independent set of patients with S aureus (n = 21) and E coli (n = 19) infections, which were again selected without regard to age or type of antibiotic treatment (S aureus, 9 years [10 months to 18 years]; E coli, 2 months [2 weeks to 5 months]). The 30 genes correctly classified 34 of the 40 samples (85% accuracy; Figure 3C). Two samples (INF175 and INF206) were misclassified and 4 samples were indeterminate in their classification (INF168, INF220, INF281, and INF315). The greater heterogeneity of clinical disease and severity represented by the patients with S aureus infections may contribute to the lower predictive accuracy for this group, although no specific pattern of misclassification was evident.
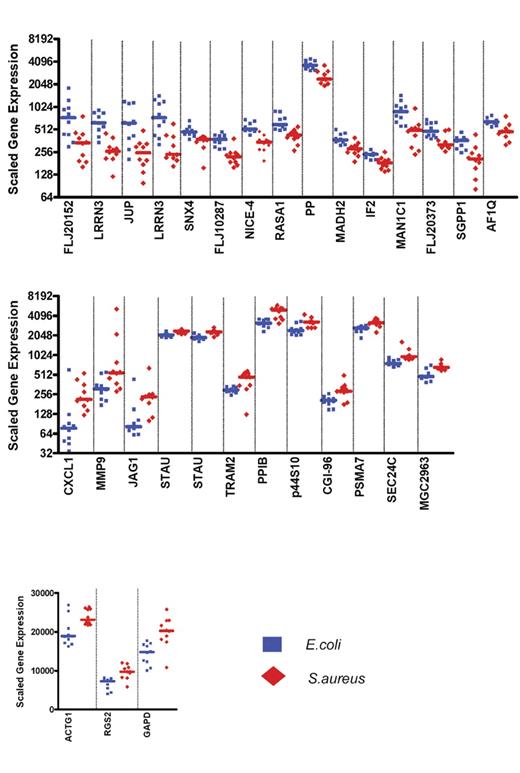
Expression levels of the 30 classifier genes discriminating patients with E coli infections from patients with S aureus infections. Scaled gene expression values (average difference intensity) are plotted for the 30 classifier genes represented Figure 3B that discriminate between samples from patients with E coli (10 samples, blue squares) and S aureus infections (10 samples, red diamonds). Each plot represents one sample, lines represent median expression.
Expression levels of the 30 classifier genes discriminating patients with E coli infections from patients with S aureus infections. Scaled gene expression values (average difference intensity) are plotted for the 30 classifier genes represented Figure 3B that discriminate between samples from patients with E coli (10 samples, blue squares) and S aureus infections (10 samples, red diamonds). Each plot represents one sample, lines represent median expression.
Thus, these results demonstrate that blood leukocyte transcriptional signatures distinguish disease etiology in patients with acute infections caused by S aureus or by E coli. Furthermore, notable functional convergence among discriminatory signatures were identified; IFN-inducible genes were found among genes overexpressed in patients with influenza A, whereas genes associated with neutrophils were expressed at higher levels in S aureus compared to E coli groups.
Classifier genes discriminating samples from patients with acute influenza A, E coli, S aureus, or S pneumoniae infections show minimal overlap
We have defined sets of classifier genes that discriminate patients with influenza A versus bacterial infections and patients with E coli versus S aureus infections. To complete our panel of classifier genes we performed additional pair-wise comparisons and identified sets of genes discriminating patients with S pneumoniae infections. Comparison of E coli (n = 11) and S pneumoniae (n = 11) infection groups yielded 264 significantly differently expressed genes (P < .01) and 45 classifier genes (Figure S2A-B; Tables S7–S8). Sample class was assigned correctly for 20 of 22 samples (91% accuracy) in leave-one-out cross-validation of the training set. Comparison of S aureus (n = 12) and S pneumoniae (n = 11) infection groups yielded 127 differently expressed genes (P < .01) and 30 classifier genes (Figure S2C-D; Tables S9–S10). Sample class was assigned correctly for 19 of 23 samples (83% accuracy) in leave-one-out cross-validation of the training set.
The sets of classifier genes obtained for each pair-wise analysis were systematically compared and found to be almost mutually exclusive (Figure 5A). Furthermore, none of the 102 genes that discriminated one bacterial species from the other was necessary to distinguish influenza A from bacterial infections (Figure 5B). Thus, we find that multiple infectious disease etiologies can be distinguished using independent sets of transcriptional signatures.
