Key Points
Mutational loss of PIK3R1 induces resistance to NOTCH1 inhibition in T-ALL.
Pharmacological Notch inhibition synergizes with CDK4/6 inhibitors in T-ALL.
Abstract
Gain-of-function mutations in NOTCH1 are among the most frequent genetic alterations in T-cell acute lymphoblastic leukemia (T-ALL), highlighting the Notch signaling pathway as a promising therapeutic target for personalized medicine. Yet, a major limitation for long-term success of targeted therapy is relapse due to tumor heterogeneity or acquired resistance. Thus, we performed a genome-wide CRISPR-Cas9 screen to identify prospective resistance mechanisms to pharmacological NOTCH inhibitors and novel targeted combination therapies to efficiently combat T-ALL. Mutational loss of phosphoinositide-3-kinase regulatory subunit 1 (PIK3R1) causes resistance to Notch inhibition. PIK3R1 deficiency leads to increased PI3K/AKT signaling, which regulates cell cycle and the spliceosome machinery, both at the transcriptional and posttranslational level. Moreover, several therapeutic combinations have been identified, in which simultaneous targeting of the cyclin-dependent kinases 4 and 6 (CDK4/6) and NOTCH proved to be the most efficacious in T-ALL xenotransplantation models.
Introduction
T-cell acute lymphoblastic leukemia (T-ALL) is an aggressive hematological malignancy caused by genetic alterations during T-cell development. The 5-year overall survival rate in pediatric patients with T-ALL improved considerably over the past 30 years, whereas it stagnated in adults.1,2 The increased survival rate is largely attributed to improved risk-based stratification and administration of aggressive combination chemotherapies.3 However, classical chemotherapy treatment proves inadequate when treating relapsed and refractory T-ALL,4 requiring the development of novel therapeutic strategies.
Next-generation sequencing and related genomic diagnostics of patient with ALL samples have not only provided unprecedented insight into different T-ALL subgroups associated with different mutation and gene expression signatures but also identified actionable targets.5,6 Using this approach, oncogenic gain-of-function mutations in NOTCH1 have been identified in >55% of T-ALL cases.5-7 Identification of NOTCH1 as one of the most frequently mutated genes in T-ALL5-7 and the finding that NOTCH genes are also mutated in other cancers8 have boosted the development of a spectrum of therapeutics. These include antibodies neutralizing Notch receptors or ligands, or γ-secretase inhibitors (GSIs) preventing NOTCH activation.8 In this context, we identified and preclinically validated a novel orally active small molecule (CB-103) that efficiently blocks the Notch transcription activation complex without causing dose-limiting intestinal toxicities.9 CB-103 has successfully been evaluated in a recent phase 1/2 clinical trial (https://clinicaltrials.gov/ct2/show/NCT03422679), and complete response was observed in a patient with relapsed and refractory T-ALL.10
Although novel therapeutics blocking Notch signaling show promising outcomes, the use of monotherapies will likely result in relapse due to tumor heterogeneity and acquired resistance. Thus, a better understanding of resistance mechanisms to Notch inhibitors and development of novel combination therapies will facilitate effective treatment of patients with T-ALL.
Here, we screened human T-ALL cells using genome-wide CRISPR-Cas9. We identified the phosphoinositide-3-kinase regulatory subunit 1 (PIK3R1) as a key player in NOTCH treatment response. Mutational loss of PIK3R1 activity confers resistance to pharmacological Notch inhibition. Unbiased transcriptomic and proteomic analyses in PIK3R1-deficient T-ALL cells revealed PI3K-AKT–mediated upregulation of prosurvival and proliferation pathways along with alterations of the spliceosome machinery in response to Notch inhibition. Moreover, our screen led to the identification of pathways that can be pharmacologically targeted synergistically with Notch inhibitors, resulting in prolonged survival in a preclinical xenograft T-ALL model. Overall, our study identified novel resistance mechanisms to pharmacological Notch inhibition and combination strategies bearing the potential to treat patients with refractory or relapsed T-ALL more efficiently than current options.
