Key Points
A defined 3D in vitro model, under hypoxic condition, sustains long-term propagation of JMML cells with specific hallmarks.
In vitro low oxygen levels drive a metabolic switch that redirect JMML cells toward self-renewal.
Abstract
Juvenile myelomonocytic leukemia (JMML) is a rare clonal stem cell disorder that occurs in early childhood and is characterized by the hyperactivation of the RAS pathway in 95% of the patients. JMML is characterized by a hyperproliferation of granulocytes and monocytes, and little is known about the heterogeneous nature of leukemia-initiating cells, as well as of the cellular hierarchy of the JMML bone marrow. In this study, we report the generation and characterization of a novel patient-derived three-dimensional (3D) in vitro JMML model, called patient-derived JMML Atypical Organoid (pd-JAO), sustaining the long-term proliferation of JMML cells with stem cell features and patient-specific hallmarks. JMML cells brewed in a 3D model under different microenvironmental conditions acquired proliferative and survival advantages when placed under low oxygen tension. Transcriptomic and microscopic analyses revealed the activation of specific metabolic energy pathways and the inactivation of processes leading to cell death. Furthermore, we demonstrated the pd-JAO–derived cells’ migratory, propagation, and self-renewal capacities. Our study contributes to the development of a robust JMML 3D in vitro model for studying and defining the impact of microenvironmental stimuli on JMML disease and the molecular mechanisms that regulate JMML initiating and propagating cells. Pd-JAO may become a promising model for compound tests focusing on new therapeutic interventions aimed at eradicating JMML progenitors and controlling JMML disease.
Introduction
Juvenile myelomonocytic leukemia (JMML) is a rare and aggressive myeloproliferative/myelodysplastic clonal stem cell disorder of early childhood,1-3 characterized by driver mutations that occur in RAS-related pathway genes.4,5 To date, the only curative treatment for patients with JMML is allogeneic hematopoietic stem cell transplantation (HSCT); however, it is followed by a high relapse rate.6 The use of demethylating agents has been shown to offer clinical benefits to control the disease before HSCT.7 Although it has been demonstrated that poor prognosis is related to additional mutations,8-10 specific gene expression or methylation signatures,11-13 very little is known about the hierarchy of bone marrow (BM) cells that sustain leukemia relapse.
It is known that leukemia stem cells reside mainly in the stem cell niche of the BM, where oxygen tension levels are physiological regulators of stem cell self-renewal, maintenance, and fate, through activation of hypoxia-signaling pathways driven by the hypoxia-inducible factor (HIF) complex.14 In normal hematopoiesis, hematopoietic stem and progenitor cells (HSPCs) display a hypoxic phenotype in both the endosteal and the perivascular niches, although different oxygen concentrations are present in the entire BM cavity.15 Severe hypoxia was demonstrated to be fundamental in the selection of hematopoietic progenitors with stem cell potential in BM cell cultures of patients with myelodysplastic syndrome (MDS).16 The canonical two-dimensional in vitro cell culture usually used for preclinical studies guarantees survival of JMML cells for a few days only and does not account for the natural three-dimensional (3D) cellular environment and the continuous cross talk among cells, the extracellular matrix, (ECM) and other resident cells.17-19
To our knowledge, this is the first report of an optimized 3D in vitro JMML model, called the patient-derived JMML Atypical Organoid (pd-JAO). This system mimics the key features of the BM physiological environment and allows the long-term survival and proliferation of immature primary JMML cell progenitors under hypoxic conditions. Furthermore, we have highlighted the biological features of potentially propagating JMML cells that can survive chemotherapy in the BM niche.
Methods
Isolation of mesenchymal stromal cells
Human mesenchymal stromal cells from healthy donors (HD-hMSCs) were derived from BM aspirates. In detail, 1 × 106 BM cells were plated (40 000 cells per cm2) in StemMACS (MSC Expansion Media XF, Miltenyi Biotech, BergischGladbach, Germany) medium supplemented with 100 U/mL penicillin/streptomycin (Gibco, Life Technologies, CA) and incubated at 37°C. The adherent cells were expanded to 90% confluence in T75 flasks before their use. HD-hMSCs were trypsinized (Trypsin/EDTA phosphate-buffered saline 1:250 Solution, BioConcept, Allschwill, Switzerland) at 37°C for 5 minutes, washed with Hanks balanced salt solution (BioConcept, Allschwill, Switzerland), and resuspended in an appropriate medium. HD-hMSCs between passages 3 to 5 were used. Immunophenotype analysis (hCD45-ECD-Beckman Coulter; hCD29-PE and hCD73-PE, Thermo Fisher Scientific) and osteogenic/adipocyte differentiation assay (PromoCell, Heidelberg, Germany) were carried out to characterize derived HD-hMSCs before use (supplemental Figure 1A-B). Project ID 281/2015-PR was approved by the local Ethical Committee of Padua University. The study was conducted following the Declaration of Helsinki.
