Key Points
Detection of mutations using different NGS methods shows over 95% agreement for genes used in therapeutic assignment on the BAMT.
Minimal discrepancies were due to VUS/sample quality, indicating that using different NGS methods for clinical decisions may be acceptable.
Abstract
Next-generation sequencing (NGS) to identify pathogenic mutations is an integral part of acute myeloid leukemia (AML) therapeutic decision-making. The concordance in identifying pathogenic mutations among different NGS platforms at different diagnostic laboratories has been studied in solid tumors but not in myeloid malignancies to date. To determine this interlaboratory concordance, we collected a total of 194 AML bone marrow or peripheral blood samples from newly diagnosed patients with AML enrolled in the Beat AML Master Trial (BAMT) at 2 academic institutions. We analyzed the diagnostic samples from patients with AML for the detection of pathogenic myeloid mutations in 8 genes (DNMT3A, FLT3, IDH1, IDH2, NPM1, TET2, TP53, and WT1) locally using the Hematologic Neoplasm Mutation Panel (50-gene myeloid indication filter) (site 1) or the GeneTrails Comprehensive Heme Panel (site 2) at the 2 institutions and compared them with the central results from the diagnostic laboratory for the BAMT, Foundation Medicine, Inc. The overall percent agreement was over 95% each in all 8 genes, with almost perfect agreement (κ > 0.906) in all but WT1, which had substantial agreement (κ = 0.848) when controlling for site. The minimal discrepancies were due to reporting variants of unknown significance (VUS) for the WT1 and TP53 genes. These results indicate that the various NGS methods used to analyze samples from patients with AML enrolled in the BAMT show high concordance, a reassuring finding given the wide use of NGS for therapeutic decision-making in AML.
Introduction
The diagnosis, detailed molecular characterization, and treatment of myeloid malignancies, specifically acute myeloid leukemia (AML), has rapidly evolved over the past 10 years with the incorporation of next-generation sequencing (NGS) results into therapeutic decision-making in everyday clinical practice.1 The rapid development and clinical use of this methodology, using a variety of NGS platforms and mutational panels, has allowed for more specific risk-stratification, incorporation into targeted treatment algorithms (especially for precision oncology master trials), and detection of measurable residual disease in those with an otherwise heterogeneous hematological malignancy.2-5
One such ground-breaking, large-scale, umbrella clinical trial is the Beat AML Master Trial (BAMT), in which patients aged ≥60 years with newly diagnosed AML are enrolled and assigned to precision oncology targeted substudies based, in part, on molecular mutations.6 The pathology, cytogenetics, and fluorescence in situ hybridization results are provided by the participating institution, whereas the molecular mutation profile is provided by a central laboratory, Foundation Medicine, Inc (FMI). Assignment of these patients to a substudy is based on application of an algorithm (using a combination of cytogenetic and molecular mutation data) within 7 days after sample collection and return of results by the central laboratory.6,7
Historically, clinical trials have used central laboratory diagnostic testing to ensure uniformity of methodology for sample analysis and test results. The limitation of this approach is the increased turnaround time and potential delay in sample analysis. These factors are particularly important in precision oncology trials in which starting therapy expediently is critical for patients with high-risk disease. The challenge of doing such testing at local institutional laboratories is that the participating institutions may have their own different NGS methodology, using different technical platforms and interpretive criteria, which would result in potentially heterogenous results.2,8-11
Several studies have compared the sensitivity, specificity, and clinical use of various heterogeneous NGS platforms for patients with solid tumors and demonstrated some level of interlaboratory variability.12,13 These discordances are attributed to many possible factors including sample type, operational processes, and technical or interpretive aspects of sequencing. These studies demonstrate the challenge in balancing the use of a homogeneous central laboratory diagnostic assay compared with other assays using different NGS methods for clinical decision-making for patients with solid tumors.
As a comparison of NGS mutational results across various laboratories and platforms has not been extensively studied in myeloid malignancies, we used diagnostic AML samples collected from newly diagnosed older patients with AML enrolled in the BAMT from 2 participating institutions, Oregon Health and Science University (OHSU) and Ohio State University (OSU), that collectively enrolled over 50% of the patients from 2016 to 2019. We compared identification of pathogenic mutations on diagnostic samples among the institutional laboratories enrolling the study patients with those made by the central laboratory (FMI) to determine the agreement in identification of molecular mutations among different NGS platforms and methodologies.