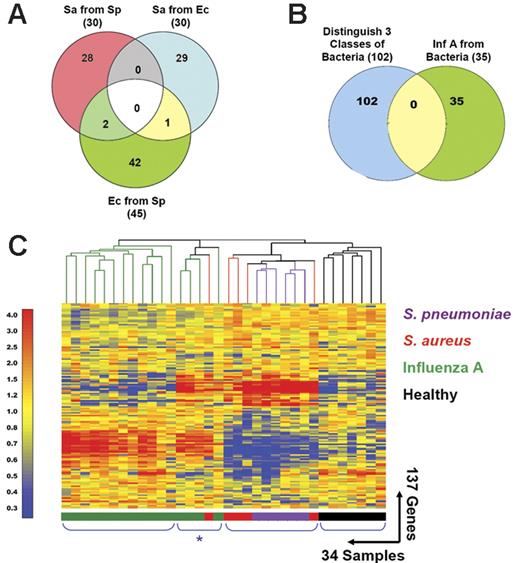
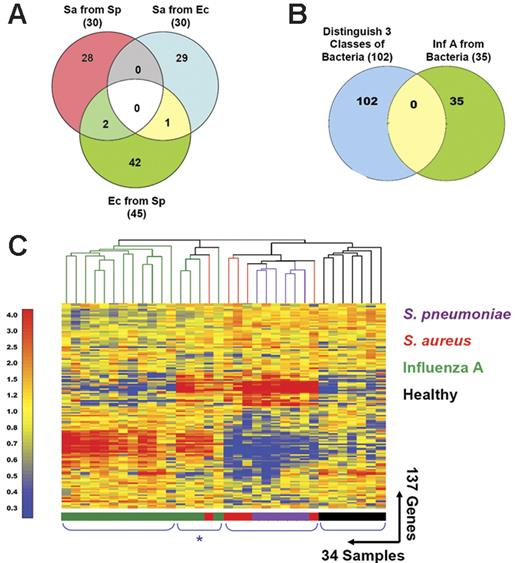
Distinctive patterns of gene expression in circulating leukocytes obtained from patients with acute respiratory infections. (A) In addition to the 30 classifier genes found to discriminate S aureus from E coli (Venn diagram, right: Sa from Ec; Figure 2; Table S6), we identified 30 genes that distinguish S aureus from S pneumoniae (Venn diagram, left: Sa from Sp; Figure S2; Table S10) and 45 genes that distinguish E coli from S pneumoniae (Venn diagram, bottom: Ec from Sp; Figure S2; Table S8). Only 3 genes were shared between either of these groups. (B) The 3 groups of genes found to discriminate samples from patients with bacterial infections shown in panel A were merged (102 unique genes, Venn diagram, left) and compared to the classifier genes used to discriminate influenza A from bacterial infections (35 genes, Venn diagram, right; Figure 1; Table S3). No genes were shared between these 2 groups. (C) The 137 classifier genes that discriminate influenza A from bacterial infections and the 3 groups of patients with different bacterial infections were merged and used to generate discriminatory patterns of expression among 27 patients with respiratory infections and 7 healthy volunteers. Values were normalized to the median expression of each gene across all donors. Clustering of conditions partitioned samples into 4 major groups. Four samples belonging to the influenza A group and one from the S aureus formed a distinct subgroup characterized by a mixed signature (*) and are listed in Table 1 (Figure 5C*).
Distinctive patterns of gene expression in circulating leukocytes obtained from patients with acute respiratory infections. (A) In addition to the 30 classifier genes found to discriminate S aureus from E coli (Venn diagram, right: Sa from Ec; Figure 2; Table S6), we identified 30 genes that distinguish S aureus from S pneumoniae (Venn diagram, left: Sa from Sp; Figure S2; Table S10) and 45 genes that distinguish E coli from S pneumoniae (Venn diagram, bottom: Ec from Sp; Figure S2; Table S8). Only 3 genes were shared between either of these groups. (B) The 3 groups of genes found to discriminate samples from patients with bacterial infections shown in panel A were merged (102 unique genes, Venn diagram, left) and compared to the classifier genes used to discriminate influenza A from bacterial infections (35 genes, Venn diagram, right; Figure 1; Table S3). No genes were shared between these 2 groups. (C) The 137 classifier genes that discriminate influenza A from bacterial infections and the 3 groups of patients with different bacterial infections were merged and used to generate discriminatory patterns of expression among 27 patients with respiratory infections and 7 healthy volunteers. Values were normalized to the median expression of each gene across all donors. Clustering of conditions partitioned samples into 4 major groups. Four samples belonging to the influenza A group and one from the S aureus formed a distinct subgroup characterized by a mixed signature (*) and are listed in Table 1 (Figure 5C*).
