Abstract
Signal transducer and activator of transcription 1 (STAT1) is a critical mediator of interferon-γ (IFN-γ)–induced transcription that is regulated through posttranslational modifications and through transacting proteins such as protein inhibitor of activated STAT1 (PIAS1). PIAS proteins have been shown to function as E3-type small ubiquitin-like modifier (SUMO) ligases, and sumoylation has been identified as a modulatory mechanism for several transcription factors. Here we show that STAT1 is subject to SUMO-1 modification, and sumoylation occurs in vivo and in vitro at a single, evolutionary conserved amino acid residue Lys703. Members of the PIAS family of proteins were found to strongly stimulate sumoylation of STAT1. Furthermore, activation of STAT1 by IFN-γ or pervanadate induced SUMO-1 conjugation. Mutation of Lys703 in STAT1 resulted in increased IFN-γ–mediated transactivation, suggesting a negative regulatory function for sumoylation. These results indicate that STAT1 is covalently modified by SUMO-1 in cytokine signaling and that PIAS proteins promote SUMO-1 conjugation to STAT1.
Introduction
Regulation of signal transducer and activator of transcription 1 (STAT1) involves posttranslational modifications such as phosphorylation of Tyr701 and Ser727 as well as arginine methylation.1,2 STAT1 signaling is negatively regulated by protein tyrosine phosphatases (PTPs), suppressors of cytokine signaling (SOCS), and protein inhibitor of activated STAT1 (PIAS) proteins.1-3 The family of PIAS proteins consists of 5 members that have been implicated in the regulation of several nuclear proteins. PIAS1 and PIAS3 were identified as interaction partners for STAT1 and STAT3, respectively, and they were found to inhibit the DNA-binding activity of activated STATs.3-5 The other members, PIASxα/ARIP3 (androgen receptor–interacting protein 3), PIASxβ/Miz1, and PIASy, function as transcriptional coregulators for steroid hormone receptors.6-8 Recently, several PIAS proteins have been shown to function as E3-type small ubiquitin-like modifier (SUMO) ligases.9-12 SUMO-1, -2, and -3 are small modifier proteins that are covalently conjugated to specific lysine residues of target proteins.13 A number of nuclear proteins, such as transcription factors AR, p53, and c-Jun, become modified by SUMO-1 conjugation, and this reversible posttranslational modification has been implicated in regulation of protein-protein interactions, protein stability, localization, or activity.9,13,14 However, it is currently unknown whether STAT factors are modified through sumoylation.
Study design
Reagents
Antibodies used include anti–SUMO-1 (mouse anti–GMP-1) (Zymed, San Francisco, CA); anti-HA (clone 16B12) (Berkeley-Antibody, Richmond, CA); anti-Flag (anti-Flag M2) (Sigma Aldrich, St Louis, MO); anti-STAT1 (N terminus) (Transduction Laboratories, BD Biosciences); anti-STAT3 (Santa Cruz Biotechnology, Santa Cruz, CA) biotinylated antimouse (Dako, Glustrup, Denmark) and streptavidin-biotin horseradish peroxidase conjugate (Amersham Pharmacia Biotech, Buckinghamshire, United Kingdom). Human interferon-γ (huIFN-γ) was purchased from Immugenex, Los Angeles, CA.
Plasmid constructs
SUMO-1–Flag and SUMO-1–Flag–Flag were a kind gift from Dr H. Yasuda.15 The SUMO-1, Flag-PIAS1, Flag-PIAS1mut (PIAS1Δ310-407), Flag-PIAS3, Flag-ARIP3 Flag-ARIP3mut (ARIP3Δ347-418) plasmids4,9 as well as STAT1-WT-HA and GAS-luc reporter construct have been previously described.16 The STAT1 Lys703Arg mutation was created from STAT1-WT-HA using direct polymerase chain reaction (PCR) mutagenesis with the following primers: 5′GGAACTGGATATATCAGGACTGAGTTGATTTCTGTGTCTG-3′ and 5′-CAGACACAGAAATCAACTCAGTCCTGATATATCCAGTTCC-3′.