Methods
CRISPR-Cas9 screen and analysis
Cas9-expressing DND-41 cells were transduced with human GeCKOv2 library (Addgene, 1000000048 and 1000000049) at a multiplicity of infection of ∼0.3 and selected with 1 μg/mL of puromycin for 6 days. Cells were cultured and treated with either dimethyl sulfoxide (DMSO), GSI (DAPT, 10 μM), or CB-103 (5 μM) for 3 weeks, in triplicates. Minimally, 3 × 107 cells per replicate were harvested at the starting and ending points for genomic DNA isolation. Single-guide RNA (sgRNA) sequences were amplified and sequenced using Illumina NextSeq 500. MAGeCK (version 0.5.9.2) was used for data analysis.
In vivo xenografts
All animal experiments were approved by cantonal veterinary service (VD3323 and VD3665) and performed in accordance with Swiss national guidelines. Female NSG mice were IV injected with 1 × 106 luciferase-expressing RPMI-8402 cells. Tumor growth was measured twice a week using in vivo imaging system, and upon tumor establishment, mice were randomized and treated with vehicle or drugs (refer to detailed information in supplemental Data) for 2 weeks. Tumor growth was monitored until the end point.
Proteomics assay
Peptides (250 μg) were labeled with tandem mass tags, pooled, and acidified. Phosphopeptides were further enriched using titanium dioxide and ferric nitrilotriacetate. Raw data were processed via Proteome Discoverer (version 2.4), using the Uniprot Human reference proteome.
Results
Genome-wide CRISPR screen identifies PIK3R1 associated with resistance to pharmacological Notch inhibition in T-ALL
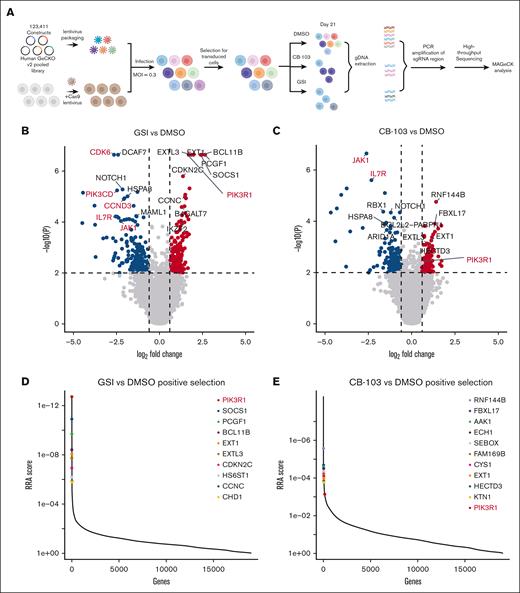
The efficacy of targeted therapies for the treatment of patients with cancer is often limited by the development of drug resistance.11 Potential resistance mechanisms to pharmacological Notch1 inhibition mediated by GSI or CB-103 in T-ALL are currently unclear. Thus, we performed a genome-wide loss-of-function (LoF) CRISPR/Cas9 screen12 to identify genes responsible for the resistance to Notch inhibition and novel combination therapies for the efficient treatment of human T-ALL. We used a Notch-dependent human T-ALL cell line, DND-41, which responds moderately to both GSI and CB-103 treatment in vitro.9 DND-41 cells stably expressing Cas9 were infected with human GeCKO version 2 CRISPR libraries, containing 123 411 sgRNAs targeting 19 050 genes and treated with either vehicle, GSI, or CB-103 for 21 days, enabling both positive and negative selection of sgRNAs (Figure 1A). Two different pharmacological Notch inhibitors were used with the aim of identifying common targets as the most likely candidates for causing resistance to or working in synergy with the Notch blockade.
Functional genome-wide CRISPR screen identifies PIK3R1 as being associated with resistance to Notch inhibition and druggable candidate pathways for combination therapies in T-ALL. (A) Schematic representation of the genome-wide LoF CRISPR screen using DND-41 T-ALL cells. (B) Volcano plots depicting genes targeted by sgRNAs that were negatively or positively selected, comparing GSI vs DMSO treatment. Red, adjusted P < .01 and log2 fold change (FC) > 0.6; blue, adjusted P < .01 and log2FC < −0.6. (C) Volcano plots showing genes targeted by sgRNAs that were negatively or positively selected, comparing CB-103 vs DMSO treatment. Red, adjusted P < .01 and log2FC > 0.6; blue, adjusted P < .01 and log2FC < −0.6. (D) Robust rank aggregation (RRA) plots displaying the top 10 enriched sgRNAs, comparing GSI vs DMSO treatment. (E) RRA plots displaying top enriched sgRNAs, comparing CB-103 vs DMSO treatment.