Materials and methods
Sample collection
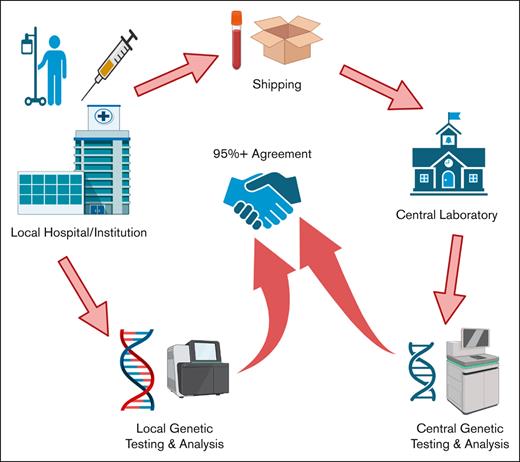
We analyzed diagnostic samples collected from patients with AML who consented to the BAMT per declaration of Helsinki and institutional review board guidelines at 2 academic sites, OSU (defined as site 1) and OHSU (defined as site 2), from 15 November 2016 to 15 April 2019. For this analysis, 194 diagnostic samples collected from patients with AML, who wereenrolled on the BAMT, were included (125 at site 1 and 69 at site 2). DNA was extracted from bone marrow (BM; 77 at site 1 and 57 at site 2) and/or peripheral blood (PB; 48 at site 1 and 12 at site 2) and were analyzed at the screening institution. At the same time, additional BM and/or PB samples were collected and then sent to FMI (defined as central laboratory) for analysis using the Beat AML clinical trial assay panel (Figure 1). Type of tissue (BM vs PB) analyzed for variants between the institutional and central site was the same in all but 6 cases (all from site 2, 5 BM, and 1 PB). All diagnostic samples analyzed at institutional and central laboratory were obtained either on the same day or within 3 days of previous sample collection and before any treatment was initiated. Samples sent to both laboratories >3 days apart were excluded to minimize variability in samples from the same patient.
Schematic for the analysis of patient samples between central and institutional laboratories.
Schematic for the analysis of patient samples between central and institutional laboratories.
NGS methods
Each sample was analyzed at the central (FMI) laboratory and at one of the institutional laboratories. The central laboratory clinical trial assay platform used capture-based sequencing of the complete coding region of 406 genes with median depth of unique coverage of 500×. The sensitivity was >99% for base substitutions ≥5% minor allele frequency and >97% for insertions/deletions up to 40 base pairs at ≥10% minor allele frequency. Turnaround time for this Beat AML clinical trial assay platform was ≤7 days. Per specifications in the Beat AML Master Protocol, all samples received were processed regardless of tumor purity, and all mutations detected (eg, known/likely pathogenic, variant of unknown significance) were reported.
The platform used at site 1 was the OSU’s Hematologic Neoplasm Mutation Panel (50-gene myeloid indication filter). This panel used an amplicon–based NGS strategy using gene-specific primers followed by sequencing on the Illumina MiSeq. The panel focuses on base substitutions and small indels with full coding coverage for 36 genes and hotspot coverage for 14 additional genes. The mutant allele sensitivity is 100% at variant allele frequency (VAF) ≥5% (99% at VAF ≥2%) with an average read depth of 2100×.
The platform used at site 2 was the GeneTrails Comprehensive Heme Panel. This assay used amplification-based NGS with gene-specific primers (GSPs) and unique molecular indexes (UMIs) followed by sequencing on the Illumina NextSeq 500. The panel identified genomic alterations including base substitutions and small indels in selected exons of CARD11 and the entire coding region 75 other genes. The average read depth after UMI deduplication was 2200×; the mutant allele sensitivity was 100% at VAF ≥5% for base substitutions and ≥2% for small indels. Further details on all assays have been provided in the supplemental Materials.
We evaluated the ability to identify short variant mutations (eg, missense mutations and small indels) in following 8 genes: DNMT3A, FLT3, IDH1, IDH2, NPM1, TET2, TP53, and WT1 as used in treatment assignment in the BAMT.