Transfections, luciferase assay, and immunodetection
COS-7 cells were electroporated using a Bio-Rad (Hercules, CA) gene pulser at 260 V and 960 microfarads (μF) and lysed in Triton X lysis buffer supplemented with 5 mM N-ethylmaleimide (NEM) (Sigma Aldrich). HeLa cells were transfected using a calcium phosphate method. For luciferase assay, U3A cells were transfected using a calcium phosphate method as described.16 Immunodetection was performed as described.16
In vitro sumoylation reaction
GST–SUMOGG-1, GST–Ubc-9, GST-SAE2, and GST-SAE1 were produced in Escherichia coli BL21-CodonPlus bacteria (Stratagene, La Jolla, CA) and purified with glutatione Sepharose 4B (Amersham Pharmacia Biotech). Purified proteins were eluted in buffer containing 50 mM HEPES (N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid) (pH 7.8), 100 mM NaCl, 10 mM MgCl2, 10% glycerol, and 20 mM glutathione. The conjugation reaction was carried out as described.9
Results and discussion
We have previously analyzed PIAS proteins in regulation of steroid receptor and cytokine receptor–mediated transcription.6,17 The SUMO E3 ligase function of PIAS proteins prompted us to examine whether STAT1 is subject to sumoylation. The level of free SUMO-1 in cells is low and apparently rate limiting, because ectopic expression of SUMO-1 increases the level of sumoylation of target proteins. To this end, COS-7 cells were transfected with HA-tagged STAT1 (STAT1-WT-HA) alone or together with SUMO-1, Flag-tagged SUMO-1 (Flag–SUMO-1), or tandem Flag–Flag–SUMO-1 (Figure 1A, lanes 1-4). STAT1 was immunoprecipitated with anti-HA antibody and analyzed by immunoblotting. Both anti–SUMO-1 and anti-HA antibodies reacted with a major band of about 110 kDa in cells transfected with STAT1-WT-HA and SUMO-1 (Figure 1A, lanes 1-2). The molecular mass of the recognized protein agrees with the covalent attachment of one SUMO-1 moiety to STAT1. In cells transfected with STAT1-WT-HA and Flag–SUMO-1 or Flag–Flag–SUMO-1, the mobility of the anti–SUMO-1 and anti-HA detected bands was shifted according to the presence of Flag sequences in the SUMO-1 expression constructs (Figure 1A, lanes 3-4). This conclusion was verified by immunoprecipitating the same lysates with anti-Flag antibody, where STAT1 was detected only in cells transfected with Flag-tagged SUMO-1 constructs (data not shown).
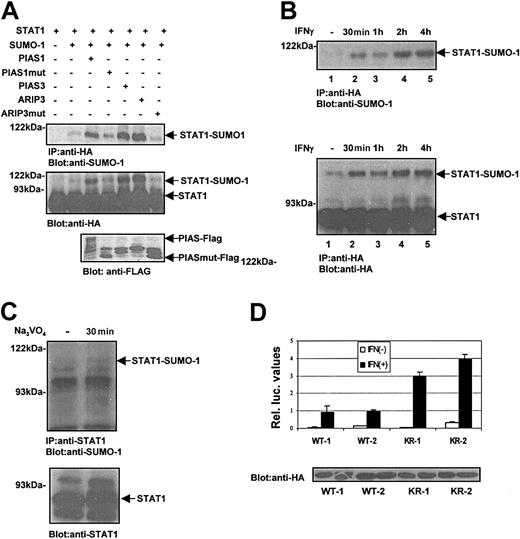
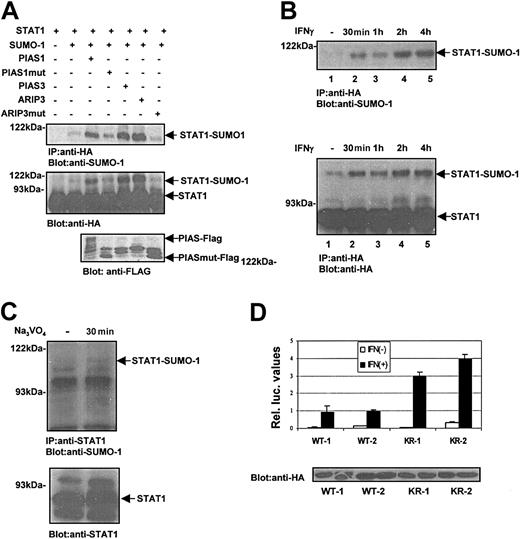
SUMO-1 conjugation to STAT1 in vivo and in vitro. (A) Sumoylation of STAT1-WT and STAT1-KR (Lys703Arg mutant) in COS-7 cells. COS-7 cells were transfected with 2 μg STAT1-WT-HA (lanes 1-4) or STAT1-KR-HA (lanes 5-8) together with different SUMO-1 constructs (2 μg) as indicated. After 36 hours the cells were lysed in Triton X lysis buffer (50 mM Tris [tris(hydroxymethyl)aminomethane]–HCl, pH 7.4; 150 mM NaCl; 1 mM EDTA [ethylenediaminetetraacetic acid]; 50 mM NaF; 5 mM NEM; 1% Triton X-100; and 10% glycerol and protease inhibitors), and equal