Distinct expression patterns in patients with acute respiratory infections caused by different pathogens
Next, we examined gene expression patterns in a mixed cohort of patients presenting with the same clinical manifestations. Because lower respiratory infections represent the most common infection leading to hospitalization, we focused our analysis in this particular group of patients. Sets of classifier genes identified throughout this study (Figure 5A-B) were merged and used to generate expression patterns in the group of patients with either influenza or bacterial infections who presented with clinical evidence of lower respiratory infection (27 samples listed Table 1; used in Figure 5C). Seven samples collected from healthy volunteers were used as a reference (Table 3). Hierarchical clustering of genes and samples identified 4 prototypical expression signatures; healthy controls were clearly distinguishable from all the infectious disease groups based on PBMC expression profiles. This finding is in itself remarkable because none of the training sets used to generate the classifiers included samples from healthy volunteers. A second signature was associated with samples from patients with influenza A infection (including IFN-inducible genes) and was clearly different from a third signature, which characterized infections caused by S aureus and S pneumoniae (including neutrophil-associated genes). Distinctions between these 2 Gram-positive bacteria were minimized by the overt dominance of signatures differentiating the 3 major classes of samples. Interestingly, 4 samples belonging to the influenza A group and one from the S aureus group were characterized by a fourth signature, which combined elements of the previous ones (IFN-inducible and neutrophil-associated genes: Figure 5C, indicated by the asterisk). This finding suggests one of at least 2 possibilities: (1) the mixed signatures arise as the result of coinfections that could not be detected by routine diagnostic methods, or (2) the analysis of PBMC transcriptional signatures can reveal the existence of distinct patient subgroups. Further review of the medical records of the 5 patients with mixed signature, identified 3 patients with influenza (nos. 101, 128, and 132) who had radiologic evidence of pneumonia and white blood cell differential counts with 11%, 16%, and 28% bands, respectively. Thus, although not proven, the evidence suggests the possibility of bacterial coinfections in these 3 cases. A larger patient cohort will be necessary to investigate these possibilities and identify potential clinical implications.
These results clearly demonstrate that discriminative blood leukocyte transcriptional patterns that identify the different microbial pathogens can be obtained in patients presenting with similar clinical findings.
Results can be reproduced in a distinct set of samples and across microarray platforms
Our study design includes a training set for the identification of classifiers (Figure 1B, influenza versus bacteria; n = 23 samples) and 2 test sets to independently validate our findings (Figure 1C, influenza versus bacteria; n = 37 samples; and Figure 1D an additional 31 patients infected with S aureus). These data, obtained from a total of 91 patients, were generated using Affymetrix U133A GeneChips. To take data validation one step further to unequivocally confirm our findings, we carried out a similar analysis on additional sets of patients using different microarray platforms. PBMC expression profiles from a new set of patients were analyzed using the most recent version of Affymetrix GeneChips (U133 plus 2.0); 16 new patients with acute influenza and bacterial infections were recruited and the remaining RNA from 6 patients with bacterial infections used in a prior analysis was relabeled. Infections caused by S aureus or S pneumoniae could again be distinguished almost perfectly from infections caused by influenza (Figure 6A; one influenza sample grouped in the bacterial infection cluster). A discriminative signature was also obtained in patients with acute respiratory infection (Figure 6B). The mixed signature that we have described initially was not observed for any of the samples in this new set of patients that segregated perfectly. This fact can be attributed to the smaller number of patients recruited or possibly to the fact that the most recent samples (Figure 6B) were collected during the 2005-2006 flu season, which was particularly mild. Earlier samples (Figure 5C), on the other hand, were collected during the 2003-2004 flu season, which produced an unusually large number of severe cases as well as bacterial superinfections.
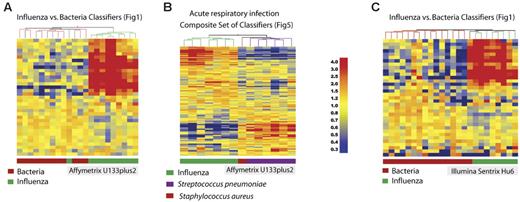
Independent confirmation and validation across microarray platforms. (A) A new set of data obtained from patients with acute influenza (n = 10) and bacterial infection (S aureus: n = 6; S pneumoniae: n = 6) was analyzed using Affymetrix U133 plus 2.0 GeneChips. The original classifier genes found to discriminate influenza A from bacterial infections (35 genes, Venn diagram, right; Figure 1; Table S3) were used to cluster this new set of samples. (B) A subset of 14 samples from patients with acute respiratory infection included in panel A were clustered using the list of 137 transcripts from Figure 5. (C) Another independent set of samples was obtained from a new set of patients with acute influenza (n = 8) or bacterial infection (S aureus: n = 13; S pneumoniae: n = 3) analyzed using Illumina Sentrix Hu6 whole genome BeadChips. Classifier genes used to discriminate influenza A from bacterial infections (35 genes, Venn diagram, right; Figure 1; Table S3) were used to cluster this new set of samples. Transformed expression levels are indicated by color scale, with red representing relative high expression and blue indicating relative low expression compared to the median expression for each gene across all donors.