Patients and generation of the 3D pd-JAO system
Fresh or frozen cells obtained from total BM or peripheral blood of 15 patients with JMML were studied (supplemental Table 1). Written informed consent was obtained from the patients’ parents or legal guardians before sample collection, following the Declaration of Helsinki. The 3D pd-JAO system was generated by mixing 1 × 105 of JMML primary cells with 2.5 × 104 HD-hMSCs (4:1 ratio), embedded in 10 μL of Matrigel (Corning, NY) and plated on a sheet of sterile parafilm for 1 hour at 37°C. The pd-JAO model was gently moved in enriched medium composed of serum-free RPMI 1640 medium supplemented with 10% BIT9500 serum substitute (StemCell Technologies, Canada Inc), 2 mM L-glutamine (Life Technologies, Carlsbad, CA) and 0.5% Penicillin/Streptomycin antibiotics (Life Technologies) containing stem cell factor (SCF) (50 ng/mL), thrombopoietin (TPO) (50 ng/mL), FLT3L (50 ng/mL), interleukin 3 (IL-3) (20 ng/mL), and interleukin 6 (20 ng/mL) cytokines (Cell guidance system, Cambridge, United Kingdom). The system was placed in an atmosphere containing 1.5% oxygen (PROOX/PROCO2 Chamber, Biospherycs) or 20% oxygen, 5% carbon dioxide, and balanced nitrogen. For continuous expansion, half of the enriched medium was replaced every week. When cells sprouted out from the 3D system (pd-JAO–derived cells [pd-JAO-dc]) covering at least half of the well, we harvested and plated them in a new 24-well plate in the same enriched medium and oxygen atmosphere of the 3D system they originated from.
Different media conditions were assessed to maintain the JMML cells: (1) plain medium (RPMI 1640 medium) supplemented with 10% BIT9500, 2 mM L-glutamine, and 0.5% penicillin/streptomycin; (2) plain medium removing cytokine one by one; and (3) enriched medium containing all cytokines (supplemental Figure 1C-D).
To assess the influence of Matrigel on the proliferation and selection of cell populations obtained from the 3D system, we generated a pd-JAO model with 2 different additional matrix gels: (1) Cultrex Reduced Growth Factor Basement Membrane Matrix Type R1 and (2) Cultrex Reduced Growth Factor Basement Membrane Matrix, Type 2, Trevigen, Gaithersburg, MD (supplemental Figure 2A-C). 3D pd-JAOs were also generated without Matrigel support (for specific rationale and methods see supplemental data; supplemental Figure 2C). For all the experiments reported in the manuscript, pd-JAO optimized with JMML primary cells, HD-hMSCs (4:1 ratio), and Matrigel (Corning, NY) were used.
Gene expression profiling and data analysis
Transcriptomic data analysis of pd-JAO-dc (ie, normoxia, hypoxia, and CD34+) was performed using 3’IVT Pico Kit and Human Genome U133 Plus 2.0 arrays (Affymetrix, Santa Clara, CA).
CEL files were normalized using the robust multiarray averaging expression measure of the Affy-R package (www.r-project.org). Gene Set Enrichment Analysis (GSEA)20 was performed using GSEA version 2.0, with genes ranked by signal-to-noise ratio and statistically significant gene sets determined by 1000 permutations and phenotype permutations. For GSEA, the false discovery rate (FDR) cutoff < 0.25 and P < .05 were considered for the analysis. The probe sets were collapsed to genes for maxima probe. GSEA was visualized and connected using the enrichment map plug-in of Cytoscape version 7.5 to create network overlap. Only gene sets with FDR < 0.05 derived from the c5 gene ontology biological process MSigDB were used to build the network. Gene set relationships were generated using Overlap Coefficient. To portray the landscape of the cellular heterogeneity and report the cell type enrichment analysis in pd-JAO-dc, we used a deconvolution approach based on the xCell Software.21 Microarray data are available at Gene Expression Omnibus under accession number GSE188608.
Immunophenotype
For immunophenotype analysis, 1×105 cells were stained with the conjugated antibodies reported in supplemental Table 2. The cells were harvested, incubated for 20 minutes with primary antibodies, and washed with Hanks solution. Fluorescence was acquired with a CytoFLEX Flow Cytometer (Beckman Coulter, Brea, CA) and analyzed using Kaluza Analysis Software (version 2.1).
Electron microscopy
pd-JAOs after 5 days of generation were fixed with 2.5% glutaraldehyde in 0.1 M sodium cacodylate buffer pH 7.4 for 1 hour at 4°C. The samples were postfixed with 1% osmium tetroxide plus potassium ferrocyanide 1% in 0.1 M sodium cacodylate buffer for 1 hour at 4°C. After 3 water washes, the samples were dehydrated in a graded ethanol series and embedded in an epoxy resin (Sigma-Aldrich). Ultrathin sections (60-70 nm) were obtained using an Ultrotome V (LKB) ultramicrotome, counterstained with uranyl acetate and lead citrate, and viewed with a Tecnai G2 (FEI) transmission electron microscope (TEM), operating at 100 kV. The images were captured using a Veleta digital camera (Olympus Soft Imaging System).
Statistics
Results
3D pd-JAO system sustains long-term proliferation of JMML myeloid progenitors
We have developed a robust 3D in vitro model, the pd-JAO, consisting of the generation of an atypical hematologic organoid that can support and promote proliferation and self-renewal of primary JMML cells. Based on several studies22,23 that highlight the importance of hypoxia in the leukemic BM niche to promote leukemic initiating cells’ self-renewal and invasion, we cultured our pd-JAO system under hypoxic (oxygen <2 %) conditions (Figure 1A).