amounts of total cell lysates were immunoprecipitated with anti-HA antibodies and subjected to sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblotting with anti–SUMO-1 and anti-HA antibodies. (B) Sequence comparison of consensus SUMO-1 attachment sites in RanGAP1, PML, Sp100, p53, IκBα, AR, and STAT1. The consensus residues are marked in italics. The asterisk indicates the lysine that serves as a potential SUMO-1 attachment site. STAT1 sequence comparison starts at amino acid 698. (C) SUMO-1 conjugation of STAT1 in vitro. STAT1-WT (lanes 1-2) and STAT1-KR (lanes 3-4) were in vitro translated in the presence of [35S]methionine using TNT-coupled transcription/translation system (Promega, Madison, WI). The in vitro–translated products were then used for an in vitro SUMO-1 conjugation reaction. The samples were resolved by SDS-PAGE gel and visualized by fluorography. (D) Sumoylation of endogenous STAT1. HeLa cells were untransfected or transfected with SUMO-1 (2 μg) plasmid using a calcium phosphate method. After 36 hours the cells were lysed as described in panel A. Sumoylation of endogenous STAT1 was analyzed after immunoprecipitation using anti-STAT1 antibody and Western blotting with anti–SUMO-1.
SUMO-1 conjugation to STAT1 in vivo and in vitro. (A) Sumoylation of STAT1-WT and STAT1-KR (Lys703Arg mutant) in COS-7 cells. COS-7 cells were transfected with 2 μg STAT1-WT-HA (lanes 1-4) or STAT1-KR-HA (lanes 5-8) together with different SUMO-1 constructs (2 μg) as indicated. After 36 hours the cells were lysed in Triton X lysis buffer (50 mM Tris [tris(hydroxymethyl)aminomethane]–HCl, pH 7.4; 150 mM NaCl; 1 mM EDTA [ethylenediaminetetraacetic acid]; 50 mM NaF; 5 mM NEM; 1% Triton X-100; and 10% glycerol and protease inhibitors), and equal amounts of total cell lysates were immunoprecipitated with anti-HA antibodies and subjected to sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblotting with anti–SUMO-1 and anti-HA antibodies. (B) Sequence comparison of consensus SUMO-1 attachment sites in RanGAP1, PML, Sp100, p53, IκBα, AR, and STAT1. The consensus residues are marked in italics. The asterisk indicates the lysine that serves as a potential SUMO-1 attachment site. STAT1 sequence comparison starts at amino acid 698. (C) SUMO-1 conjugation of STAT1 in vitro. STAT1-WT (lanes 1-2) and STAT1-KR (lanes 3-4) were in vitro translated in the presence of [35S]methionine using TNT-coupled transcription/translation system (Promega, Madison, WI). The in vitro–translated products were then used for an in vitro SUMO-1 conjugation reaction. The samples were resolved by SDS-PAGE gel and visualized by fluorography. (D) Sumoylation of endogenous STAT1. HeLa cells were untransfected or transfected with SUMO-1 (2 μg) plasmid using a calcium phosphate method. After 36 hours the cells were lysed as described in panel A. Sumoylation of endogenous STAT1 was analyzed after immunoprecipitation using anti-STAT1 antibody and Western blotting with anti–SUMO-1.
