Key Points
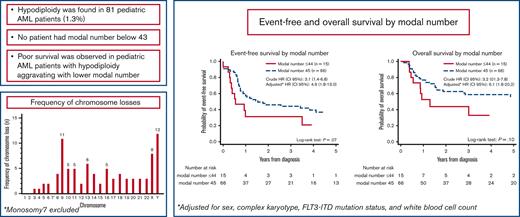
Hypodiploidy is found in <2% of pediatric AML. No patient had modal number below 43.
Poor survival is observed in hypodiploid AML with aggravating prognosis in patients with modal number 43 to 44.
Abstract
Hypodiploidy, defined as modal numbers (MNs) 45 or lower, has not been independently investigated in pediatric acute myeloid leukemia (AML) but is a well-described high-risk factor in pediatric acute lymphoblastic leukemia. We aimed to characterize and study the prognostic impact of hypodiploidy in pediatric AML. In this retrospective cohort study, we included children below 18 years of age with de novo AML and a hypodiploid karyotype diagnosed from 2000 to 2015 in 14 childhood AML groups from the International Berlin-Frankfurt-Münster (I-BFM) framework. Exclusion criteria comprised constitutional hypodiploidy, monosomy 7, composite karyotype, and t(8;21) with concurring sex chromosome loss. Hypodiploidy occurred in 81 patients (1.3%) with MNs, 45 (n = 66); 44 (n = 10) and 43 (n = 5). The most frequently lost chromosomes were chromosome 9 and sex chromosomes. Five-year event-free survival (EFS) and overall survival (OS) were 34% and 52%, respectively, for the hypodiploid cohort. Children with MN≤44 (n = 15) had inferior EFS (21%) and OS (33%) compared with children with MN = 45 (n = 66; EFS, 37%; OS, 56%). Adjusted hazard ratios (HRs) were 4.9 (P = .001) and 6.1 (P = .003). Monosomal karyotype or monosomy 9 had particular poor OS (43% and 15%, respectively). Allogeneic stem cell transplantation (SCT) in first complete remission (CR1) (n = 18) did not mitigate the unfavorable outcome of hypodiploidy (adjusted HR for OS was 1.5; P = .42). We identified pediatric hypodiploid AML as a rare subgroup with an inferior prognosis even in the patients treated with SCT in CR1.
Introduction
Pediatric acute myeloid leukemia (AML), comprising 15% to 20% of childhood leukemias,1,2 is a rare and highly heterogenous disease entity. A variety of morphological, immunological, and genetic features have been incorporated within the World Health Organization (WHO) classification of hematological disorders.3,4 Although there is general agreement on favorable prognostic factors of recurrent genetic lesions such as (8;21)(q22;q22), t(15;17)(q21;q21), and inv(16)(p13;q22)/t(16;16)(p13;q22), data regarding high-risk features for pediatric AML are conflicting.4-8 Cytogenetic aberrations provide a robust prognostic parameter in risk-adapted treatment regimens, and around 25% of children with AML harbor chromosomal abnormalities considered as high-risk markers.5,6 Risk stratified treatment protocols refined by international collaborations have prominently improved the outcome in pediatric AML with 5-year event-free survival (EFS) and overall survival (OS) reaching 55% and 75%, respectively.9-15
The International Berlin-Frankfurt-Münster AML Study Group (I-BFM-AML SG) has previously studied genetic subsets (eg, t(6;9)/DEK::NUP214, t(16;21), monosomy 7/del7q, 11q23 rearrangements).16-19 Hypodiploid pediatric acute lymphoblastic leukemia (ALL) occurring in ∼5% of patients may be categorized in high hypodiploidy (40-44 chromosomes), low hypodiploidy (30-39 chromosomes), and near haploidy (24-29 chromosomes) with especially low hypodiploidy and near haploidy portending inferior survival rates.20-23 However, hypodiploidy in AML has not been independently investigated owing to its rarity but may constitute a high-risk factor.24,25
Biological and clinical characteristics of hypodiploidy in pediatric AML remain poorly defined but may add information for future guidance of optimal treatment stratification.14 This study aimed to elucidate the occurrence, genetic characteristics, and prognostic impact of hypodiploidy in a large international cohort of pediatric patients with AML.
Methods
Patients
All study groups affiliated with the I-BFM-AML SG were invited to participate in this retrospective cohort analysis. In addition to data regarding patient characteristics, laboratory results (cytogenetic and molecular tests including karyotype and fluorescence in situ hybridization [FISH]), and outcome, the study groups were requested to provide the number of patients with AML with full karyotyping within the study period to assess the frequency of hypodiploidy. Patients from study groups with no information on full number of karyotyped patients were excluded from calculation of hypodiploidy frequency (n = 7).
Children below the age of 18 years diagnosed with de novo AML between January 2000 and December 2015 and a hypodiploid karyotype were considered eligible for the study. Children with either acute promyelocytic leukemia, Down syndrome, juvenile myelomonocytic leukemia, AML secondary to bone marrow failure syndromes or therapy-related AML were excluded.