Characterization of JMML progenitors generated in pd-JAO system. (A) Schematic representation of the pd-JAO 3D system generation. (B) Time course of pd-JAO 3D structure (upper panel) and pd-JAO derived cells (lower panel) in in vitro culture. Stereomicroscopic images at original magnification 2× and 6.3×. (C) Hematoxylin/eosin staining of fixed pd-JAO 3D structure. Notably, JMML cells are incorporated in the ECM. Original magnification 40×. (D) Violin plot of immunophenotype analysis of pd-JAO-dc at 15 days of the 3D system generation in 13 of 15 patients tested. (E) May-Grünwald-Giemsa (MGG) staining (20× magnification) of pd-JAO-dc after 15 days of culture. (F) Immunophenotype analysis of pd-JAO-dc collected at 3 different time points from the 3D generation (ie, T1: 1-20 days; T2: 20-60 days; T3: >60 days) For each marker, we reported the percentage of positive live cells. HD-hMSC, healthy donor human mesenchymal stromal cells; tBM, total BM bulk.
Characterization of JMML progenitors generated in pd-JAO system. (A) Schematic representation of the pd-JAO 3D system generation. (B) Time course of pd-JAO 3D structure (upper panel) and pd-JAO derived cells (lower panel) in in vitro culture. Stereomicroscopic images at original magnification 2× and 6.3×. (C) Hematoxylin/eosin staining of fixed pd-JAO 3D structure. Notably, JMML cells are incorporated in the ECM. Original magnification 40×. (D) Violin plot of immunophenotype analysis of pd-JAO-dc at 15 days of the 3D system generation in 13 of 15 patients tested. (E) May-Grünwald-Giemsa (MGG) staining (20× magnification) of pd-JAO-dc after 15 days of culture. (F) Immunophenotype analysis of pd-JAO-dc collected at 3 different time points from the 3D generation (ie, T1: 1-20 days; T2: 20-60 days; T3: >60 days) For each marker, we reported the percentage of positive live cells. HD-hMSC, healthy donor human mesenchymal stromal cells; tBM, total BM bulk.
This optimized pd-JAO (“Methods”) expanded between days 2 and 10 and was transparent, with a perfectly round shape. From day 10, the JMML cells began sprouting from the pd-JAO structure and continued to proliferate in suspension. This phenomenon progressively caused size reduction and shape remodeling of the 3D system, resulting in complete jagged edges and a color shift from transparent-white to brown, due to cell death, starting from day 60 (Figure 1B). The architecture of the 3D structure, internal location, and attachment of cells to the ECM inside the model was observed using hematoxylin/eosin staining (Figure 1C) on day 5 of culture.
The pd-JAO model typically sprouted out ∼0.5 × 105 to 1.5 × 105 total cells every 15 days, and these cells in suspension were able to maintain the proliferation boost in a new bottom plate, without the 3D pd-JAO support (supplemental Figure 3A-B) for long-term culture.
Interestingly, the pd-JAO system was successful in 92% (14/15) of the tested primary samples (supplemental Table 1). Immunophenotype analysis of cells sprouted from the pd-JAO system (ie, pd-JAO-dc) and inside the pd-JAO system (after 5 days in culture) showed expanded, well-defined, and immature CD33+, CD99+, CD117+, CD14dim, and CD15dim cell population (Figure 1D; supplemental Figure 3C-D). Notably, in 3 out of 14 patient samples used to generate our in vitro pd-JAO system, we noticed a substantial expansion of the CD34+ population after long-term culture (ie, #6, day 60; #13, day 50; and #14, day 30), reaching a median value of 50.5% in hypoxia (supplemental Figure 3E; Figure 1D-F). The immature nature of pd-JAO-dc was also demonstrated by its morphology (Figure 1E), which was maintained over time (Figure 1F).
However, these cells did not show a unique population over time. According to the immunophenotype, a tiny counterpart of mature cells showed a fluctuating trend, with constant regeneration of immature bulk cells.
Moreover, pd-JAO-dc had an increased ability to form colonies than cells at diagnosis, probably because of their immature nature combined to high proliferation rate (supplemental Figure 4A). Remarkably, pd-JAO-dc harbored the same driver and secondary additional mutations, chromosome abnormalities (ie, monosomy 7), concordant hypersensitivity to granulocyte-macrophage colony-stimulating factor, and preserved LIN28B gene expression present at diagnosis (supplemental Figure 4B-E).
Overall, these results define an in vitro model that can sustain immature JMML myeloid cells, while preserving disease hallmarks.
Hypoxic environment sustains proliferation of immature JMML cells
To investigate the factors that sustain JMML immature cell proliferation more closely, we analyzed the microenvironmental conditions of our 3D model system. It has been extensively reported that hypoxia promotes cell stemness and aggressiveness through the activation of HIFs.24,25 Therefore, we decided to culture pd-JAO-dc in hypoxic (oxygen < 2%) and normoxic (oxygen ≅ 20%) environments. Interestingly, pd-JAO-dc proliferated under both normoxic and hypoxic conditions (Figure 2A; supplemental Figure 5A). Hypoxia significantly enhanced JMML cell growth, with more than double the proliferation rate (P < .0001) (Figure 2B). Furthermore, the time of cell sprouting was shorter in hypoxia than in normoxia (median time of 4.7 days compared with 7 days, respectively, P = .0386, Figure 2C). The collapse of the system occurred later in hypoxia than in normoxia (ie, median value 92 days vs 57 days, P = .0102, Figure 2D). Furthermore, the phenotypic profile of the cell populations was similar in the 2 conditions, with only a tendency toward a slightly more mature immunophenotype in normoxia (Figure 2E; supplemental Figure 5B).