Independent confirmation and validation across microarray platforms. (A) A new set of data obtained from patients with acute influenza (n = 10) and bacterial infection (S aureus: n = 6; S pneumoniae: n = 6) was analyzed using Affymetrix U133 plus 2.0 GeneChips. The original classifier genes found to discriminate influenza A from bacterial infections (35 genes, Venn diagram, right; Figure 1; Table S3) were used to cluster this new set of samples. (B) A subset of 14 samples from patients with acute respiratory infection included in panel A were clustered using the list of 137 transcripts from Figure 5. (C) Another independent set of samples was obtained from a new set of patients with acute influenza (n = 8) or bacterial infection (S aureus: n = 13; S pneumoniae: n = 3) analyzed using Illumina Sentrix Hu6 whole genome BeadChips. Classifier genes used to discriminate influenza A from bacterial infections (35 genes, Venn diagram, right; Figure 1; Table S3) were used to cluster this new set of samples. Transformed expression levels are indicated by color scale, with red representing relative high expression and blue indicating relative low expression compared to the median expression for each gene across all donors.
Microarray data are notoriously difficult to compare across different platforms.20–22 Our initial results obtained with Affymetrix GeneChips were nevertheless reproduced in an entirely new set of 24 samples using the Illumina whole genome Sentrix Hu6 BeadChips (Figure 6C; only one sample from the bacterial infection group clustered with influenza samples). In this cohort, only 2 patients belonging to the S aureus or S pneumoniae group presented with acute respiratory infection.
Altogether 144 microarray analyses have been carried out in the context of this study, including 137 on samples collected from 131 patients with acute infections. Along with the confirmation obtained by real-time PCR (Figure S1 and Table S5), the independent data validation carried out across microarray platforms attests to the robustness of our findings.
Discussion
A number of studies have shown that different transcriptional programs could be triggered on exposure of immune cells to various pathogens in vitro,23–26 and more recently transcriptional signatures have been identified in the blood of patients presenting with infections.27–29
The comparative analysis of a compendium of host-pathogen microarray data sets (encompassing 32 studies) identified both common host transcriptional response to infections and pathogen-specific signatures.30 Broad similarities exist, with, for instance, dynamic cascades of cytokines and chemokines involved in the activation and recruitment of immune cells observed in the context of fungal, bacterial, or viral infections.31–35 However, 2 factors contribute to the specificity of transcriptional responses to infections: (1) the diversity of the molecular mechanisms involved in pathogen recognition, and (2) alterations of host responses by pathogens. On activation, Toll-like receptor (TLR) family members trigger signaling pathways that share common components while retaining unique characteristics accounting for the specificity of transcriptional responses.36 Hence, qualitative and quantitative differences in the responses to Gram-positive and Gram-negative bacteria, respectively, recognized by TLR2 and TLR4, have been observed.24,25 Furthermore, responses measured in dendritic cells exposed to influenza virus (through TLR3), E coli (through TLR4), and Candida (through TLR2/TLR4) were also found to be markedly different.26 Reprogramming of host cells by pathogens also contributes significantly to the diversification of transcriptional responses to infection. As measured by microarrays, mycobacterial products are, for instance, able to inhibit IFN-γ–induced gene regulation in macrophages.37 Similarly, microarray studies have demonstrated the ability of herpes virus, pseudorabies virus, hepatitis C, varicella-zoster virus, or rhinovirus to limit the ability of the host to develop effective antiviral responses by a variety of mechanisms.38–42 Altogether the vast body of in vitro experimental data accumulated over recent years suggests that hosts can mount pathogen-specific transcriptional responses to infections.
Here, we have demonstrated that blood leukocyte gene expression patterns can be used to distinguish patients with acute infections caused by 4 different pathogens: influenza A virus, the Gram-negative bacterium, E coli, and Gram-positive bacteria S aureus and S pneumoniae, which are among the most common infections leading to hospitalization of children.