Cytogenetics
Karyotyping was performed according to standard chromosome banding techniques and at least 10 metaphases were analyzed per case.26 Karyotype strings were made according to International System for Human Cytogenomic Nomenclature (ISCN).27 Hypodiploidy is defined as a modal number (MN) between 35 and 45 (both numbers included) when fulfilling criteria for clonality (hypodiploidy present in minimum 3 metaphases). Ploidy was assigned according to the lowest MN for karyotypes with multiple clones. Hypodiploidy can result from 3 mechanisms: (1) pure numerical, ie, whole chromosome loss; (2) unbalanced structural rearrangements (derivatives/dicentric [Der/dic] chromosomes); or (3) a combination of the 2 mechanisms. Hypodiploid karyotypes irrespective of underlying mechanism were included. Monosomal karyotype (MK) was defined as either at least 2 autosomal monosomies or 1 single autosomal monosomy in combination with at least 1 structural abnormality and absence of core-binding factor (CBF) abnormalities.28,29 For complex karyotype (CK), varying definitions exists.5,6,30-32 We used a commonly agreed definition of CK being characterized by the presence of at least 3 independent chromosomal aberrations irrespective of being of structural or numerical origin and absence of recurrent aberrations as defined by the WHO.33,34 The clone with the highest number of aberrations defined the complexity status.
Composite karyotype and constitutional hypodiploidy including Robertsonian translocations of constitutional origin were excluded. Because sex chromosome loss along with t(8;21) does not affect the superior prognosis of the CBF abnormality, t(8;21) with sex chromosome loss was excluded (regardless of other monosomies).35,36 Loss of chromosome 7 (regardless of other aberrations) was also excluded as monosomy 7 is a well-recognized high-risk marker with or without other aberrations.17
All karyotype reports were centrally reviewed according to ISCN (2016) by a hematological cytogenetic specialist (E.K) and a trained researcher (A.S.H). Results from FISH and molecular tests were considered and included, when available, as the molecular landscape of pediatric AML has previously been investigated and molecular genetics is becoming increasingly important.37 All clonal aberrations of included patients are listed in supplemental Table 1.
Statistical analysis
Complete remission (CR) was defined as <5% blasts in the bone marrow, with hematopoietic regeneration of normal hematopoiesis and absence of extramedullary disease. Relapse was defined as ≥5% bone marrow blasts, reappearance of blasts in peripheral blood or development of extramedullary disease in patients who had reached CR. For comparison of clinical characteristics, the Wilcoxon rank sum test, χ2 test, or Fisher exact tests were used. EFS was calculated from either date of diagnosis to first occurring event (ie, death in remission, relapse, evidence of refractory disease, or secondary malignancies) or date of last follow-up. Refractory disease was considered an event at day 0. OS was calculated from either the date of diagnosis to the date of death because of any cause or date of last follow-up. Cumulative incidence of relapse (CIR) was defined as the probability of a relapse occurring within a given time after diagnosis and estimated using pseudovalues with death as a competing risk.
Survival estimates (EFS and OS) with 95% confidence intervals (95% CI) and impact of prognostic factors were calculated by the Kaplan-Meier method and compared by the log-rank test. The Cox proportional hazard regression was used for multivariate analyses with FLT3-ITD, sex, CK, and white blood cell (WBC) count as covariates. To investigate the characteristics and outcome of decreasing MN, we divided our cohort into 2 groups: children with MN 45 and children with MNs 43 to 44. Missing data for FLT3-ITD was not addressed by multiple imputation because all patients with FLT3-ITD mutation had MN 45. Multivariate analyses were thus performed on cases with available FLT3-ITD information only.
The effect of stem cell transplantation (SCT) on OS was estimated by the Mantel-Byar method considering SCT in first remission a time-dependent event.38 P-values were 2-sided and statistical significance level was set at 0.05. All analyses were performed using Stata/IC 15.1 for Mac (StataCorp, College Station, TX).
Results
Patient characteristics
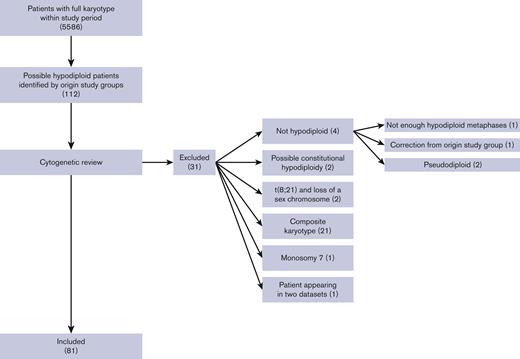
Fourteen collaborative study groups (supplemental Table 2) covering 20 countries (Austria, Czech Republic, Denmark, Finland, France, Germany, Greece, Hong Kong, Hungary, Iceland, Israel, Italy, Japan, Lithuania, The Netherlands, Norway, Poland, Slovenia, Sweden, and the United States) participated. Clinical and cytogenetic data from 81 children fulfilled the inclusion criteria, shown in Figure 1. Seven patients were excluded from the frequency analysis owing to missing information on total number of patients from the study groups. Thus, 74 of 5586 pediatric patients with AML harbored a hypodiploid karyotype defined according to our inclusion criteria, yielding a frequency of 1.3%.
Flowchart of included patients. Clinical and cytogenetic data from 112 children were provided. After cytogenetic review, 81 patients were included. The main exclusion reason was composite karyotype followed by no hypodiploidy. Numbers in () denotes number of patients.