SUMO-1 is conjugated to targeted proteins at the ϵ-amino group of lysine residues.13 The attachment of SUMO-1 to its substrates involves a minimal consensus sequence, ψKxE (Figure 1B). The consensus SUMO-1 target sequence was identified around Lys110 and Lys703 in STAT1. The C-terminal sumoylation sequence is well conserved across species in STAT1, but it is not present in the transactivation domains of other STATs. To investigate whether Lys703 is a target for sumoylation, we constructed a STAT1 Lys703Arg mutation (STAT1-KR) and performed similar experiments as described above. The STAT1-KR mutant did not show any SUMO-1 conjugation (Figure 1A, lanes 5-8). Furthermore, we performed the sumoylation experiments under cell-free conditions (Figure 1C). In vitro sumoylation of STAT1–wild type (STAT1-WT) was readily observed, whereas STAT1-KR was not sumoylated. Together, these results indicate that STAT1 is subject to conjugation of a single SUMO-1 molecule at Lys703. Furthermore, ectopic expression of SUMO-1 induced sumoylation of endogenous STAT1 in HeLa cells (Figure 1D). While this paper was under review, sumoylation of STAT1 on Lys703 was reported also by Rogers et al.18
To investigate the regulation of STAT1 sumoylation and the role of PIAS proteins, the experimental conditions were modified by reducing the amount of exogenous SUMO-1. COS-7 cells were transfected with STAT1-WT-HA with or without different Flag-tagged PIAS proteins, PIAS1, PIAS3, ARIP3, and really interesting new gene (RING) fingerlike domain-deficient PIAS mutant (PIAS1mut, ARIP3mut) (Figure 2A). Cotransfection of different PIAS proteins strongly enhanced STAT1 sumoylation in a RING finger-dependent manner (Figure 2A). These results suggest functional redundancy of PIAS proteins as SUMO E3 ligases toward STAT1. Previously PIAS1 has been shown to promote sumoylation of several target proteins such as p53, AR, c-Jun.9,14,15 Thus, various PIAS proteins can interact with several proteins and enhance sumoylation, and the specificity of these interactions and SUMO modification may also involve cell type–specific components.