Functional genome-wide CRISPR screen identifies PIK3R1 as being associated with resistance to Notch inhibition and druggable candidate pathways for combination therapies in T-ALL. (A) Schematic representation of the genome-wide LoF CRISPR screen using DND-41 T-ALL cells. (B) Volcano plots depicting genes targeted by sgRNAs that were negatively or positively selected, comparing GSI vs DMSO treatment. Red, adjusted P < .01 and log2 fold change (FC) > 0.6; blue, adjusted P < .01 and log2FC < −0.6. (C) Volcano plots showing genes targeted by sgRNAs that were negatively or positively selected, comparing CB-103 vs DMSO treatment. Red, adjusted P < .01 and log2FC > 0.6; blue, adjusted P < .01 and log2FC < −0.6. (D) Robust rank aggregation (RRA) plots displaying the top 10 enriched sgRNAs, comparing GSI vs DMSO treatment. (E) RRA plots displaying top enriched sgRNAs, comparing CB-103 vs DMSO treatment.
sgRNAs targeting 197 or 154 genes were identified as significantly depleted (P < .01; log2–fold change < −0.6) in GSI- or CB-103–treated T-ALL cells, respectively, compared with those in the vehicle control (Figure 1B-C; supplemental Table 1). Negatively selected sgRNAs indicate genes that, when inhibited, might function synergistically with Notch inhibition to effectively eradicate T-ALL cells. Pathway enrichment analysis revealed that significantly depleted genes were associated with MYC and E2F signaling as well as G2M checkpoint and mTOR signaling pathways (supplemental Figure 1).
Conversely, sgRNAs targeting 194 or 139 genes were identified as significantly enriched (P < .01; log2 fold change > 0.6) in GSI- or CB-103–treated cells compared with those in the vehicle control, indicating that loss of these genes could confer resistance to Notch inhibition. Robust rank aggregation method was used to identify genes preferentially lost in response to Notch inhibition (Figure 1D-E; supplemental Table 1). Among these genes, PIK3R1 was identified at the top of the list in the GSI vs DMSO screen and was also identified as preferentially lost in the screen of CB-103– vs DMSO-treated T-ALL cells. The PIK3R1 gene encodes for the p85α regulatory subunit of PI3K, which contains an SH2 domain that binds to and inhibits the catalytic subunit (p110) of PI3Ks. Interestingly, PIK3R1 mutations were identified as drivers of tumorigenesis in ovarian,13 endometrial,14 and breast cancer.15PIK3R1 hotspot mutations in the SH2 domain were also recently reported in pediatric patients with T-ALL.5,6 In addition, we noticed that the positive regulatory subunit of PI3K (PIK3CD, leukocyte-restricted catalytic p110δ subunit) was depleted in Notch inhibitor–treated cells. Thus, these observations suggest that PI3K signaling plays a key role in acquired resistance to Notch1 inhibition.