Flowchart of included patients. Clinical and cytogenetic data from 112 children were provided. After cytogenetic review, 81 patients were included. The main exclusion reason was composite karyotype followed by no hypodiploidy. Numbers in () denotes number of patients.
The clinical, morphological, cytogenetic, and genetic characteristics for the cohort are presented in Table 1. The cohort had an even sex distribution and a median age of 6 years (range, 0-17). The median WBC count at diagnosis was 16 × 109/L (range, 0.9-353).
Baseline characteristics
| . | Modal number ≤ 44, (n = 15) . | Modal number = 45, (n = 66) . | Total (n = 81) . |
|---|---|---|---|
| n (%) . | n (%) . | n (%) . | |
| Sex (male/female) | 10/5 (67/33) | 32/34 (48/52) | 42/39 (52/48) |
| Median age (range) | 9 (1-17) | 6 (0-17) | 6 (0-17) |
| Median WBC at diagnosis (109/L) (range) | 13.8 (2.4-315) | 16.6 (0.9-353) | 16.13 (0.9-353) |
| Median PB blast % at diagnosis, (range, n) | 46.5 (0-90, 13) | 45 (0-100, 59) | 46 (0-100, 73) |
| Median BM blast % at diagnosis, (range, n) | 80 (38-99, 13) | 74 (3-100, 57) | 75.5 (3-100, 70) |
| Median platelet count at diagnosis (109/L) (range, n) | 73 (34-121, 14) | 74 (5-359, 60) | 74 (5-369, 74) |
| FAB type | |||
| M0 | 4 (27) | 5 (8) | 9 (11) |
| M1 | 3 (20) | 6 (9) | 9 (11) |
| M2 | 2 (13) | 17 (26) | 19 (23) |
| M4 | 1 (7) | 11 (17) | 12 (5) |
| M5 | 2 (13) | 13 (20) | 15 (9) |
| M6 | 0 (0) | 1 (2) | 1 (1) |
| M7 | 1 (7) | 4 (6) | 5 (6) |
| Other/missing | 1/1 (7/7) | 4/5 (6/8) | 5/6 (6/7) |
| CNS involvement | |||
| Yes | 5 (33)∗ | 4 (6)∗ | 9 (11) |
| No | 10 (67) | 60 (91) | 70 (86) |
| No data | 0 (0) | 2 (3) | 2 (2) |
| Extramedullary disease | |||
| Yes | 4 (27) | 7 (11) | 11(14) |
| No | 11 (73) | 56 (85) | 67 (83) |
| No data | 0 (0) | 3 (4) | 3 (4) |
| Stem cell transplantation | |||
| CR1 | 5 (33) | 13 (20) | 18 (22) |
| After relapse | 2 (13) | 13 (20) | 15 (19) |
| No data (not CR1) | 0 (0) | 4 (6) | 4 (5) |
| Cytogenetics† and genetics | |||
| t(8;21) | 0 (0) | 1 (2) | 1 (1) |
| inv(16)/t(16;16) | 0 (0) | 2 (3) | 2 (1) |
| t(6;9) | 0 (0) | 1 (2) | 1 (1) |
| t(9;11) | 1 (7) | 1 (2) | 2 (1) |
| Other 11q23‡ | 1 (7) | 5 (8) | 7 (8) |
| Complex karyotype | 13 (87) | 35 (53) | 48 (59) |
| Monosomal karyotype | 14 (93) | 44 (67) | 58 (72) |
| FLT3-ITD (n %, 54 tested) | 0 (0, 10 tested) | 4 (9, 44 tested) | 4 (7, 54 tested) |
| TP53 (n %, 29 tested) | 0 (0, 5 tested) | 1 (4, 24 tested) | 1 (3, 29 tested) |
| Event status | |||
| No event | 4 (27) | 27 (41) | 31 (38) |
| Induction death, death in CR | 2 (13) | 2 (3) | 4 (5) |
| Refractory disease | 1 (7) | 6 (9) | 7 (9) |
| Relapse | 8 (53) | 30 (45) | 38 (47) |
| Secondary malignancy | 0 (0) | 1 (2) | 1 (1) |
| Outcome, n (%) (95% CI) | |||
| 5-year EFS | 21 (4-46) | 37 (24-49) | 34 (23-45) |
| 5-year OS | 33 (10-59) | 56 (42-68) | 52 (40-63) |
| . | Modal number ≤ 44, (n = 15) . | Modal number = 45, (n = 66) . | Total (n = 81) . |
|---|---|---|---|
| n (%) . | n (%) . | n (%) . | |
| Sex (male/female) | 10/5 (67/33) | 32/34 (48/52) | 42/39 (52/48) |
| Median age (range) | 9 (1-17) | 6 (0-17) | 6 (0-17) |
| Median WBC at diagnosis (109/L) (range) | 13.8 (2.4-315) | 16.6 (0.9-353) | 16.13 (0.9-353) |
| Median PB blast % at diagnosis, (range, n) | 46.5 (0-90, 13) | 45 (0-100, 59) | 46 (0-100, 73) |
| Median BM blast % at diagnosis, (range, n) | 80 (38-99, 13) | 74 (3-100, 57) | 75.5 (3-100, 70) |
| Median platelet count at diagnosis (109/L) (range, n) | 73 (34-121, 14) | 74 (5-359, 60) | 74 (5-369, 74) |