PIAS proteins and activation of STAT1 promote SUMO-1 conjugation. (A) PIAS proteins promote SUMO-1 conjugation to STAT1-WT in vivo. COS-7 cells were transiently transfected with plasmids encoding STAT1-WT-HA (2 μg) and SUMO-1 (0.6 μg) together with Flag-PIAS1 (2.5 μg), Flag-PIAS1mut (1 μg), PIAS3 (4 μg), ARIP3 (4 μg), and ARIP3mut (0.5 μg) as indicated. The plasmid amounts were adjusted to yield similar expression levels. After 36 hours the cells were lysed in Triton X lysis buffer, and STAT1 protein was immunoprecipitated using anti-HA antibody and analyzed by immunoblotting as indicated. PIAS and PIASmut protein levels were analyzed by immunoblotting of 15 μg total cell lysates with anti-Flag antibody. (B) IFN-γ stimulation enhances SUMO-1 conjugation to STAT1. COS-7 cells were transfected with plasmids encoding STAT1-WT-HA (2 μg) and SUMO-1 (1 μg). After 36 hours the cells were serum starved and stimulated with 100 ng/mL human IFN-γ for different time points as indicated. STAT1 was immunoprecipitated and analyzed by immunoblotting. (C) Pervanadate-induced phosphorylation of STAT1 enhances SUMO-1 conjugation. HeLa cells were starved overnight and left unstimulated or stimulated with 50 μM pervanadate for 30 minutes. Total cell lysates were prepared as described in Figure 1A, and STAT1 was immunoprecipitated with anti-STAT1 antibody, and immunoblotting was performed with anti–SUMO-1 or anti-STAT1 antibody. (D) The effect of Lys703Arg mutation on STAT1-mediated gene activation in response to IFN-γ stimulation. U3A clones stably expressing STAT1-WT-HA (U3A-WT1 and U3A-WT2) or STAT1-KR-HA (U3A-KR1 and U3A-HA-KR2) were transiently transfected with SUMO-1 (1 μg), GAS-luc reporter plasmid (0.5 μg), and pCMV-βGal. After 24 hours the cells were starved in 0.5% serum and left unstimulated or stimulated with 10 ng/mL human IFN-γ for 6 hours followed by luciferase measurement. The mean relative luciferase units ± SD from 3 independent experiments are shown. The lower panel shows the STAT1 levels in different clones.
PIAS proteins and activation of STAT1 promote SUMO-1 conjugation. (A) PIAS proteins promote SUMO-1 conjugation to STAT1-WT in vivo. COS-7 cells were transiently transfected with plasmids encoding STAT1-WT-HA (2 μg) and SUMO-1 (0.6 μg) together with Flag-PIAS1 (2.5 μg), Flag-PIAS1mut (1 μg), PIAS3 (4 μg), ARIP3 (4 μg), and ARIP3mut (0.5 μg) as indicated. The plasmid amounts were adjusted to yield similar expression levels. After 36 hours the cells were lysed in Triton X lysis buffer, and STAT1 protein was immunoprecipitated using anti-HA antibody and analyzed by immunoblotting as indicated. PIAS and PIASmut protein levels were analyzed by immunoblotting of 15 μg total cell lysates with anti-Flag antibody. (B) IFN-γ stimulation enhances SUMO-1 conjugation to STAT1. COS-7 cells were transfected with plasmids encoding STAT1-WT-HA (2 μg) and SUMO-1 (1 μg). After 36 hours the cells were serum starved and stimulated with 100 ng/mL human IFN-γ for different time points as indicated. STAT1 was immunoprecipitated and analyzed by immunoblotting. (C) Pervanadate-induced phosphorylation of STAT1 enhances SUMO-1 conjugation. HeLa cells were starved overnight and left unstimulated or stimulated with 50 μM pervanadate for 30 minutes. Total cell lysates were prepared as described in Figure 1A, and STAT1 was immunoprecipitated with anti-STAT1 antibody, and immunoblotting was performed with anti–SUMO-1 or anti-STAT1 antibody. (D) The effect of Lys703Arg mutation on STAT1-mediated gene activation in response to IFN-γ stimulation. U3A clones stably expressing STAT1-WT-HA (U3A-WT1 and U3A-WT2) or STAT1-KR-HA (U3A-KR1 and U3A-HA-KR2) were transiently transfected with SUMO-1 (1 μg), GAS-luc reporter plasmid (0.5 μg), and pCMV-βGal. After 24 hours the cells were starved in 0.5% serum and left unstimulated or stimulated with 10 ng/mL human IFN-γ for 6 hours followed by luciferase measurement. The mean relative luciferase units ± SD from 3 independent experiments are shown. The lower panel shows the STAT1 levels in different clones.
Next we investigated whether sumoylation of STAT1 is regulated through endogenous IFN-γ receptor. As shown in Figure 2B, STAT1 was not detectably sumoylated in unstimulated COS-7 cells, but IFN-γ treatment readily induced sumoylation of STAT1 within 30 minutes with a peak at 2 hours. Furthermore, sumoylation of endogenous STAT1 was detected in HeLa cells after pervanadate treatment (Figure 2C), suggesting that activation of STAT1 serves as a signal for SUMO-1 conjugation.