Loss of PIK3R1 renders T-ALL cells resistant to pharmacological Notch inhibition
To validate the screening results, we generated multiple PIK3R1 knockout (KO) clones in 2 different NOTCH1-driven T-ALL cell lines (DND-41 and RPMI-8402) using the top 2 enriched sgRNAs from the screen and stable knockdown (KD) clones in the NOTCH3-driven cell line TALL-1 (supplemental Figure 2A-C), because TALL-1 cells were refractory to single-cell CRISPR targeting. Loss of PIK3R1 in several T-ALL cell lines led to no or only a mild growth advantage compared with nontargeting (NT) control sgRNA clones or scrambled (scr) short hairpin RNA controls. In contrast, cell growth of all GSI- or CB-103–treated PIK3R1 KO or KD clones was significantly enhanced compared with that of NT or scr controls (Figure 2A; supplemental Figure 2D). We observed a significant decrease in the percentage of cells in S phase in NT or scr T-ALL clones when treated with GSI, confirming that GSI induces cell cycle arrest in T-ALL cells.7 However, this effect was alleviated in all PIK3R1 KO and KD cell lines under the same treatment conditions (Figure 2B; supplemental Figure 2E). Interestingly, we also observed a cell cycle arrest in RPMI-8402 and TALL-1 control lines treated with CB-103, and the effect was significantly decreased when PIK3R1 was lost (Figure 2C; supplemental Figure 2F). Previously, we showed that CB-103 induces apoptosis in T-ALL cells.9 Consistently, CB-103 treatment for 3 days induced substantial apoptosis in all 3 control cell lines. However, the loss of PIK3R1 ablated this effect (Figure 2D; supplemental Figure 2G). Altogether, these results suggest that the loss of PIK3R1 confers resistance of T-ALL cells to Notch inhibition, protecting them from drug-induced apoptosis and cell cycle arrest.
Loss of PIK3R1 leads to resistance to Notch inhibition in T-ALL cells. (A) Cell proliferation assays of T-ALL PIK3R1 KO cell lines under DMSO, CB-103, or GSI treatment conditions. Gray connected dots, NT control; and colored dots, representative PIK3R1 KO cell lines. (B) Cell cycle analyses of PIK3R1 KO cell lines performed 6 days after DMSO or GSI treatment at indicated concentrations. (C) Cell cycle analyses of PIK3R1 KO cell lines 24 hours after DMSO or CB-103 treatment at indicated concentrations. (D) Apoptosis assays of PIK3R1 KO cell lines performed 3 days after DMSO or CB-103 treatment at indicated concentrations. Experiments shown here were performed with 2 independent T-ALL cell lines, DND-41 and RPMI-8402. The values shown are mean ± standard deviation (SD) (n = 3 biologically independent samples, 2 independent experiments). One-way analysis of variance (ANOVA); nonsignificant (ns); ∗P < .0332; ∗∗P < .0021; ∗∗∗P < .0002; ∗∗∗∗P < .0001.
Loss of PIK3R1 leads to resistance to Notch inhibition in T-ALL cells. (A) Cell proliferation assays of T-ALL PIK3R1 KO cell lines under DMSO, CB-103, or GSI treatment conditions. Gray connected dots, NT control; and colored dots, representative PIK3R1 KO cell lines. (B) Cell cycle analyses of PIK3R1 KO cell lines performed 6 days after DMSO or GSI treatment at indicated concentrations. (C) Cell cycle analyses of PIK3R1 KO cell lines 24 hours after DMSO or CB-103 treatment at indicated concentrations. (D) Apoptosis assays of PIK3R1 KO cell lines performed 3 days after DMSO or CB-103 treatment at indicated concentrations. Experiments shown here were performed with 2 independent T-ALL cell lines, DND-41 and RPMI-8402. The values shown are mean ± standard deviation (SD) (n = 3 biologically independent samples, 2 independent experiments). One-way analysis of variance (ANOVA); nonsignificant (ns); ∗P < .0332; ∗∗P < .0021; ∗∗∗P < .0002; ∗∗∗∗P < .0001.