| FAB type | |||
| M0 | 4 (27) | 5 (8) | 9 (11) |
| M1 | 3 (20) | 6 (9) | 9 (11) |
| M2 | 2 (13) | 17 (26) | 19 (23) |
| M4 | 1 (7) | 11 (17) | 12 (5) |
| M5 | 2 (13) | 13 (20) | 15 (9) |
| M6 | 0 (0) | 1 (2) | 1 (1) |
| M7 | 1 (7) | 4 (6) | 5 (6) |
| Other/missing | 1/1 (7/7) | 4/5 (6/8) | 5/6 (6/7) |
| CNS involvement | |||
| Yes | 5 (33)∗ | 4 (6)∗ | 9 (11) |
| No | 10 (67) | 60 (91) | 70 (86) |
| No data | 0 (0) | 2 (3) | 2 (2) |
| Extramedullary disease | |||
| Yes | 4 (27) | 7 (11) | 11(14) |
| No | 11 (73) | 56 (85) | 67 (83) |
| No data | 0 (0) | 3 (4) | 3 (4) |
| Stem cell transplantation | |||
| CR1 | 5 (33) | 13 (20) | 18 (22) |
| After relapse | 2 (13) | 13 (20) | 15 (19) |
| No data (not CR1) | 0 (0) | 4 (6) | 4 (5) |
| Cytogenetics† and genetics | |||
| t(8;21) | 0 (0) | 1 (2) | 1 (1) |
| inv(16)/t(16;16) | 0 (0) | 2 (3) | 2 (1) |
| t(6;9) | 0 (0) | 1 (2) | 1 (1) |
| t(9;11) | 1 (7) | 1 (2) | 2 (1) |
| Other 11q23‡ | 1 (7) | 5 (8) | 7 (8) |
| Complex karyotype | 13 (87) | 35 (53) | 48 (59) |
| Monosomal karyotype | 14 (93) | 44 (67) | 58 (72) |
| FLT3-ITD (n %, 54 tested) | 0 (0, 10 tested) | 4 (9, 44 tested) | 4 (7, 54 tested) |
| TP53 (n %, 29 tested) | 0 (0, 5 tested) | 1 (4, 24 tested) | 1 (3, 29 tested) |
| Event status | |||
| No event | 4 (27) | 27 (41) | 31 (38) |
| Induction death, death in CR | 2 (13) | 2 (3) | 4 (5) |
| Refractory disease | 1 (7) | 6 (9) | 7 (9) |
| Relapse | 8 (53) | 30 (45) | 38 (47) |
| Secondary malignancy | 0 (0) | 1 (2) | 1 (1) |
| Outcome, n (%) (95% CI) | |||
| 5-year EFS | 21 (4-46) | 37 (24-49) | 34 (23-45) |
| 5-year OS | 33 (10-59) | 56 (42-68) | 52 (40-63) |
Baseline characteristics have been listed for entire cohort and by modal number.
BM, bone marrow; CNS, central nervous system; CR1, first complete remission; inv, inversion; PB, peripheral blood; FAB, French-American-British.
P < .05.
Only occurring WHO aberrations listed.
Not all FISH verified.
Two MN groups were defined: children with MN 45 (66 patients, 81%) and children with MNs 43 to 44 (15 patients, 19%; of whom 10 patients had MNs 44 and 5 had MN 43). No patient had a MNs of 42 or lower. Children with MN 45 predominantly displayed FAB classification morphology M2 (n = 17, 26%), M4 (n = 11, 17%), and M5 (n = 13, 20%), whereas children with MNs 43 to 44 showed preponderance of the minimally differentiated M0 (n = 4, 27%) and M1 (n = 3, 20%) (Table 1). For further clinical characteristics regarding MN groups, refer to Table 1.
Nine children (11%) had verified CNS involvement. Children with MNs 43 to 44 were significantly more likely to have CNS involvement (33% vs 6%; P = .01).
Cytogenetics
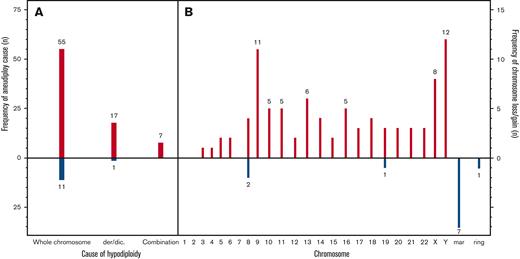
Hypodiploidy resulted from whole chromosome losses in 57 (70%) patients, of whom 8 (10%) had a chromosome loss as the sole abnormality. Figure 2 presents the frequency of specific chromosome losses. Of patients with hypodiploidy purely because of whole chromosome loss, 49 of 57 (86%) had additional structural abnormalities and 15 of 57 (26%) had >1 chromosome loss. In total, 17 of 81 (21%) patients were hypodiploid because of derivative or dicentric chromosomes and 7 of 81 (8%) patients had hypodiploidy caused by both whole chromosome loss and derivative/dicentric rearrangements. Twelve patients (15%) had gain of chromosomal material, Figure 2B.
Frequency of specific chromosome losses. Causes of aneuploidy (A). Frequency of specific chromosome losses (B). der/dic, derivative/dicentric, mar, marker chromosome.