The effect of sumoylation on STAT1-mediated transcriptional activity was assessed in U3A cell clones that stably express STAT1-WT or STAT1-KR mutant. The clones expressing the STAT1-KR showed enhanced transcriptional response to IFN-γ stimulation when compared with STAT1-WT clones (Figure 2D). Also in transient transfection system STAT1-KR was a more potent transactivator than STAT1-WT, but the difference was smaller (data not shown). These results suggest that sumoylation has a negative regulatory function for STAT1 and bears clear resemblance to the effects of sumoylation in AR, p53, and IRF-1–mediated gene activation.6,14,15,19
In summary, we show that STAT1 is subject to sumoylation in IFN-γ signaling through a single Lys703 residue. Interestingly, several proteins involved in modulation of IFN responses, such as interferon regulatory factor 1 (IRF-1)19 as well as IFN-inducible proteins PML and SP100,13 are subject to SUMO-1 conjugation. However, posttranslational modifications of STAT1 have been shown to mediate highly specific functions. Phosphorylation of Ser727 in STAT1 regulates only selected gene responses,20 and ubiquitination of STAT1 is detected only in certain cell types.21 The functions of PIAS proteins in cytokine and steroid receptor–mediated transcriptional regulation have been shown to involve both promoter- as well as cell type–specific components.6 Likewise, SUMO-1 modification also may modulate highly specific and even cell type–restricted functions of STAT1.
Prepublished online as Blood First Edition Paper, July 10, 2003; DOI 10.1182/blood-2002-12-3816.
Supported by grants from the Medical Research Council of the Academy of Finland, the Medical Research Foundation of Tampere University Hospital, the Finnish Foundation for Cancer Research, and the Sigrid Jusélius Foundation.
D.U. and S.V. contributed equally to this work.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank H. Yasuda and I. Kerr for reagents and P. Kosonen and M. Lehtinen for excellent technical assistance.

![Figure 1. SUMO-1 conjugation to STAT1 in vivo and in vitro. (A) Sumoylation of STAT1-WT and STAT1-KR (Lys703Arg mutant) in COS-7 cells. COS-7 cells were transfected with 2 μg STAT1-WT-HA (lanes 1-4) or STAT1-KR-HA (lanes 5-8) together with different SUMO-1 constructs (2 μg) as indicated. After 36 hours the cells were lysed in Triton X lysis buffer (50 mM Tris [tris(hydroxymethyl)aminomethane]–HCl, pH 7.4; 150 mM NaCl; 1 mM EDTA [ethylenediaminetetraacetic acid]; 50 mM NaF; 5 mM NEM; 1% Triton X-100; and 10% glycerol and protease inhibitors), and equal amounts of total cell lysates were immunoprecipitated with anti-HA antibodies and subjected to sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblotting with anti–SUMO-1 and anti-HA antibodies. (B) Sequence comparison of consensus SUMO-1 attachment sites in RanGAP1, PML, Sp100, p53, IκBα, AR, and STAT1. The consensus residues are marked in italics. The asterisk indicates the lysine that serves as a potential SUMO-1 attachment site. STAT1 sequence comparison starts at amino acid 698. (C) SUMO-1 conjugation of STAT1 in vitro. STAT1-WT (lanes 1-2) and STAT1-KR (lanes 3-4) were in vitro translated in the presence of [35S]methionine using TNT-coupled transcription/translation system (Promega, Madison, WI). The in vitro–translated products were then used for an in vitro SUMO-1 conjugation reaction. The samples were resolved by SDS-PAGE gel and visualized by fluorography. (D) Sumoylation of endogenous STAT1. HeLa cells were untransfected or transfected with SUMO-1 (2 μg) plasmid using a calcium phosphate method. After 36 hours the cells were lysed as described in panel A. Sumoylation of endogenous STAT1 was analyzed after immunoprecipitation using anti-STAT1 antibody and Western blotting with anti–SUMO-1.