Enhanced proliferation of JMML progenitor cells under hypoxic condition. (A) Representative microscopic images (6.3× magnification) of pd-JAO-dc maintained under hypoxic and normoxic conditions on day 15 derived from 3 patients diagnosed with JMML. (B) pd-JAO-dc cell count (day 15, normoxia N = 14, hypoxia N = 33), (C) time of cell sprouting from the 3D system (normoxia N = 11, hypoxia N = 13), and (D) pd-JAO system death (normoxia N = 11, hypoxia N = 12) in hypoxia and normoxia. ∗P ≤ .05; ∗∗P ≤ .01;∗∗∗∗P ≤ .0001 by unpaired t test with Welch's correction. Bars are shown as mean ± standard error of the mean (SEM). (E) Immunophenotype analysis of pd-JAO-dc maintained in the 2 tested conditions. Pt, patient.
Enhanced proliferation of JMML progenitor cells under hypoxic condition. (A) Representative microscopic images (6.3× magnification) of pd-JAO-dc maintained under hypoxic and normoxic conditions on day 15 derived from 3 patients diagnosed with JMML. (B) pd-JAO-dc cell count (day 15, normoxia N = 14, hypoxia N = 33), (C) time of cell sprouting from the 3D system (normoxia N = 11, hypoxia N = 13), and (D) pd-JAO system death (normoxia N = 11, hypoxia N = 12) in hypoxia and normoxia. ∗P ≤ .05; ∗∗P ≤ .01;∗∗∗∗P ≤ .0001 by unpaired t test with Welch's correction. Bars are shown as mean ± standard error of the mean (SEM). (E) Immunophenotype analysis of pd-JAO-dc maintained in the 2 tested conditions. Pt, patient.
These data indicate that the growth, long-term maintenance, and expansion of an immature myeloid population of JMML cells from pd-JAO systems are strictly dependent on hypoxic conditions.
Myeloid progenitors originate from stem cell clones
It is known that the BM of patients with JMML at diagnosis is characterized by the hyperproduction of monocytic and granulocytic cells, reduction in immature progenitor cells, and disrupted hierarchy of myeloid commitment. At diagnosis, CD34+ cells in the BM were ∼5% (1%-16%),26 and CD99+ and CD117+ cells were <20% (supplemental Figure 5C).
To track which JMML BM population could generate a clone with long-term maintenance, we sorted different cell populations from samples of 5 patients collected at disease onset and used them to generate new 3D pd-JAO systems maintained in hypoxic conditions. We isolated hematopoietic stem cells (HSCs) (CD34+), monocytes (CD34–CD33+CD14+CD15–) granulocytes (CD34–CD33+CD14–CD15+), and the rest of the BM (CD34–CD33–CD14–CD15–). After 15-day culture, only 3D systems generated from sorted CD34+ cells (CD34+-pd-JAO) were able to sprout cells with a high proliferation rate (CD34+-pd-JAO-dc) (Figure 3A; supplemental Figure 6A-B).
Stem cells origin of JMML propagating cells. (A) Cell count of pd-JAO-dc from isolated subpopulations (ie, CD34+, CD34–CD33–CD14–CD15–, CD34–CD33+CD14+CD15–, and CD34–CD33+CD14–CD15+) on day 15 derived from 5 patients at diagnosis of JMML; ∗∗P ≤ .01. (B) pd-JAO-dc cell count on day 15 (tBM pd-JAO-dc N = 33, CD34+-pd-JAO-dc N = 12), (C) time of sprouting from the 3D system (tBM pd-JAO-dc N = 13, CD34+-pd-JAO-dc N = 5), (D) death of the pd-JAO (tBM pd-JAO-dc N = 12, CD34+-pd-JAO-dc N = 3), and (E) immunophenotype analysis of pd-JAO-dc obtained from total BM and CD34+ enrichment maintained under hypoxia. Bars are shown as mean ± SEM. ∗∗P ≤ .01; ∗∗∗∗P ≤ .0001; ns, not significant by unpaired t test with Welch's correction. (F) Portrayal of the cellular heterogeneity landscape of pd-JAO-dc whole transcriptomic profiling. The heat map shows the cell type Enrichment Score based on the xCell software results. A deconvolution approach was applied to compare the gene expression profile data of tBM and CD34+pd-JAO-dc maintained under normoxia and hypoxia to gene signature of pure HSCs, progenitors, and mature myeloid derived cells. (G) Kaplan-Meier estimate survival analysis for 3D systems generated from different cell sources: CD34+ enriched cells and tBM of patients with JMML at diagnosis in hypoxia and normoxia, patients with acute myeloid leukemia (AML) at diagnosis, tBM, and CB-34+ from healthy donors. Log-rank (Mantel Cox) P values were calculated across all the subtypes.