Statistical comparison of assay results
A detection cutoff of 2% was used to define the presence or absence of a mutation. Overall percent agreement was defined as the number of times the site and central laboratories made the same call divided by the total number of patients. Sensitivity was defined as the number of present calls made at each site divided by the number of present calls made centrally, and specificity as the number of absent calls made at each site divided by the number of absent calls made centrally. The κ statistic provides a measure of chance-corrected agreement when controlling for site. Values <0 indicate less than a chance agreement and sometimes systematic disagreement, 0.01 to 0.20 slight agreement, 0.21 to 0.40 fair agreement, 0.41 to 0.60 moderate agreement, 0.61 to 0.80 substantial agreement, 0.81 to 0.99 almost perfect agreement, and 1.0 perfect agreement. Overall percent agreement, sensitivity and specificity using central reads as the reference, and κ statistics have been provided with 95% confidence intervals. All analyses were performed using SAS v9.4.
Results
Agreement between institutional and central laboratories
The overall percent agreement between institutional and central laboratories ranged from 96.4% (TP53) to 100% (NPM1). The remaining genes’ overall percent agreement were 97.9% (WT1, DNMT3A, and TET2), 99.0% (FLT3 and IDH2), and 99.5% (IDH1). The sensitivity was at least 94% for TET2 (97.1%), DNMT3A (94.4%), FLT3 (94.6%), IDH1 (95.5%), IDH2 (97.8%), and NPM1 (100%), with the exception of TP53 (88.9%) and WT1 (72.7%) (Table 1). The specificity was at least 98% for all genes, with 3 genes showing 100% specificity (NPM1, FLT3, and IDH1). The κ statistic, adjusted for site, showed substantial agreement for WT1 (0.848), nearly perfect agreement for 6 genes including TP53 (0.906), DNMT3A (0.906), TET2 (0.932), FLT3 (0.945), IDH1 (0.948), and IDH2 (0.973), and perfect agreement for NPM1 (1.0) (Table 1).
Statistical analysis of the 8 genes compared between central and institutional laboratories
| Feature . | Overall agreement∗ (%, 95% CI) . | Sensitivity† (%, 95% CI) . | Specificity‡ (%, 95% CI) . | Overall κ§ (95% CI) . |
|---|---|---|---|---|
| DNMT3A | 190/194 (97.9, 94.8-99.4) | 51/54 (94.4, 84.6-98.8) | 139/140 (99.3, 96.1-100) | 0.906 (0.815-0.996) |
| FLT3 | 192/194 (99.0, 96.3-99.9) | 35/37 (94.6, 81.8-99.3) | 157/157 (100, 97.7-100) | 0.945 (0.869-1) |
| IDH1 | 193/194 (99.5, 97.2-100) | 21/22 (95.5, 77.2-99.9) | 172/172 (100, 97.9-100) | 0.948 (0.847-1) |
| IDH2 | 192/194 (99.0, 96.3-99.9) | 44/45 (97.8, 88.2-99.9) | 148/149 (99.3, 96.3-100) | 0.973 (0.934-1) |
| NPM1 | 194/194 (100, 98.1-100) | 45/45 (100, 92.1-100) | 149/149 (100, 97.6-100) | 1 |
| TET2 | 190/194 (97.9, 94.8-99.4) | 33/34 (97.1, 84.7-99.9) | 157/160 (98.1, 94.6-99.6) | 0.932 (0.865-0.999) |
| TP53 | 187/194 (96.4, 92.7-98.5) | 40/45 (88.9, 75.9-96.3) | 147/149 (98.7, 95.2-99.8) | 0.906 (0.834-0.978) |
| WT1 | 190/194 (97.9, 94.8-99.4) | 8/11 (72.7, 39.0-94.0) | 182/183 (99.5, 97.0-100) | 0.848 (0.679-1.017) |
| Feature . | Overall agreement∗ (%, 95% CI) . | Sensitivity† (%, 95% CI) . | Specificity‡ (%, 95% CI) . | Overall κ§ (95% CI) . |
|---|---|---|---|---|
| DNMT3A | 190/194 (97.9, 94.8-99.4) | 51/54 (94.4, 84.6-98.8) | 139/140 (99.3, 96.1-100) | 0.906 (0.815-0.996) |
| FLT3 | 192/194 (99.0, 96.3-99.9) | 35/37 (94.6, 81.8-99.3) | 157/157 (100, 97.7-100) | 0.945 (0.869-1) |
| IDH1 | 193/194 (99.5, 97.2-100) | 21/22 (95.5, 77.2-99.9) | 172/172 (100, 97.9-100) | 0.948 (0.847-1) |
| IDH2 | 192/194 (99.0, 96.3-99.9) | 44/45 (97.8, 88.2-99.9) | 148/149 (99.3, 96.3-100) | 0.973 (0.934-1) |
| NPM1 | 194/194 (100, 98.1-100) | 45/45 (100, 92.1-100) | 149/149 (100, 97.6-100) | 1 |
| TET2 | 190/194 (97.9, 94.8-99.4) | 33/34 (97.1, 84.7-99.9) | 157/160 (98.1, 94.6-99.6) | 0.932 (0.865-0.999) |
| TP53 | 187/194 (96.4, 92.7-98.5) | 40/45 (88.9, 75.9-96.3) | 147/149 (98.7, 95.2-99.8) | 0.906 (0.834-0.978) |
| WT1 | 190/194 (97.9, 94.8-99.4) | 8/11 (72.7, 39.0-94.0) | 182/183 (99.5, 97.0-100) | 0.848 (0.679-1.017) |
CI, confidence interval.