Frequency of specific chromosome losses. Causes of aneuploidy (A). Frequency of specific chromosome losses (B). der/dic, derivative/dicentric, mar, marker chromosome.
Of the 15 children with MNs 43 to 44, 8 (53%) was hypodiploid solely because of whole chromosome loss, 3 (20%) because of derivative/dicentric rearrangements, and the remaining 4 (27%) harbored both whole chromosome loss and derivative/dicentric rearrangements.
Numerical and structural aberrations
Chromosome Y was the most frequently lost chromosome (n = 12), and 3 patients had a missing chromosome Y as the only abnormality. Other frequently lost chromosomes were chromosome 9 (n = 11) and chromosome X (n = 8, all females) (Figure 2). Loss of chromosome 9 was significantly associated with MNs 43 to 44 (P = .01). No patients showed a loss of chromosome 1 or 2. Loss of chromosomes X, Y, 8, 19, and 21 occurred unaccompanied by structural aberrations. Of structural aberrations, del7q and del9q were each found in 4 patients (5%). With the exception of 1 patient with MN 44 and t(9;11)(p12;q23), all WHO recurrent aberrations (listed in Table 1) occurred in children with MN 45. One karyotype (45, XY, -10, -11, +mar) was observed in 2 patients. In addition, 1 other patient had concurring loss of chromosome 10 and 11. Notably, no FISH was available for such cases to rule out a KMT2A rearrangement.
Complex and monosomal karyotype
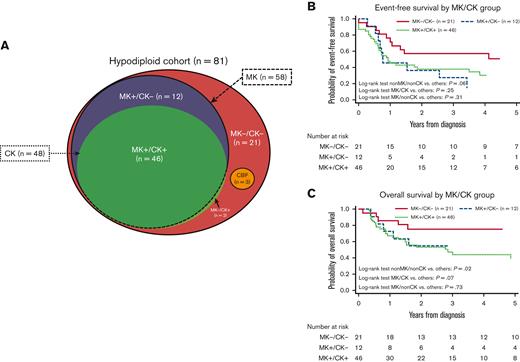
Fifty-eight patients (71%) were classified as MK and 48 (59%) as CK, of which 46 patients (57%) qualified for both MK and CK (CK/MK). Figure 3 shows the distribution of patients according to CK/MK status as well as the extensive overlap between the groups. Twenty-one patients (26%) displayed neither CK nor MK (non-CK/non-MK), 12 patients showed MK without CK (non-CK/MK) and 2 patients (2%) had CK without MK (CK/non-MK). Of the 15 children with MNs 43 to 44, 13 (85%) harbored CK.
Distribution and survival of monosomal karyotype (MK) and complex karyotype (CK) within the cohort. Distribution of MK/CK within the cohort (A). Event-free survival by MK/CK group (B). Dichotome log-rank tests: MK−/CK−: P = .06; MK+/CK−: P = .31; MK+/CK+: P = .32. OS by MK/CK group (C) Dichotome log-rank tests: MK−/CK−: P = .02; MK+/CK−: P = .73; MK+/CK+: P = .07.
Distribution and survival of monosomal karyotype (MK) and complex karyotype (CK) within the cohort. Distribution of MK/CK within the cohort (A). Event-free survival by MK/CK group (B). Dichotome log-rank tests: MK−/CK−: P = .06; MK+/CK−: P = .31; MK+/CK+: P = .32. OS by MK/CK group (C) Dichotome log-rank tests: MK−/CK−: P = .02; MK+/CK−: P = .73; MK+/CK+: P = .07.
Molecular genetics
We investigated available molecular data (NRAS, KRAS, KIT, FLT3-ITD, and TP53) and correlation to MN. Data regarding NRAS, KRAS, and KIT were sparse (supplemental Table 3). Fifty-four patients (67%) had available FLT3-ITD information. Four of these patients (7%) harbored a FLT3-ITD mutation, all with MN 45. Regarding TP53, 29 patients (36%) had available information. The only TP53 mutation was found in a child with MN 45. Unfortunately, it was not possible to establish a germline status.
Prognosis
Median follow-up time for patients alive was 4.5 years (range, 0.2-15.3). Seventy-four children (91%) achieved CR after induction therapy. Thirty-eight children relapsed, yielding a CIR of 50% with a median of 9.2 months from diagnosis to relapse. Twenty-seven of the relapses (71%) occurred within the first year of diagnosis. Twelve children (32%) experiencing leukemia relapse were alive at the end of follow-up. The 5-year EFS and OS for the entire cohort (n = 81) were 34% (95% CI, 23-45) and 52% (95% CI, 40-63), respectively. The 5-year EFS and OS for patients with available FLT3-ITD status (n = 54) were 35% (95% CI, 22-49) and 52% (95% CI, 37-67), respectively.