Stem cells origin of JMML propagating cells. (A) Cell count of pd-JAO-dc from isolated subpopulations (ie, CD34+, CD34–CD33–CD14–CD15–, CD34–CD33+CD14+CD15–, and CD34–CD33+CD14–CD15+) on day 15 derived from 5 patients at diagnosis of JMML; ∗∗P ≤ .01. (B) pd-JAO-dc cell count on day 15 (tBM pd-JAO-dc N = 33, CD34+-pd-JAO-dc N = 12), (C) time of sprouting from the 3D system (tBM pd-JAO-dc N = 13, CD34+-pd-JAO-dc N = 5), (D) death of the pd-JAO (tBM pd-JAO-dc N = 12, CD34+-pd-JAO-dc N = 3), and (E) immunophenotype analysis of pd-JAO-dc obtained from total BM and CD34+ enrichment maintained under hypoxia. Bars are shown as mean ± SEM. ∗∗P ≤ .01; ∗∗∗∗P ≤ .0001; ns, not significant by unpaired t test with Welch's correction. (F) Portrayal of the cellular heterogeneity landscape of pd-JAO-dc whole transcriptomic profiling. The heat map shows the cell type Enrichment Score based on the xCell software results. A deconvolution approach was applied to compare the gene expression profile data of tBM and CD34+pd-JAO-dc maintained under normoxia and hypoxia to gene signature of pure HSCs, progenitors, and mature myeloid derived cells. (G) Kaplan-Meier estimate survival analysis for 3D systems generated from different cell sources: CD34+ enriched cells and tBM of patients with JMML at diagnosis in hypoxia and normoxia, patients with acute myeloid leukemia (AML) at diagnosis, tBM, and CB-34+ from healthy donors. Log-rank (Mantel Cox) P values were calculated across all the subtypes.
Remarkably, CD34+-pd-JAO-dc showed a higher proliferation capacity at day 15 (P < .0001) and an anticipated timing of germinating cells (P = .0159) compared with cells sprouted from pd-JAO cells generated from tBM (Figure 3B-C; supplemental Figure 6C-E), probably owing to an enrichment of cells with stem-like potential. Furthermore, the life span of the system was slightly prolonged, and death occurred after a medium time of 4 months (Figure 3D).
CD34+-pd-JAO-dc showed a predominant expression of CD99+, CD117+, and CD33+ cells and a low percentage of CD14+ and CD15+ cells (Figure 3E; supplemental Figure 6F-G), similar to pd-JAO-dc resulting from tBM. Interestingly, CD34+ enriched cells derived from patient #7 were able to successfully proliferate and generated pd-JAO, which was not the case when using the patient’s total BM cells (supplemental Figure 6B).
Data supporting the commitment of pd-JAO-dc toward progenitor cells were also observed at the transcriptome level by a deconvolution approach.21 Myeloid progenitors were enriched with high confidence in most pd-JAO-dc (Figure 3F), and a similar myeloid progenitor enrichment was also identified in pd-JAO-dc under normoxic conditions, except for 2 patients who showed a transcriptome profile overlapping that of more mature myeloid cells (Figure 3F).
Collectively, these data suggest that both pd-JAO-dc starting from the BM bulk and CD34+ sorted cells originate from a stem cell population undergoing maturation followed by a block in a committed progenitor state.
3D model is suitable only for JMML cells
To test the ability of our system to support the proliferation of other types of primary cells, we generated the 3D model using 5 AML samples at diagnosis and 2 healthy BM donor samples. For the AML samples, the 3D system sprouted living cells; however, on day 12, the model turned brown, and the cells full of debris underwent apoptosis and died within a maximum of 30 days (supplemental Figure 7A). Interestingly, the tendency for higher cell proliferation under hypoxic rather than in normoxic conditions was maintained also for AML cells (except for the first 5 days of culture), albeit cell viability was lower than that of the pd-JAO system, and long-term proliferation was not supported (supplemental Figure 7B). Moreover, in cells sprouted from the 3D system, we observed a reduction in immature cell surface markers (ie, CD34, CD99, and CD117) that were highly expressed in cells at diagnosis (supplemental Figure 7C). Furthermore, the 3D system generated from healthy tBM donors revealed enrichment of sprouted cells with a typical immunophenotype BM maturation curve and the absence of a pure immature cell expansion (supplemental Figure 7D). On day 15, both the 3D model and cells in the suspension died. Notably, in cell derived from both healthy donors’ tBM and cord blood CD34+-enriched cells, we observed a decreased colony-forming unit cell potential (supplemental Figure 7E), in contrast to that observed for JMML cells. In summary, our pd-JAO model strongly prompted the exclusive expansion, proliferation, and survival of an immature cell population in JMML cells (Figure 3G).
Transcriptome modulation induced by hypoxia in derived immature JMML cells
The data presented thus far provide strong evidence that hypoxia plays a key role in sustaining pd-JAO-dc proliferation. In this respect, the HIF-1α protein, a well-known master transcriptional regulator in response to low oxygen tension levels, was expressed in the cells and stabilized in cell nuclei of pd-JAO-dc maintained under hypoxia (supplemental Figure 8A-B). We also showed the expression of HIF-2α protein in pd-JAO-dc and a higher expression of the CA9 gene in hypoxia than in normoxia, which is a known downstream target of HIF protein stabilization (supplemental Figure 8C-D; Figure 4).