Number concordant divided by the total number of pairs.
Number present by local determination divided by number present by central determination.
Number absent by local determination divided by number absent by central determination.
Controlling for site (OSU/Oregon).
Discrepancies between institutional and central laboratories
Twenty-four discrepancies were identified in 21 of 194 (10.8%) paired samples analyzed (Figure 1), with 3 patients harboring 2 discrepancies. With the exception of NPM1, discrepancies were identified for all genes, such as TP53 (7), TET2 (4), WT1 (4), DNMT3A (4), FLT3 (2), IDH2 (2), and IDH1 (1) (Table 2). Out of the 21 paired samples with discrepancies, 12 were BM, 8 were PB, and 1 was a PB/BM paired sample. Most discrepancies arose because of alternative approaches in variant reporting by the central laboratory including those of unknown significance (VUS), as requested by the medical monitors of Beat AML, although certain VUS were not captured or included on local institutional panels and reports. Another common cause for discrepancies was differences in the reportable range for low VAF variant calls.
Variances between institutional and central laboratories
| Gene . | Tissue type . | Mutation present or absent (VAF % ∗) . | VUS, SS, and other comments specific to the mutation identified . | ||
|---|---|---|---|---|---|
| Institution . | Central . | Institution . | Central . | ||
| FLT3 | |||||
| 1 | BM | BM | A | P (14.41) | FLT3 N676K, not ITD or TKD, not pathogenic and not in OSU panel |
| 2 | PB | PB | A | P (3.66) | FLT3 S543F, mutation not clearly pathogenic, not in OSU design |
| IDH1 | |||||
| 1 | PB | PB | A | P (49.71) | IDH1 F32V, VUS |
| IDH2 | |||||
| 1 | BM | BM | P (40.3) | A | IDH2 R172_H173delinsSA, complex indel |
| 2 | BM | BM | A | P (2.29) | IDH2 R140W, observed but below cutoff (2%) at OHSU laboratory |
| DMNT3A | |||||
| 1 | PB | PB | A | P (44.20) | DNMT3A SS 855+1G>A, SS calls are excluded in OSU panel |
| 2 | BM | BM | A | P (50.00) | DNMT3A G156E, VUS |
| 3 | PB | PB | A | P (44.26) | DNMT3A SS 2478+1G>A, SS calls are excluded in OSU panel |
| 4 | BM | BM | P (20.2) | A | DNMT3A R379C, VUS, low tumor purity at central laboratory |
| TET2 | |||||
| 1 | BM | BM | P (49.6) | A | TET2 S1039L, benign SNP |
| 2 | BM | BM | A | P (82.78) | TET2 SS 4044+1G>A, SS calls are excluded in OSU panel |
| 3 | BM | BM | P (47.0) | A | TET2 I1873T, reported as a somatic pathogenic variant |
| 4 | BM | BM | P (45.0, 49.0) | A | TET2 I1873T, reported as a somatic pathogenic variant (45%); R814C is likely germ line variant (49%) |
| WT1 | |||||
| 1 | BM | BM | A | P (4.20) | WT1 A382fs, VUS |
| 2 | PB | PB | A | P (53.56) | WT1 A5V, benign SNP |
| 3 | BM | BM | A | P (52.82) | WT1 G6OR, area not covered on OSU panel |
| 4 | BM | PB | P (3.00) | A | WT1 R462Q, tissue mismatch |
| TP53 | |||||
| 1 | BM | BM | A | P (10.37) | TP53 P153fs, likely artifact owing to large insertion |