PIK3R1 deficiency leads to elevated gene expression of proliferation and prosurvival pathways in response to Notch inhibition
To gain insight into how the loss of PIK3R1 confers resistance to pharmacological Notch inhibition in T-ALL cells, we performed gene expression analysis (supplemental Figure 3A). We chose to compare RPMI-8402 and corresponding PIK3R1 KO cell lines because they exhibit the largest difference in sensitivity profiles to Notch inhibitors. Treatment of RPMI-8402 cells for 24 hours with CB-103 resulted in significant downregulation of genes associated with hallmark pathways, including NOTCH signaling, MYC targeting, and E2F targeting (supplemental Figure 3B; supplemental Table 2), as previously reported,9 whereas GSI treatment resulted in significant downregulation of MYC targets and MTOR signaling (supplemental Figure 3B; supplemental Table 2). We did not observe significant enrichment of hallmark pathways when analyzing the gene expression differences between RPMI-8402 PIK3R1 KO and NT cells, albeit a moderate trend of increased expression of PI3K-AKT and KRAS hallmark pathway genes was observed (supplemental Figure 3C; supplemental Table 2). This might explain why only a mild growth advantage was observed under normal culture conditions after the loss of PIK3R1 in RPMI-8402 cells (Figure 2A). Interestingly, gene set enrichment analysis of CB-103–treated KO vs NT cells revealed enrichment of multiple hallmarks, including E2F and MYC targeting, PI3K-AKT-MTOR signaling, G2M checkpoint, and apoptosis pathways (Figure 3A; supplemental Table 2). Increased expression of MYC target genes was also observed in GSI-treated KO vs that in NT cells (supplemental Figure 3D; supplemental Table 2). Specifically, upregulation of key E2F family transcriptional activators, including E2F1, E2F2, and E2F3 and cell cycle regulators CCND2 and CCND3, and downregulation of the transcriptional repressor E2F5 were observed. In addition, we detected significant upregulation of antiapoptotic genes, such as BCL2 and BCL-xL (Figure 3B-C; supplemental Table 2). In contrast, typical Notch target genes including MYC, HES1, or DTX1 were similarly downregulated in CB-103– and GSI-treated PIK3R1 KO and NT cells (supplemental Figure 3E). These results are consistent with the increased proliferation and survival observed in drug-treated PIK3R1 KO vs NT cells (Figure 2B-D) and provide some mechanistic insight for Notch inhibitor resistance.
RNA-seq analysis of PIK3R1 KO cells reveals responses to Notch inhibition at transcriptional level. (A) Top significantly enriched hallmark pathways from a gene set enrichment analysis using differential gene expression results of PIK3R1 KO CB-103–treated RPMI-8402 cells compared with NT control CB-103–treated RPMI-8402 cells. (B) Heatmap showing unbiased clustering of changes in gene expression level, comparing PIK3R1 KO CB-103–treated vs NT CB-103–treated RPMI-8402 cells, highlighting key genes involved in E2F targets, PI3K-AKT-mTOR signaling, and apoptosis pathway. (C) Expression of a subset of differentially expressed genes measured as TPM comparing PIK3R1 KO CB-103–treated vs NT CB-103–treated RPMI-8402 cells. Values shown are mean ± SD. One-way ANOVA test; ∗P < .0332; ∗∗P < .0021; ∗∗∗P < .0002. TPM, transcripts per million.
RNA-seq analysis of PIK3R1 KO cells reveals responses to Notch inhibition at transcriptional level. (A) Top significantly enriched hallmark pathways from a gene set enrichment analysis using differential gene expression results of PIK3R1 KO CB-103–treated RPMI-8402 cells compared with NT control CB-103–treated RPMI-8402 cells. (B) Heatmap showing unbiased clustering of changes in gene expression level, comparing PIK3R1 KO CB-103–treated vs NT CB-103–treated RPMI-8402 cells, highlighting key genes involved in E2F targets, PI3K-AKT-mTOR signaling, and apoptosis pathway. (C) Expression of a subset of differentially expressed genes measured as TPM comparing PIK3R1 KO CB-103–treated vs NT CB-103–treated RPMI-8402 cells. Values shown are mean ± SD. One-way ANOVA test; ∗P < .0332; ∗∗P < .0021; ∗∗∗P < .0002. TPM, transcripts per million.