Hypoxia modulation on pd-JAO-dc. (A) Heat map showing the top 50 upregulated and downregulated genes in differential gene expression profile analysis of pd-JAO-dc maintained in hypoxic vs normoxic conditions in 5 pairs of samples. (B) Enrichment map shows the positive enriched biological process (derived from c5 GSEA-MSigDB) in pd-JAO-dc in normoxic vs hypoxic conditions. Nodes represent the functional gene sets, and the size is proportional to the size of the gene set. Edge thickness is proportional to the overlap between gene sets. Only gene sets that are enriched with an FDR < 0.05 are shown. Nodes with common biological functions were clustered by Auto Annotate Cytoscape plug-in using the MCL cluster. (C) Absolute expression of AK4, GLUT1, MYC, PROM1, and CA9 genes in pd-JAO-dc by droplet digital polymerase chain reaction analysis in both conditions. Heat map columns show the mean value of absolute copies per μL for all samples for each condition as log2. (D) Bars showing the enriched clusters of GSEA of top gene sets positively enriched in normoxic conditions compared with hypoxia. X-axis normalized enriched score (NES) value. (E) TEM images of fixed tBM pd-JAO systems after 5 days from the generation for 2 patients. Degraded mitochondria showed characteristic swellings, large portions of empty matrix, inside it are recognizable residues of cristae, and the double membrane is clearly visible. An autophagic body can be larger than a mitochondrion and inside of it, we can appreciate reticulum residues membranes and vesicles. Red arrow indicates an example of AVs in the cells. Scale bar, 2 μm and 500 nm, left and right respectively for both hypoxia and normoxia. (F) Number of AVs counted in the pd-JAO TEM section (hypoxia, N = 12 for patient #6 and N = 12 for patient #8, normoxia N = 11 for patient #6 and N = 7 for patient #8). Mean and SEM are shown. ∗∗P ≤ .01, ∗∗∗∗P ≤ .0001 by 2-tailed unpaired t test. AV: autophagic vesicles; M: mitochondria; N: nuclei.
Hypoxia modulation on pd-JAO-dc. (A) Heat map showing the top 50 upregulated and downregulated genes in differential gene expression profile analysis of pd-JAO-dc maintained in hypoxic vs normoxic conditions in 5 pairs of samples. (B) Enrichment map shows the positive enriched biological process (derived from c5 GSEA-MSigDB) in pd-JAO-dc in normoxic vs hypoxic conditions. Nodes represent the functional gene sets, and the size is proportional to the size of the gene set. Edge thickness is proportional to the overlap between gene sets. Only gene sets that are enriched with an FDR < 0.05 are shown. Nodes with common biological functions were clustered by Auto Annotate Cytoscape plug-in using the MCL cluster. (C) Absolute expression of AK4, GLUT1, MYC, PROM1, and CA9 genes in pd-JAO-dc by droplet digital polymerase chain reaction analysis in both conditions. Heat map columns show the mean value of absolute copies per μL for all samples for each condition as log2. (D) Bars showing the enriched clusters of GSEA of top gene sets positively enriched in normoxic conditions compared with hypoxia. X-axis normalized enriched score (NES) value. (E) TEM images of fixed tBM pd-JAO systems after 5 days from the generation for 2 patients. Degraded mitochondria showed characteristic swellings, large portions of empty matrix, inside it are recognizable residues of cristae, and the double membrane is clearly visible. An autophagic body can be larger than a mitochondrion and inside of it, we can appreciate reticulum residues membranes and vesicles. Red arrow indicates an example of AVs in the cells. Scale bar, 2 μm and 500 nm, left and right respectively for both hypoxia and normoxia. (F) Number of AVs counted in the pd-JAO TEM section (hypoxia, N = 12 for patient #6 and N = 12 for patient #8, normoxia N = 11 for patient #6 and N = 7 for patient #8). Mean and SEM are shown. ∗∗P ≤ .01, ∗∗∗∗P ≤ .0001 by 2-tailed unpaired t test. AV: autophagic vesicles; M: mitochondria; N: nuclei.
We investigated the extent to which the transcriptional program of pd-JAO-dc was influenced by HIF-1α stabilization. To this end, the gene expression profiles of cells under hypoxic conditions were compared with the profiles of paired cells under normoxic conditions (Figure 4A). GSEA revealed a metabolic adaptation of the cells cultured in hypoxia, showing a negatively enriched gene set related to oxidative phosphorylation and the electron transport chain, mainly due to oxygen deprivation and their protective mechanisms to maintain oxygen homeostasis by reducing oxidative metabolism and consumption (Figure 4B). By contrast, specific genes involved in glycolysis and directly modulated by hypoxia, such as glucose transporter 1 (GLUT1),27 adenylate kinase isozyme 4 (AK4),28MYC,29 and prominin1 (PROM1),30 were upregulated in JMML cells maintained under hypoxic conditions (Figure 4C).
Nevertheless, in JMML cells cultured under normoxic conditions, we observed a positive enrichment of biological processes involved in Golgi vesicle transport and budding, vacuole and autophagosome formation, and activation of the macro-autophagy process (Figure 4D). Autophagy activation, observed at the transcriptional levels, was further investigated using TEM images of the pd-JAO maintained at different oxygen tensions. Interestingly, on day 5, JMML cells inside pd-JAO and incubated under normoxia showed a significantly brimful of autophagic bodies (Figure 4E-F), whereas those maintained under hypoxic conditions showed only sporadic autophagic bodies. We also observed impaired quality control of mitochondria in normoxic cells, resulting in damage, changes in size, and loss of internal structure (Figure 4E).