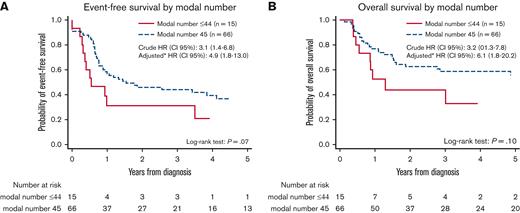
Children with MNs 43 to 44 showed inferior EFS (21% [95% CI, 4-46] vs 37% [95% CI, 24-49]; P = .07) and OS (33% [95 % CI, 10-59] vs 56% [95 % CI, 42-68]; P = .1) compared with children with MN 45, though the differences did not reach statistical significance (Figure 4). Fifty-four patients with FLT3-ITD data were included in the multivariate analysis (44 with MN 45, 5 with MN 44, and 5 with MN 43). All 5 children with MN 43 experienced relapse, subsequently leading to death of 4 children. MNs 43 to 44 were independent predictor of an inferior EFS (crude HREFS for patients with information on FLT3-ITD, 3.1 [95% CI, 1.4-6.8], P = .005; and adjusted HREFS, 4.9 [95% CI, 1.8-13.0], P = .001) and OS (crude HROS for patients with information on FLT3-ITD, 3.2 [95% CI, 1.3-7.8], P = .01; and adjusted HROS, 6.1; 95% CI, 1.8-20.2; P = .003). Crude HREFS and HROS for the entire cohort were 1.8 (95% CI, 0.9-3.6) and 1.9 (95% CI, 0.9-4.0), respectively.
Event-free and overall survival (OS) by modal number (MN). Event-free survival by MN 45 vs 43 to 44 (A). OS by MN 45 vs 43 to 44 (B). HRs represent analyses on patients with complete FLT3-ITD. ∗Adjusted for sex, complex karyotype, FLT3-ITD, and WBC. HR, hazard ratio.
Event-free and overall survival (OS) by modal number (MN). Event-free survival by MN 45 vs 43 to 44 (A). OS by MN 45 vs 43 to 44 (B). HRs represent analyses on patients with complete FLT3-ITD. ∗Adjusted for sex, complex karyotype, FLT3-ITD, and WBC. HR, hazard ratio.
Patients with loss of chromosome Y (n = 12) tended to have a better prognosis compared with other male patients (supplemental Figure 1). Loss of chromosome X (n = 8, all female) showed inferior EFS compared with other female patients (supplemental Figure 2), however not reaching statistical significance. Poor survival rates were observed in patients with monosomy 9, monosomy 10, and monosomy 16. Survival estimates for the most common chromosome losses are shown in Table 2.
Survival estimates for specific chromosome losses
| Group (n) . | EFS, % (95% CI) . | OS, % (95% CI) . |
|---|---|---|
| Loss of chromosome Y (12) | 58 (27-80; P= .19) | 75 (41-91; P= .23) |
| Loss of chromosome X (8) | 19 (1-54; P= .87) | 31 (2-72; P= .89) |
| Loss of chromosome 9 (11) | 14 (1-43; P= .12) | 15 (1-47; P= .12) |
| Loss of chromosome 10 (5) | 20 (1-58; P= .24) | 20 (1-58; P= .054) |
| Loss of chromosome 16 (5) | 25 (1-67; P= .36) | 25 (1-67; P= .16) |
| Group (n) . | EFS, % (95% CI) . | OS, % (95% CI) . |
|---|---|---|
| Loss of chromosome Y (12) | 58 (27-80; P= .19) | 75 (41-91; P= .23) |
| Loss of chromosome X (8) | 19 (1-54; P= .87) | 31 (2-72; P= .89) |
| Loss of chromosome 9 (11) | 14 (1-43; P= .12) | 15 (1-47; P= .12) |
| Loss of chromosome 10 (5) | 20 (1-58; P= .24) | 20 (1-58; P= .054) |
| Loss of chromosome 16 (5) | 25 (1-67; P= .36) | 25 (1-67; P= .16) |
Children with CNS involvement had significantly worse EFS (11% [95% CI, 1-39] vs 40% [95% CI, 28-52]; P = .04), primarily because of relapse (n = 7).
Comparable EFS was observed in patients with CK (31%; 95% CI, 17-46) and patients with non-CK (38%; 95% CI, 21-55; P = .28). OS was lower for patients with CK (39%, 95% CI, 23-55) compared with patients with non-CK (68%; 95% CI, 49-81; P = .058). MK showed nonsignificantly worse EFS (27%; 95% CI, 14-40; P = .052) compared with patients with non-MK (50%; 95% CI, 28-69; P = .052). OS was significantly worse in patients with MK (43%; 95% CI, 28-56) compared with patients with non-MK (75%; 95% CI, 50-87; P = .03). Outcomes for non-CK/non-MK, CK/MK, and MK/non-CK were dichotomously compared, and survival curves are presented in Figure 3B,3C. Only 2 patients displayed CK/non-MK and were not independently evaluated. Children with non-CK/non-MK (n = 21) had better EFS (51% [95% CI, 28-70] vs 27% [95% CI, 15-41]; P = .06). OS for patients with non-CK/non-MK was significantly superior to that of the remaining patients (76% [95% CI, 51-89] vs 43% [95% CI, 29-56]; P = .02). Children with MK/CK (n = 46) had EFS similar to the rest of the cohort (30% [95% CI, 16-45] vs 38% [95% CI, 22-55]; P = .25) but displayed a dismal OS (39% [95% CI, 23-55] vs 67% [95% CI, 48-80]; P = .07). Children with MK/non-CK (n = 12) displayed poor EFS compared with the remaining cohort (14% [95% CI, 1-43] vs 37% [95% CI, 25-49]; P = .31). This did not translate into inferior OS (55% [95% CI, 23-78] vs 51% [95% CI, 38-63]; P = .73).