Notch-inhibited PIK3R1-mutant T-ALL cells reveal major phosphorylation changes in the cell cycle and spliceosome machinery
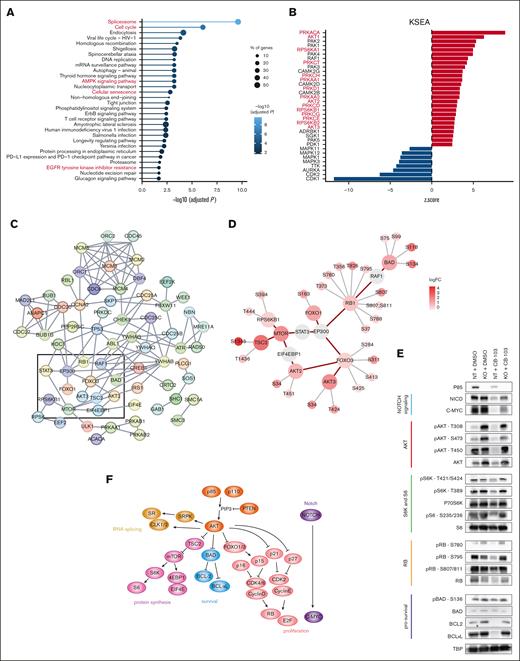
The p85 protein, which is encoded by PIK3R1, is an essential component of a pivotal kinase signaling complex. Its loss may lead to immediate altered signaling events. Therefore, we performed total- and phospho-proteome analysis of RPMI-8402 NT and PIK3R1 KO cells treated with DMSO or CB-103 (supplemental Figure 4A-B). Across samples, we quantified 29 904 peptides corresponding to 7886 protein groups and 25 221 phosphopeptides, of which 21 601 were categorized as class I phosphosites16 originating from 5531 phosphoproteins (supplemental Figure 4C; supplemental Table 3). At the total protein level, we observed 54 (NT CB-103–treated vs vehicle-treated), 215 (PIK3R1 KO vs NT), and 206 (PIK3R1 KO CB-103–treated vs NT CB-103-treated) significant changes (supplemental Figure 4D; supplemental Table 3). The comparisons at the phosphorylation level revealed 2983 (NT CB-103–treated vs vehicle-treated), 2636 (PIK3R1 KO vs NT), and 3731 (PIK3R1 KO CB-103–treated vs NT CB-103–treated) significant changes (supplemental Figure 4E; supplemental Table 3). Thus, changes occurring at the level of phosphorylation profiles are much more pronounced compared with changes of the total proteome. KEGG analysis of total protein changes of CB-103–treated PIK3R1 KO vs NT cells identified cell cycle regulation as the most significantly affected pathway (supplemental Figure 4F; supplemental Table 3), which corroborated observations from the RNA sequencing (RNA-seq) data. Similar analysis at the phosphoproteome level pointed to the alterations in the cell cycle and spliceosome as the most significant ones (Figure 4A; supplemental Table 3).
Phosphoproteomics analysis of PIK3R1 KO cells reveals signaling responses to Notch inhibition. (A) Significantly enriched KEGG pathways of proteins with altered phosphorylation sites, comparing PIK3R1 KO CB-103–treated vs NT control CB-103–treated RPMI-8402 cells. Top 30 pathways are shown, solid line color scale indicates adjusted P value, dot size of leading edge displays percentage of genes enriched in corresponding pathways. (B) Kinase substrate–enrichment analysis (KSEA) of phosphorylation profiles, comparing PIK3R1 KO CB-103–treated vs NT CB-103–treated RPMI-8402 cells. Red, kinases with positive z score; blue, kinases with negative z score. (C) Interactions among phosphoproteins within 4 of the top enriched KEGG pathways in panel A: assessing cell cycle, AMPK signaling, cellular senescence, and EGFR tyrosine kinase inhibitor resistance pathways. Line color indicates the strength of interaction (“Confidence” from the STRING database). Key nodes are gated with black rectangle. (D) Detailed plots of key phosphoproteins with annotated phosphosites and corresponding FCs from rectangle area in panel C. Red circle indicates phosphoproteins with phosphorylation changes as log2FC > 1; false discovery rate < 0.05. Gray circle indicates phosphoproteins with phosphorylation sites omitted. Red connecting line, protein interaction from STRING database. Gray radiating line, detailed phosphorylation sites associated with phosphoproteins. (E) Total protein and phosphorylation level of indicated phosphosites by western blotting for key proteins involved in the indicated nodes or pathways: Notch signaling (light blue); AKT (red); S6K and S6 (green); RB (orange); prosurvival signaling (dark blue) in RPMI-8402 NT and PIK3R1 KO cells in response to CB-103. TBP was used as loading control. (F) Model summarizing the key nodes of Notch signaling–inhibition resistance mechanism caused by the loss of PIK3R1. TBP, TATA box-binding protein.