| 2 | PB | PB | A | P (46.6) | TP53 R205Q, VUS |
| 3 | PB | PB | P (2.3) | A | TP53 C124R, below cutoff at central laboratory |
| 4 | PB | PB | A | P (3.44) | TP53 L194R, below cutoff at institution laboratory (OSU) |
| 5 | BM | BM | P (10.8) | A | TP53 G245S, suboptimal tumor purity and sample quality at central laboratory |
| 6 | PB | PB | A | P (69.55) | TP53 C215G, area not covered on OSU panel |
| 7 | BM | BM | A | P (49.85) | TP53 T125T, detected by local laboratory (OHSU) but filtered as a synonymous variant. |
| Gene . | Tissue type . | Mutation present or absent (VAF % ∗) . | VUS, SS, and other comments specific to the mutation identified . | ||
|---|---|---|---|---|---|
| Institution . | Central . | Institution . | Central . | ||
| FLT3 | |||||
| 1 | BM | BM | A | P (14.41) | FLT3 N676K, not ITD or TKD, not pathogenic and not in OSU panel |
| 2 | PB | PB | A | P (3.66) | FLT3 S543F, mutation not clearly pathogenic, not in OSU design |
| IDH1 | |||||
| 1 | PB | PB | A | P (49.71) | IDH1 F32V, VUS |
| IDH2 | |||||
| 1 | BM | BM | P (40.3) | A | IDH2 R172_H173delinsSA, complex indel |
| 2 | BM | BM | A | P (2.29) | IDH2 R140W, observed but below cutoff (2%) at OHSU laboratory |
| DMNT3A | |||||
| 1 | PB | PB | A | P (44.20) | DNMT3A SS 855+1G>A, SS calls are excluded in OSU panel |
| 2 | BM | BM | A | P (50.00) | DNMT3A G156E, VUS |
| 3 | PB | PB | A | P (44.26) | DNMT3A SS 2478+1G>A, SS calls are excluded in OSU panel |
| 4 | BM | BM | P (20.2) | A | DNMT3A R379C, VUS, low tumor purity at central laboratory |
| TET2 | |||||
| 1 | BM | BM | P (49.6) | A | TET2 S1039L, benign SNP |
| 2 | BM | BM | A | P (82.78) | TET2 SS 4044+1G>A, SS calls are excluded in OSU panel |
| 3 | BM | BM | P (47.0) | A | TET2 I1873T, reported as a somatic pathogenic variant |
| 4 | BM | BM | P (45.0, 49.0) | A | TET2 I1873T, reported as a somatic pathogenic variant (45%); R814C is likely germ line variant (49%) |
| WT1 | |||||
| 1 | BM | BM | A | P (4.20) | WT1 A382fs, VUS |
| 2 | PB | PB | A | P (53.56) | WT1 A5V, benign SNP |
| 3 | BM | BM | A | P (52.82) | WT1 G6OR, area not covered on OSU panel |
| 4 | BM | PB | P (3.00) | A | WT1 R462Q, tissue mismatch |
| TP53 | |||||
| 1 | BM | BM | A | P (10.37) | TP53 P153fs, likely artifact owing to large insertion |
| 2 | PB | PB | A | P (46.6) | TP53 R205Q, VUS |
| 3 | PB | PB | P (2.3) | A | TP53 C124R, below cutoff at central laboratory |
| 4 | PB | PB | A | P (3.44) | TP53 L194R, below cutoff at institution laboratory (OSU) |
| 5 | BM | BM | P (10.8) | A | TP53 G245S, suboptimal tumor purity and sample quality at central laboratory |
| 6 | PB | PB | A | P (69.55) | TP53 C215G, area not covered on OSU panel |
| 7 | BM | BM | A | P (49.85) | TP53 T125T, detected by local laboratory (OHSU) but filtered as a synonymous variant. |
A, absent; SS, splice site; P, present.
†VAF % cutoff value for OSU is 2.0 and for FMI is 2.0.
Assume absent of mutation is equivalent to 0.00 VAF % unless otherwise specified.