Three of 4 patients with FLT3-ITD did not survive, 2 experienced relapse and subsequently died, and 1 died of SCT-related causes. The patient with TP53 mutation experienced relapse and died of SCT-related causes.
Thirty-seven children (46%) were treated with SCT, either in CR1 (n = 18) or after relapse (n = 19). SCT in CR1 did not translate into superior survival with HR 1.4, (95% CI, 0.65-3.02; P = .39) and 1.5 (95% CI, 0.54-4.33; P = .42) in crude and adjusted analyses, respectively. Exclusion of children never reaching CR1 (n = 7) did not significantly change HR (HR, 1.8; 95% CI, 0.62-5.47; P = .27). Children with MNs 43 to 44 and MN 45 were equally likely to receive SCT in CR1 (P = .4). A total of 9 children died after SCT in CR1, 3 died of SCT-related toxicities, and 6 suffered from relapse and subsequently died of disease progression.
Discussion
In this large I-BFM based series of pediatric patients with AML with complete karyotyping, hypodiploidy defined as an MN between 35 and 45 and absence of monosomy 7 and t(8;21) with sex chromosome loss occurred infrequently (1.3%) but was associated with an inferior outcome, especially with MNs 43 to 44.
Patients with MNs of 45 or lower had inferior survival rates and a higher CIR as compared with the same outcome measures reported from pediatric AML cohorts within the last 2 decades.39 In particular, children with MNs 43 to 44 had a poor prognosis, which remained inferior compared with MN 45 in multivariate analyses. The proportion of excluded patients because of missing FLT3-ITD information was comparable between the 2 MN groups. In the cohort with complete mutation status, the HR of an inferior outcome in patients with MNs 43 to 44 was increased and remained statistically significant after adjustment for relevant covariates and was thus independently associated with an inferior outcome. Consolidation with SCT in CR1 did not improve the outcome. However, because we did not have detailed information on SCT indication, there might be a selection of poor responders or poor genetics among patients treated with SCT, and the study is small. Reduced EFS and OS were also observed in hypodiploid ALL with decreasing survival with lower MN20 and similar to AML SCT, did not improve survival.40,41
Despite being the largest study of hypodiploidy in pediatric AML to date, the cohort may be too small to demonstrate significant differences in outcome. The size of the study limits the number of covariates applicable for multivariate analyses, and hence the dataset neither allowed adjustment for all high-risk features nor did it allow us to stratify the patients based on all established co-occurring translocations. Furthermore, the patients were treated with 12 different protocols over a 16-years timespan and were subjected to various risk stratification and treatments that may influence the prognosis. However, in addition to our findings of hypodiploidy, this study also demonstrates the importance of successful international collaboration while investigating rare entities in pediatric AML.
Although being a standardized method to investigate aberrant clonality, the use of standard G-band karyotyping may have missed relevant rearrangements. Molecular aberrations detectable by next-generation sequencing or copy number aberrations detected by aCGH/SNP analyses with possible influence on prognosis were not included in this study because of lack of information but may be associated with chromosome loss.42 Cytogenetic investigation does, however, remain relevant in risk stratification.43 Furthermore, it was not always possible to distinguish partial chromosome loss from complete chromosome loss without elaborate array data. Especially for derivative or dicentric rearrangements this presented a challenge. Thus, some monosomies may only represent partial chromosome loss.
Hypodiploidy has previously been observed in ∼10% of pediatric patients with AML6,24,36 limited to MN 45 and less frequently 44 and 43,6,24 though lower MNs have been reported (MN, 41 and 387,44). We excluded hypodiploidy caused by the most frequent chromosome losses, namely monosomy 7 and sex chromosome loss with t(8;21) because both are well described17,36 and would dominate the present series if included. Furthermore, we excluded composite karyotypes (n = 21) because the karyotype does not allow identification of the precise MN. We observed no cases with MNs below 43, a finding contrasting hypodiploid ALL, in which patients may harbor near-haploid karyotypes.20,21
Previous studies showed that chromosome 7 is the most frequently lost autosome and is associated with a poor prognosis in both children and adults.5,17,24,35 Monosomy 5 is considered a high-risk marker in adult AML45 but has only been reported in a limited number of pediatric patients.46 We found only 2 patients with monosomy 5, supporting previous reports of its rarity among children.8,46 A study comprising 24 patients with monosomies (monosomy 7, favorable cytogenetics and KMT2A rearrangements not investigated) reported no difference in outcome compared with the rest of their cohort5 but did not account for MN. We show that loss of the chromosomes 9 (n = 11), 10 (n = 5), and 16 (n = 5) may be associated with a poor prognosis. Two patients with monosomy 16 had additional t(16;21)(p11;q22) (FUS::ERG), which is associated to poor outcome.18 Loss of chromosome 9 was significantly associated to MNs below 45, suggesting that loss of this specific chromosome may indicate distinct genetic instability and a poor prognosis. Monosomy 9 is also observed in bladder cancer and in high–grade renal clear cell carcinomas in which it correlates to an inferior prognosis and disease recurrence.47,48 In our cohort 8 of 11 patients with monosomy 9 experienced disease relapse.