Phosphoproteomics analysis of PIK3R1 KO cells reveals signaling responses to Notch inhibition. (A) Significantly enriched KEGG pathways of proteins with altered phosphorylation sites, comparing PIK3R1 KO CB-103–treated vs NT control CB-103–treated RPMI-8402 cells. Top 30 pathways are shown, solid line color scale indicates adjusted P value, dot size of leading edge displays percentage of genes enriched in corresponding pathways. (B) Kinase substrate–enrichment analysis (KSEA) of phosphorylation profiles, comparing PIK3R1 KO CB-103–treated vs NT CB-103–treated RPMI-8402 cells. Red, kinases with positive z score; blue, kinases with negative z score. (C) Interactions among phosphoproteins within 4 of the top enriched KEGG pathways in panel A: assessing cell cycle, AMPK signaling, cellular senescence, and EGFR tyrosine kinase inhibitor resistance pathways. Line color indicates the strength of interaction (“Confidence” from the STRING database). Key nodes are gated with black rectangle. (D) Detailed plots of key phosphoproteins with annotated phosphosites and corresponding FCs from rectangle area in panel C. Red circle indicates phosphoproteins with phosphorylation changes as log2FC > 1; false discovery rate < 0.05. Gray circle indicates phosphoproteins with phosphorylation sites omitted. Red connecting line, protein interaction from STRING database. Gray radiating line, detailed phosphorylation sites associated with phosphoproteins. (E) Total protein and phosphorylation level of indicated phosphosites by western blotting for key proteins involved in the indicated nodes or pathways: Notch signaling (light blue); AKT (red); S6K and S6 (green); RB (orange); prosurvival signaling (dark blue) in RPMI-8402 NT and PIK3R1 KO cells in response to CB-103. TBP was used as loading control. (F) Model summarizing the key nodes of Notch signaling–inhibition resistance mechanism caused by the loss of PIK3R1. TBP, TATA box-binding protein.
To dissect kinase regulation in more detail, we performed kinase substrate–enrichment analysis on the differential phosphorylation profiles of our comparison groups (Figure 4B; supplemental Figure 5A; supplemental Table 3). The analysis of CB-103 vs vehicle revealed that CB-103 treatment led to decreased AKT1, MTOR, and S6K signaling, whereas the PIK3R1 vs NT comparison showed the expected reciprocal outcome, with increased PKC family and AKT signaling due to the loss of p85, (supplemental Figure 5A; supplemental Table 3). Importantly, comparison of PIK3R1 KO CB-103 treatment vs NT CB-103 treatment showed increased activating phosphorylation events for AKT 1/2/3, PKC family, and S6K, which were maintained and no longer downregulated after CB-103 treatment (Figure 4B; supplemental Table 3). This is in agreement with a recent proteomics study linking members of the PKC family and AKT signaling to GSI resistance in DND-41 cells.17
Subsequently, we examined interactions among key proteins (Figure 4A) using experimentally validated knowledge from the STRING database (Figure 4C) and highlighted phosphorylation changes in these proteins (Figure 4D). This detailed phospho-mapping provides insights regarding functionally established phosphorylation events, such as S780 for resistant cells (RBs), as well as less-examined events, including T451 on AKT2, which has previously been associated with oncogenic signaling (Figure 4D). Immunoblotting validated key phosphorylation events for AKTs, S6K, RB1, and BAD, which are important regulators of proliferation and cell survival (Figure 4E). CB-103 treatment resulted in marked downregulation of NICD and C-MYC9 along with reduced total AKT levels and more pronounced reduced phosphorylation at residues T308, S473, and T450. Yet, these effects were largely ablated in p85-deficient cells (Figure 4E, RPMI-8402 cells; supplemental Figure 5B, DND-41 cells). Similarly, the phosphorylation of ribosome protein S6 kinase (p-S6K, T389, and T421/S424) and its downstream substrate S6 (p-S6 and S235/236) were downregulated by CB-103 treatment but not in p85-deficient cells. Thus, under CB-103 treatment, the loss of PIK3R1 indeed helps maintain proteins involved in protein translation. In addition, all phosphorylation sites of RB tested (S780, S795, and S807/811) were downregulated in CB-103-sensitive cells compared with that in the RBs. The same holds true for BCL2 and BCL-xL, whereas BAD and p-BAD levels (prosurvival) remained comparable. These results indicate that p85-deficient RPMI-8402 cells are able to cope with Notch inhibition through increased AKT signaling and sustained protein translation, cell proliferation, and cell survival.