For the TP53 gene, which had 7 discrepancies, there were 1 VUS, 1 variant below reportable levels at the central site, and 1 variant below reportable levels at the institutional site. The remaining 4 discrepant cases were secondary to suboptimal sample quality, artifact presence, and unknown etiology (Table 2).
For the WT1 gene, which had 4 discrepant cases, 1 VUS and 1 variant was detected in a portion of the gene that was not covered at the institutional site (site 1). A third discrepant case had a mutation which was detected at the institutional laboratory (site 2) in a BM sample, but was not detected by the central laboratory in a concurrent PB sample. The fourth discrepant case was due to the institutional laboratory categorizing a variant as a benign single nuclear polymorphism (SNP) (Table 2).
The remaining 13 discrepancies were due to the reporting of VUS by the central laboratory and not by the institutional laboratories (Table 2).
Discussion
With the advent of multiple targeted therapies in the treatment of AML, incorporation of multigene genotyping by NGS is now integral to diagnostic assessment and to guide treatment decision-making in everyday clinical practice.1-5 The approval of targeted therapies is often accompanied by a Food and Drug Administration (FDA)–approved companion diagnostic assay, and there are a limited number of FDA–approved NGS myeloid panels that are commercially available. However, in real world clinical practice, NGS testing is done using myeloid mutational panels in Clinical Laboratory Improvement Amendments (CLIA)-approved laboratories that are easily accessible to practicing clinicians and can return results in a time-sensitive manner, usually at their own institutions or regional laboratories. Previous studies looking at discrepancies in NGS reporting from solid tumor patients have shown some variation in mutational reporting among different NGS platforms in similar samples.12,13
Because the interlaboratory concordance in NGS mutational reporting has not been well-studied in AML, we analyzed diagnostic BM and/or PB samples from newly diagnosed patients with AML aged ≥60 years enrolled on the BAMT.6,7 We then compared variant calls on these diagnostic samples made between the institutional sites and the central laboratory. We compared the overall percent agreement, sensitivity, and specificity using the central laboratory as the reference, and κ statistic when adjusted for site to determine the extent of agreement in mutational variant calls with the different NGS platforms used by these laboratories.
We identified a high level of overall percent agreement (>95%) in pathogenic myeloid mutations in all 8 genes studied (DNMT3A, FLT3, IDH1, IDH2, NPM1, TET2, TP53, and WT1) for the BAMT. These results are encouraging because these are actionable mutations that may affect treatment decision-making. In addition, the NGS platforms showed a high degree of sensitivity across all gene targets (median 95.1%; range, 72.7%-100%) in detecting pathogenic mutations and also high specificity in mutation detection (median 99.4%; range, 98.1%-100%). There was perfect agreement for NPM1 and almost perfect agreement in all other genes except for WT1, which had the lowest sensitivity (72.7%) but with only a few variants detected in this gene (n = 11). TP53 had the next lowest sensitivity (88.9%) among 45 positive cases reported by the central laboratory. In both cases, this is likely due to the range of differently reported variants and the size of the gene with larger genes having variable coverage on different panels.
The reasons and direction of the discrepancies depended largely on differences in both the preanalytic factors (assay design) and postanalytic factors, such as criteria for reportable variants between the central and institutional laboratories. An example of postanalytic factors include the 2 FLT3 mutations that were reported by the central laboratory but were not reported by the institutional laboratory because it was not part of the reporting algorithm of the institutional panel. The FLT3-ITD mutations were reported by a separate assay at the institution. Similarly, for IDH1, the pathogenic mutation reported by the central laboratory was a VUS and was not reported by the institutional laboratory. For IDH2, there were 2 discrepant cases: 1 was considered present at the institution with high VAF (40.3%), but considered absent at the central laboratory with low VAF; and the second case with lower VAF levels was identified at both the central and institutional laboratories, but with one above (2.29%) and the other below the 2% VAF sensitivity cutoff used by the laboratories. Consensus for the former discrepancy was that this variant was likely missed by the central laboratory because this mutation was a complex deletion, emphasizing that mutation type may contribute to heterogeneous results among NGS panels.