Two parameters might account for differences in gene expression levels observed in blood leukocytes: (1) changes in transcriptional activity (eg, up-regulation of IFN-inducible genes) or (2) an altered cellular composition of blood samples (eg, neutrophil signature). Changes in expression due to either one or both of these parameters may be mediated directly by pathogen-derived molecules or the action of secondary factors released by the host (eg, cytokines). We have not observed major differences in the cellular composition of blood samples obtained from the different groups of patients. Indeed, it is well established in clinical practice that the routine white blood cell and differential counts cannot distinguish between viral versus bacterial infections and much less between infections caused by Gram-positive and Gram-negative bacteria. However, studies from our group have found earlier that subtle differences might account for observed transcriptional signatures as exemplified by the neutrophil signature in SLE, which is due to enhanced efflux of low-density neutrophils present in PBMC preparations.6 The site of disease involvement may also influence expression profiles observed in blood leukocytes and reflects the predilection of certain species of pathogens for different infection sites. E coli, for example, is more likely to cause urinary tract infection, whereas the most common clinical manifestations of S aureus are skin/soft tissue infections and osteomyelitis. The results obtained in the present study suggest, however, that distinctive expression signatures can be found in the context of a single disease manifestation. Indeed, when analyzing samples from patients with lower respiratory infections, a clear separation between infections caused by the different pathogens was observed, confirming the existence of pathogen-associated transcriptional signatures. Notably, these findings have been the object of extensive validation, in multiple independent sets of patients, by PCR, and across microarray platforms. Furthermore, this extensive data set (148 patient transcriptional profiles) is made available in a public domain repository (see “Patients, materials, and methods” for details).
Our ability to identify etiologic agents responsible for acute infections remains disappointingly low in many clinical situations, and the analysis of blood leukocyte transcriptional profiles has the potential to transform our approach to diagnosis in infectious diseases.43,44 Because our goal was to establish the proof of concept, that is, that blood leukocytes carry signatures that allow discrimination among different microbial pathogens, blood samples were obtained from hospitalized patients with defined infections. Using this approach we were able to establish that leukocytes isolated from the peripheral blood of patients carry transcriptional signatures that can be used to distinguish infectious diseases of different etiologies. Additional studies will be necessary to evaluate the merits of this approach in a relevant clinical setting, that is, the emergency room. Furthermore, it will be important to determine whether transcriptional analysis of blood leukocytes can provide information that would permit following the progression of the disease and assess risks of complications.
In conclusion, this study illustrates the plasticity of immune responses to pathogens in the blood at the transcriptional level and highlights the potential value of blood leukocyte transcriptional signature analyses as an adjunctive means of diagnosis of infectious diseases.
Authorship
Contribution: O.R. designed research, contributed clinical samples/data, analyzed data, and wrote the paper; W.A. performed research and analyzed data; W.C. performed research, contributed clinical samples/data, and analyzed data; M.A. performed research, contributed clinical samples/data, and analyzed data; C.G. performed research and analyzed data; K.M.W. analyzed data; A.M. performed research, contributed clinical samples/data, and analyzed data; B.P. performed research and analyzed data; J.B. designed research and wrote the paper; A.K.P. designed research, analyzed data, and wrote the paper; and D.C. designed research, analyzed data and wrote the paper.
O.R., W.A., and W.C. contributed equally to the article. J.B., A.K.P., and D.C. codirected the presented work.
Conflict-of-interest disclosure: The authors declare no competing commercial or financial interests.
Correspondence: Octavio Ramilo, Damien Chaussabel, or Jacques Banchereau, Baylor Institute for Immunology Research, 3434 Live Oak, Dallas, TX 75204; e-mail: octavio.ramilo@utsouthwestern.edu, damienc@baylorhealth.edu, or jacquesb@baylorhealth.edu.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
This work was supported by Baylor Health Care Systems Foundation, Children's Medical Center Dallas Foundation, DANA Foundation (A.K.P.), Defense Advanced Research Planning Agency (J.B.), and the National Institutes of Health grants U19 AIO57234-02 and CA78846 (J.B.). J.B. holds the Caruth Chair in Organ Transplantation Immunology. A.K.P. holds the Ramsay Chair for Cancer Immunology Research.
We thank our patients and their parents/guardians for agreeing to participate in the study. We thank Evelyn Torres, RN, for help in sample collection; Elizabeth T. Kraus, Marylene Leogier, and Kristin Long for sample processing; Dr Virginia Pascual for flow cytometry interpretation; Steve Scholl and T. Brooke McClendon for sample hybridization; Laurence Monnet for help with data analysis; Victoria Cantrell for help with real-time RT-PCR; Carson Harrod for editorial help; and Cindy Samuelsen and Nicolas Taquet for administrative and technical help. We thank Drs Ira Mellman and Gerard Zurawski for critical reading of the manuscript. We thank Dr Michael Ramsay for continuous support.