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/102/9/10.1182_blood-2002-12-3816/6/m_h82135155001.jpeg?Expires=1766443551&Signature=j0v0Do1i1UO27lqJ0J7qbFvbNYLoiJ6gp73FKat7BJlV3nmILa3tsvZKH94qDhoe8apZjTpsAdPTvwl4ovG1LPjfHE5v9YKAqq~kxcHPGZjOAhwuh1xTVsraYoIostv28CS3av-buDMeq7TCudSzjvfhMUZ49P1epT~RKvS~-PLLWlpDeUkBLTMnltK8aPGrCTzv~04Vbhm-1bFfoShiiXZk~C9vWC~2c~AQt9Cp32c~lbCmNkOB2k~JduPLe2y1Ig~S3QoRAGi1cecTzcVBZwm3q~hpkWu3ijbqU~Ytu04SpaOIpwC-T~KC-F8ZN03IYG81bkrpxnqx677Ol7YPPQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

![Figure 1. SUMO-1 conjugation to STAT1 in vivo and in vitro. (A) Sumoylation of STAT1-WT and STAT1-KR (Lys703Arg mutant) in COS-7 cells. COS-7 cells were transfected with 2 μg STAT1-WT-HA (lanes 1-4) or STAT1-KR-HA (lanes 5-8) together with different SUMO-1 constructs (2 μg) as indicated. After 36 hours the cells were lysed in Triton X lysis buffer (50 mM Tris [tris(hydroxymethyl)aminomethane]–HCl, pH 7.4; 150 mM NaCl; 1 mM EDTA [ethylenediaminetetraacetic acid]; 50 mM NaF; 5 mM NEM; 1% Triton X-100; and 10% glycerol and protease inhibitors), and equal amounts of total cell lysates were immunoprecipitated with anti-HA antibodies and subjected to sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblotting with anti–SUMO-1 and anti-HA antibodies. (B) Sequence comparison of consensus SUMO-1 attachment sites in RanGAP1, PML, Sp100, p53, IκBα, AR, and STAT1. The consensus residues are marked in italics. The asterisk indicates the lysine that serves as a potential SUMO-1 attachment site. STAT1 sequence comparison starts at amino acid 698. (C) SUMO-1 conjugation of STAT1 in vitro. STAT1-WT (lanes 1-2) and STAT1-KR (lanes 3-4) were in vitro translated in the presence of [35S]methionine using TNT-coupled transcription/translation system (Promega, Madison, WI). The in vitro–translated products were then used for an in vitro SUMO-1 conjugation reaction. The samples were resolved by SDS-PAGE gel and visualized by fluorography. (D) Sumoylation of endogenous STAT1. HeLa cells were untransfected or transfected with SUMO-1 (2 μg) plasmid using a calcium phosphate method. After 36 hours the cells were lysed as described in panel A. Sumoylation of endogenous STAT1 was analyzed after immunoprecipitation using anti-STAT1 antibody and Western blotting with anti–SUMO-1.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/102/9/10.1182_blood-2002-12-3816/6/m_h82135155001.jpeg?Expires=1766443552&Signature=ukyCLIo2nyZfc1ttprHxjXPzDVbfdunylcTiLwMMnE0aab8CYLKll2Q22jZkgwhYEZBSTIOZh21sToZwgcTaVmgIDzkvNCBmXT4aB7-PwbZszJOJP0ZCfYzs9iFveoN4FF7xmpXTtFpF-c5tWAL0nXXnFCdLWVHjtMtvjN2~3KNNQkCJej7KvC-e12TqaXWq7F6IrO0hArETkP0hAlaAgfBH6ofSnthKgWWHS9zLer2-4ousZbW1cBo7bTPULjhvF4XZ9VyLBTWUlTavqMsUJmjwUogMvn15-vh8CjYCjq3Cva89i43Mp8vrplMLbPhJUOd1DSib8eqNoKlAs8YlXA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)