These results support the observation that JMML cells under normoxia tend to undergo premature death due to a high dose of cellular stress, which is absent under hypoxic conditions.
pd-JAO-dc retain stem potential and self-renewal ability
Malignant stem cells have long been known for their clonal expansion, cell migration, and invasion.31,32 To investigate these properties in pd-JAO-dc, we first investigated their ability to migrate through the 3D system. We generated an “organoid” with only HD-hMSCs and Matrigel placed in a suspension of primary JMML cells (pd-JAO-dc) for 3 days, showing migration ability inside of the structure. After moving this 3D model to a new well with only enriched medium, the JMML cells repopulated the entire well within 70 days, indicating their migratory ability and maintenance of their proliferative capacity (data not shown).
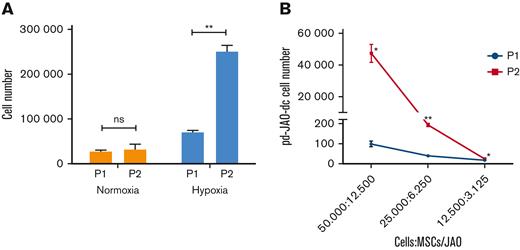
We also examined the propagating and clonal expansion capacity of pd-JAO-dc. In detail, cells sprouted from the pd-JAO system in the first generation (P1 at day 30) were used to produce a second-generation system (P2) in both hypoxic and normoxic conditions. After 30 days in culture, the data showed that high oxygen tension did not support cell propagation over time, whereas hypoxia promoted the clonal expansion of JMML cells (Figure 5A) (P = .007). Notably, the amplification boost of JMML cells at low oxygen tension was also verified using a limiting dilution assay (Figure 5B).
Self-renewal ability of pd-JAO-dc under hypoxia condition. (A) Propagating capacity of pd-JAO-dc after multiple passages in the pd-JAO system in normoxia and hypoxia conditions (N = 3 replicates). Mean and SEM are shown. ∗∗P ≤ .01 by 2-tailed unpaired t test. (B) Viability of pd-JAO-dc derived from serial dilution in 3D culture generated from P1 and P2 in the pd-JAO system (N = 3 replicates). Mean and SEM are shown. ∗P ≤ .05, ∗∗P ≤ .01 by 2-tailed paired t test.
Self-renewal ability of pd-JAO-dc under hypoxia condition. (A) Propagating capacity of pd-JAO-dc after multiple passages in the pd-JAO system in normoxia and hypoxia conditions (N = 3 replicates). Mean and SEM are shown. ∗∗P ≤ .01 by 2-tailed unpaired t test. (B) Viability of pd-JAO-dc derived from serial dilution in 3D culture generated from P1 and P2 in the pd-JAO system (N = 3 replicates). Mean and SEM are shown. ∗P ≤ .05, ∗∗P ≤ .01 by 2-tailed paired t test.
These data indicate that hypoxia is sufficient to induce migration, propagation, and self-renewal of immature JMML cells derived from the pd-JAO system. These results are in line with the typical characteristics of leukemic initiating cells, and it is to be noted that the stem cell potential is increased by multiple passages in the pd-JAO system.
Overall, these data indicated that the pd-JAO system, together with hypoxia stimuli, was able to sustain the growth and proliferation rate of JMML immature cell (CD34+/–, CD117+, and CD99+) populations with stem cell–like potential.
Discussion
In this study, we showed that a combination of multiple medium factors, a 3D spheroid structure, and low oxygen tension induces the proliferation and self-renewal of JMML progenitor cells.
In recent years, cancer research has introduced the use of 3D models, especially for solid tumors; patient-derived organoids recapitulating many structural and functional aspects of their in vivo counterpart organs are generated starting from primary tumor cells.33,34 In the hematopoietic field, organoid generation has been less frequently investigated because of the absence of a solid structure formation from HSPCs. Common alternatives to mimic cell-cell interactions in the BM niche are coculture methods,35 spheroid formation,36 and inserts37 or hydrogel38 support.
The 3D in vitro system that we have developed in the context of JMML efficiently initiated and sustained long-term primary JMML progenitor cell survival and proliferation, strictly recapitulating JMML features, which are essential for a suitable preclinical in vitro model.
The enhanced survival and proliferation of JMML cells were strongly dependent on several factors. The 3D structure and the ECM were important elements for the survival of JMML cells by facilitating intercellular communication. Several studies have reported that in addition to primary mesenchymal stromal cells releasing soluble factors,39 the ECM also plays an important role in modulating the maintenance, proliferation, self-renewal, and differentiation of stem cells40 within the stem cell niche. Here, we demonstrated that the selection of myeloid multipotent hematopoietic populations was unaffected by the type of ECM used to generate our model. Moreover, the pd-JAO system was exclusively suited for the selection and expansion of JMML cells. However, further studies are needed to better understand the persistence of immature JMML cells. Nevertheless, we could speculate that oncogenic JMML driver mutations of the RAS pathway are critical for sustaining JMML progenitor cell proliferation through HIF-1α stabilization, as has been demonstrated for colon cancer41,42 or glioblastoma.43
Our data showed that the proliferation of immature JMML cells was mainly due to hypoxic conditions, suggesting that low oxygen tension is an essential feature for the preservation of the leukemic cell status. It has been demonstrated that HIF-1α expression and its stabilization into nuclei are peculiar to stem cells in MDS, supporting leukemia cell proliferation and self-renewal44 and that this could represent a promising therapeutic target.45-47
Previous studies have shown that hypoxia induces metabolic adaptation in cancer development and progression.48,49 We demonstrated that hypoxia induces metabolic processes due to HIF-1α stabilization in proliferating JMML cells. Specifically, the overexpression of GLUT1 and reprogramming of metabolism observed in immature JMML cells could lead to the development of new therapeutic strategies. A recent study has shown that a GLUT1 inhibitor, administered either as a single agent or in combination with MEK inhibition, could be a novel therapeutic approach for targeting JMML HSPCs.50,51
Notably, cells maintained under normoxic conditions showed a shorter life span than JMML cells kept at low oxygen tension, mitochondrial impairment, and massive activation of autophagy. Nevertheless, how oxidative stress is induced in JMML cells and how its cross talk with autophagy occurs are far from being understood.