Only 3 patients with CBF aberrations [(t(8;21), t(16;16), inv(16)] and a hypodiploid karyotype were observed, indicating that hypodiploidy because of autosomal chromosome loss or derivative rearrangements in CBF leukemia is rare. All patients with CBF leukemia survived and only 1 experienced relapse. All patients with CBF had MN 45, but exclusion of the patients had minimal effect on the multivariate analyses.
Loss of a sex chromosome is associated with CBF aberration t(8;21)(q22;q22.1)7,36 and is a frequent finding not aggravating the favorable outcome among these patients.6,35 It is remarkable that loss of sex chromosome has been reported to occur in 45% of t(8;21)36 whereas it is almost completely absent in inv(16) AML with none identified in the present study. In our cohort, loss of a sex chromosome influenced prognosis in diverging directions with loss of chromosome Y displaying good outcome, whereas loss of chromosome X presenting inferior survival. Loss of chromosome Y in AML with t(8;21) may tend to have a better outcome than loss of chromosome X with t(8;21),35 supporting our findings of distinct consequences of loss of a sex chromosome in patients without CBF abnormalities.
MK has adverse effect on EFS in pediatric AML,25,32 and CK may have adverse outcome.5,6,30-32 In this study, we found MK to be a significant predictor of poor outcome for OS in patients with hypodiploidy Similarly, MK is associated with adverse prognosis in adult AML.28,29 Other studies have examined MK in pediatric AML and found low EFS and high CIR, but minimal impact on OS25,32 and hypodiploidy did not have an adverse prognosis independent of MK.25 However, none of these studies focused exclusively on MN and also included t(8;21) with sex chromosome loss, monosomy 7, and composite karyotypes. CK was common (48 patients, 59%) among hypodiploid pediatric patients, consistent with observations among adults31 and displayed inferior survival. In agreement with previous observations,32 we found that MK without CK did not affect OS, suggesting that karyotype complexity may aggravate the adverse impact of chromosome loss.
FLT3-ITD is associated with a poor outcome49,50 and is an infrequent finding in patients with chromosome loss.25,32 Our findings with only 4 of 54 patients harboring the mutation support the rare occurrence of FLT3-ITD in patients with chromosome loss.
Germline TP53 alterations have been observed in the most children with low-hypodiploid ALL.42,51,52 In adult AML, TP53 mutations occur regularly as acquired mutations but may be germline53,54 and have been associated with CK and monosomy 5.45,55,56 TP53 alterations in pediatric AML are rarely observed.37 Only 1 of our 29 patients with available information harbored a TP53 mutation, indicating that the association found between low hypodiploid pediatric ALL and TP53 mutations is not mirrored in hypodiploid pediatric AML.
Measurable residual disease (MRD) is valuable for evaluation of treatment response and is a strong indicator of prognosis useful for risk group assignment.11,57,58 An investigation of correlation between hypodiploidy and MRD could be of interest, because MRD has been proven a strong prognostic indicator in hypodiploid ALL.59 The MRD response for this cohort was, however, not available.
In conclusion, this successful international collaborative study demonstrates that hypodiploid pediatric AML, albeit rare, is a clinically relevant aberration with a dismal outcome. Patients with hypodiploidy, especially children with MNs 43 to 44, may benefit from more accurate risk stratification, ie MRD treatment response evaluation and novel therapy is needed as not all patients with hypodiploidy benefit from SCT in CR1. Loss of chromosome Y should not independently determine allocation to high-risk regimens. The strong association found between TP53 mutations and low-hypodiploid ALL was not reflected in this cohort of pediatric patients with AML. Evaluation of pediatric patients with AML with hypodiploid karyotypes in an international, prospective study supported by extended molecular analyses, and MRD techniques may provide further insights into this rare disease entity and optimal treatment schedules.
Acknowledgments
The authors thank all contributing study groups for their benevolent participation and cooperation (a full list appears in Supplemental data).
This work was presented as an award-winning poster abstract at the 60th annual meeting of the American Society of Hematology, San Diego, 1-4 December 2018.
This work was supported by grants from the Danish Cancer Society (R165-A10463) and the Else & Mogens Wedell-Wedellsborg foundation.
Authorship
Contribution: K.L.J.-D., J.D.S., and H.H. designed the study; J.A., M.C., E.D., I.H., K.J., E.A.K., G.K., G.L., F.L., R.M., U.N.-N., S.C.R., M.R., D.R., T.T., D.T., and B.Z. contributed clinical data; A.S.B.H. and E.K. performed cytogenetic review; A.S.B.H. and K.L.J.-D. performed the statistical analyses; A.S.B.H., K.L.J.-D., H.H., and E.K. wrote the manuscript; J.D.S., K.L.J.-D, H.H., and E.K. supervised the study; and all authors performed critical review of the manuscript and approved the final version of the manuscript.
A complete list of the members of the collaborators of the I-BFM Study Group appears in the supplemental Appendix.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Anne Sofie Borg Hammer, Aarhus University Hospital, Palle Juul-Jensens Blvd 99, 8200 Aarhus N, Denmark; e-mail: as.hammer@clin.au.dk.
References
Author notes
Data are available on request from the corresponding author, Anne Sofie Borg Hammer (as.hammer@clin.au.dk).
The online version of this article contains a data supplement.