Preanalytical variables related to assay design were also a factor for discrepancies. For DMNT3A, 2 mutations were detected with high VAF by the central laboratory but not at the institution, owing to these being splice-site mutations that were not incorporated into the institutional panel. A third DMNT3A mutation was detected by the central laboratory with high VAF but determined to be a VUS by the institutional laboratory. A fourth DMNT3A mutation was detected by the institution but not by the central laboratory. After consensus, it was determined that the sample provided to the central laboratory had low tumor purity with the discrepancy being related more to suboptimal sample quality.
TET2 had 4 discrepancies found between central and institutional laboratories. Three of the 4 mutations detected by the institution were not detected by the central laboratory and were, thus, concluded to be likely benign germ line polymorphisms by the institutional laboratories; a finding unlikely to have clinical implications on treatment decision-making. The remaining mutation, detected by the central laboratory but not the institution, was a splice-site variant excluded by the institutional panel.
WT1 also had 4 discrepancies between central and institutional laboratories. One of the cases was likely due to a tissue mismatch (BM sample at the institution, PB at the central laboratory); whereas, another mutation found by the central laboratory was in an area not covered on the institutional panel. The remaining mutations called by the central laboratory but absent by the institutional laboratory were a VUS (n = 1) and SNP (n = 1).
Finally, TP53 had the highest number of discrepancies (n = 7). One discrepant case was due to detection of VUS, and the others were due to a multitude of other causes including presence of artifact, differences in sensitivity cutoff (2%), and genes or areas covered on the various panels.
In summary, discrepancies in mutations identified between central and institutional laboratories were most commonly due to SNPs, VUS, and poor sample quality. Both SNPs and VUSs are common and unlikely to affect clinical decision-making and related more to pre- and postanalytic factors. Discrepancies that were due to true analytic reasons, such as sample quality were few, which emphasizes the importance of sample extraction and preparation. Together, these results point to the lack of standardization, especially in the postanalytic steps, between different laboratories and platforms. Standardization of gene panels and basic identification of benign SNPs are a few of the more relatively achievable steps, but ultimately, additional data collection and analysis with larger datasets is needed to both identify and determine how additional variables can be standardized to provide consistent reporting of actionable variants. Standardization will become more important because clinical practice for the management and treatment of AML increasingly relies on these local assays to report therapeutically actionable variants. As more targeted therapies are developed for AML, a well-validated panel approach performed by local laboratories, as opposed to individual FDA–approved companion diagnostics, may be more optimal.
This study has several limitations. The first is that it only compared institution–specific NGS platforms with a centralized laboratory platform in patients with AML. We can neither extrapolate our results to other myeloid malignancies nor to other institutional or centralized laboratories because those sites likely have their own specific platform for detecting pathogenic mutations. Unfortunately, if we had chosen to include a higher number of institutions, it may have increased variability in this study given the increasing variability in institutional panels and preparation procedures. This showcases the need for implementation of standard and well-validated panels across institutions. Furthermore, our investigation addressed the overall percent agreement and discrepancies for 8 genes, whereas some panels and studies have compared panels with 40 to 200 genes.
Overall, this study illustrates the importance of quality control and standardization as NGS continues to be widely used for clinical decision-making in AML. Based upon our findings, it is likely that different institutional and central laboratories can use different NGS platforms for the same sample and get homogeneous results for pathogenic genes specific to AML. However, although good concordance has been recognized, inherent differences, such as platform type, methodology, and detection sensitivity cutoffs, remain. To minimize those inherent differences, it would be beneficial to have more standard panels across institutions in the future.
Acknowledgments
The authors acknowledge the patients enrolled on the Beat AML Master Trial who contributed their samples to this work. The work reported in this manuscript was funded by the Leukemia & Lymphoma Society.
This work was supported by National Cancer Institute P01 CA108671 11 (R.L.L.).
Authorship
Contribution: U.B. wrote the manuscript; A.S.R. performed the data analysis; F.Y., R.P., D.J., S.C., W.Z., J.-A.V., D.C.P., and L.J. performed experiments and performed the data analysis; B.N. and T.B. performed data analysis and assisted with the writing of the manuscript; U.B., A.B., E.M.S., P.P., M.R.B., W.S., G.S., W.B., T.K., M.L., J.F., N.A.H., L.R., S.M., A.Y., M.S., B.D., J.C.B., R.L.L., and A.M. assisted with the collection of patient samples; and all authors reviewed the manuscript.