The CD cell surface markers are another important set of features that provide a functional description of cellular hierarchy. In myeloid malignancies, CD99 is shown to mark most stem cells, and CD99-antibodies reduce cell viability, colony formation, and cell migration and induce cell differentiation and death in both cell lines and primary blasts,52 whereas in xenograft mouse models, they reduce engraftment capacity.53 CD117 is a marker of stem and immature cells. Pang et al54 demonstrated that in vivo targeting of the c-kit receptor led to the eradication of MDS-HSCs, indicating that this could represent a novel therapeutic target. Moreover, Quek et al55 demonstrated that in AML CD34– fractions, leukemia stem cells activity segregated with CD117 expression, whereas Caye et al56 demonstrated that leukemia-propagating cells have a very complex and heterogeneous architecture in multiple progenitor cellular compartments in patients with JMML.
These findings are very fascinating considering our data, as we showed that cell suspension derived from the pd-JAO system was mostly CD99+ and CD117+, with a high proliferation potential and self-renewal ability, thus reflecting the immature nature of leukemia stem and progenitor cells and marking a cell population that propagates the JMML disease. This immature cell population can arise from both the BM cells bulk and enriched CD34+ cells, supporting the concept that the origin of JMML arises from stem cells, as previously concluded from studies on genomic and cell clonal data collection.
In conclusion, this study offers a novel 3D in vitro model for the long-term maintenance and expansion of patient-derived JMML progenitor cells under hypoxic conditions. Although the expansion of only immature JMML cells could be a limitation concerning mimicking the monocyte and granulocyte hyperproliferating immunophenotype of patients at diagnosis, our culture system represents an advantage for studying the JMML immature cells responsible for propagating the disease. Indeed, this system overcomes the difficulty of ex vivo modeling the disease, owing to the rapid differentiation and senescence of JMML cells. Moreover, our findings may lay the groundwork for cutting-edge research on a specific immature JMML cell population, taking advantage of the hypoxia environmental stimuli that reside in the BM niche and escape treatment, and could be responsible for relapse. In addition, the generation of a 3D model from the primary patient material could open a new perspective on the development and screening of therapeutic strategies to improve patient outcomes and to reduce risk of disease recurrence after HSCT.
Acknowledgments
The authors thank Annamaria Di Meglio and Anna Leszl of Oncohematology Laboratory for their contribution to the cytogenetic analysis. The authors especially thank Giuseppe Basso for his mentorship, support, and scientific contributions to this work. The authors also thank the staff of the Oncology Hematology Laboratory for the diagnosis of JMML, the sample biobanking of the Biobanca Oncologia Pediatrica, and the members of the Associazione Italiana Ematologia Oncologia Pediatrica (AIEOP-GdL JMML/MDS) for providing patient samples. This work was supported by Associazione Italiana Ricerca sul Cancro (MFAG #15674) (S.B.); Fondazione Cariparo, Bando di Ricerca Pediatrica (17/07 FCR and 20/16 FCR) (S.B. and L.P.); Istituto di Ricerca Pediatrica-Fondazione Città della Speranza (IRP 19/09) (S.B. and A.C.); and Umberto Veronesi Foundation #2601 and #3628 (E.R.) and #4420 and #4820 (A.C.).
This article is in memory of Giuseppe Basso.
Authorship
Contribution: S.B. and A.C. were involved in study design, data analysis, interpretation, and manuscript writing; A.C. and C.T.P. performed the experiments and collected and interpreted the data; C.F., P.S., and B.B. performed immunophenotype analysis of patients and organoids and interpreted the results; E.R., V.B., and L.P. were involved in the organoid generation and immunofluorescence procedures; F.C. acquired TEM images and performed analysis; S.F. performed morphology experiments; A.B., S.C., C.M., M.Z., L.S., M.P., R.M., and F.L. provided patient samples and patients’ clinical data; A.R. contributed to the manuscript revision; G.t.K. contributed to manuscript revision and scientific support; S.B. coordinated the study and provided funds; and all authors have read and agreed to the published version of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Silvia Bresolin, Department of Women’s and Children’s Health, Laboratory of Oncohematology, University of Padua, Padua, Italy 35127; e-mail: silvia.bresolin@unipd.it.
References
Author notes
Microarray data are available at Gene Expression Omnibus under accession number GSE188608. Other forms of data are available on request from the corresponding author, Silvia Bresolin (silvia.bresolin@unipd.it).
The full-text version of this article contains a data supplement.