Conflict-of-interest disclosure: U.B. has been a consultant for Genentech, Daiichi Sankyo, Takeda, Pfizer, AbbVie/Genentech and Novartis. A.S.R. served on an independent data monitoring committee for Telios Pharma, Inc, and is currently employed and holds stock with Eli Lilly and Company. E.M.S. has served on the advisory boards of Astellas Pharma, AbbVie, Genentech, Daiichi Sankyo, Novartis, Amgen, Seattle Genetics, Syros Pharmaceuticals, Syndax Pharmaceuticals, Agios Pharmaceuticals and Celgene; is an equity holder in Auron Therapeutics. G.S. has commercial interests in Bristol Myers Squibb, Amgen, J&J; has received fees from AbbVie, Agios, Amgen, Astellas, Bristol Myers Squibb, Incyte, Janssen, Jazz, Karyopharm, Kite, Pharmacyclics, Sanofi/Genzyme, Stemline; has received research funds from AbbVie, Actinium, Actuate, Arog, Astellas, AltruBio, AVM Bio, BMS/Celgene, Celator, Constellation, Daiichi-Sankyo, Deciphera, Delta-Fly, Forma, FujiFilm, Gamida, Genentech-Roche, GlycoMimetics, Geron, Incyte, Karyopharm, Kiadis, Kite/Gilead, Kura, Marker, Mateon, Onconova, Pfizer, PrECOG, Regimmune, Samus, Sangamo, Sellas, Stemline, Syros, Takeda, Tolero, Trovagene, Agios, Amgen, Jazz, ORCA, Ono-UK, Novartis. J.F. has received research funding from Takeda/Millennium. P.P. has served on the advisory board of Agios Pharmaceuticals and is currently an employee of Servier. J.-A.V. is an employee at Foundation Medicine Inc with equity in Roche. D.C.P. is an employee at Foundation Medicine Inc with equity in Roche. R.L.L. is on the supervisory board of Qiagen and is a scientific adviser to Imago, Mission Bio, Zentalis, Ajax, Auron, Prelude, C4 Therapeutics and Isoplexis; receives research support from and consulted for Celgene and Roche and has consulted for Incyte, Janssen, Astellas, Morphosys and Novartis; has received honoraria from Astra Zeneca, Roche, Lilly and Amgen for invited lectures and from Gilead for grant reviews. B.D. has potential competing interests in SAB: Adela Bio, Aileron Therapeutics, Therapy Architects/ALLCRON (inactive), Cepheid, Celgene, DNA SEQ, Nemucore Medical Innovations, Novartis, RUNX1 Research Program, Vivid Biosciences (inactive); SAB & Stock: Aptose Biosciences, Blueprint Medicines, Enliven Therapeutics, Iterion Therapeutics, GRAIL, Recludix Pharma; is a member of board of directors & stock for Amgen, Vincerx Pharma; board of directors: Burroughs Wellcome Fund, CureOne; joint steering committee: Beat AML LLS; advisory committee: Multicancer Early Detection Consortium; founder: VB Therapeutics; sponsored research agreement: Enliven Therapeutics, Recludix Pharma; clinical trial funding: Novartis, AstraZeneca; royalties from patent 6958335 (Novartis exclusive license) and OHSU and Dana-Farber Cancer Institute (1 Merck exclusive license, 1 CytoImage, Inc exclusive license, and 1 Sun Pharma Advanced Research Company nonexclusive license); US Patents 4326534, 6958335, 7416873, 7592142, 10473667, 10664967, 11049247. J.C.B. is a current equity holder in Vincerx Pharma Inc (a publicly traded company); holds membership on the board of directors or advisory committees of Vincerx, Newave, Orange Grove Bio; and holds consultancy, honoraria on Novartis, Trillium, Astellas, AstraZeneca, Pharmacyclics, and Syndax. A.M. has served on the advisory boards of Jazz Pharmaceuticals, AbbVie/Genentech, Astellas Pharma, PTC Therapeutics, Novartis, Agios Pharmaceuticals and Syndax Pharmaceuticals. The remaining authors declare no competing financial interests.
Correspondence: Uma Borate, Comprehensive Cancer Center, The Ohio State University, 1800 Cannon Dr, 1120G Lincoln Tower, Columbus, OH 43210; e-mail: Uma.Borate@osumc.edu.
References
Author notes
Data are available on request from the corresponding author, Uma Borate (uma.borate@osumc.edu).
The full-text version of this article contains a data supplement.