Interestingly, LoF PIK3R1 led to prominent phosphorylation changes in proteins involved in the spliceosome and RNA processing in cells treated with pharmacological Notch inhibitors (Figure 4A; supplemental Figure 6A-B; supplemental Table 3). This analysis allowed to establish changes in phosphorylation profiles of splicing factors upon altered PI3K signaling and highlighted a wide spectrum of, so far uncharacterized, phosphorylation sites. A recent report linked oncogenic PI3K signaling with splicing alterations in breast cancer at the transcriptional level.18 Thus, we reanalyzed our RNA-seq data for differentially expressed transcripts, which were indeed associated with genes involved in cell cycle and regulation of apoptosis signaling pathways (supplemental Figure 6C-D; supplemental Table 2).
Our results show that the loss of PIK3R1 in T-ALL cells led to increased PI3K-AKT signaling, causing major phosphorylation changes in the cell cycle and spliceosome machinery, changes that resulted in downstream activation of cell cycle progression, increased cell proliferation, E2F gene activation, increased protein synthesis, and cell survival. Changes in the spliceosome at phosphorylation levels also correlated with differential splicing at the transcriptional level. Consequently, these mechanisms contribute to resistance to Notch inhibition in T-ALL (Figure 4F).
Pharmacological Notch inhibitors synergize with targeted therapies in human T-ALL cells
The advantage of using a CRISPR/Cas9 screen in T-ALL cells under drug selection is that it enables the identification of not only candidate genes that mediate drug resistance, such as PIK3R1, but also genes and pathways that are crucial for cell survival under drug selection. This opens avenues to identify novel combination therapies. Preferentially depleted sgRNAs in GSI- and CB-103–treated T-ALL cells pointed to well-established signaling components within T-ALL, including components of the interleukin-7 (IL-7)/JAK pathway (IL-7 receptor [IL-7R] and JAK1), regulators of the cell cycle machinery (CDK6:CCND3), and the key gene encoding the PI3K catalytic subunit (PIK3CD) (Figure 1B-C).
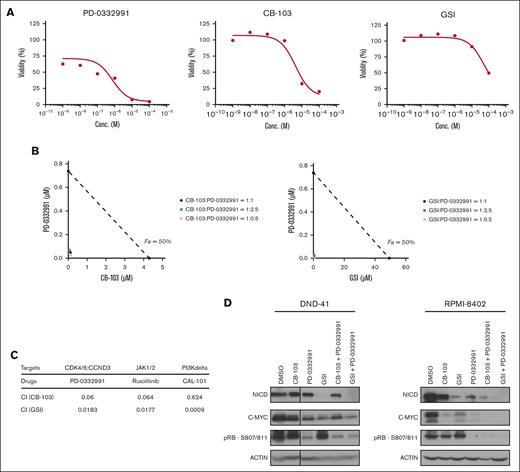
We validated these candidates using available US Food and Drug Administration (FDA)–approved inhibitors against cyclin-dependent kinases 4 and 6 (CDK4/6) (PD-0332991), JAK1/2 (Ruxolitinib), and PIK3δ (CAL-101). Firstly, we established in vitro sensitivity profiles and observed that the single-agent 50% inhibitory concentration of CB-103 for DND-41 cells was 4.3 and 0.1 μM for PD-0332991 (Figure 5A). We, then, tested a serial dilution of combination treatment administrating CB-103 and PD-0332991 starting at 3 ratios of their corresponding 50% inhibitory concentration (1:1, 1:2.5, and 1:0.5) and established isobologram curves (Figure 5B). The combination index19 was 0.06 for CB-103 plus PD-0332991, and similarly, 0.0183 for GSI plus PD-0332991, both of which were <0.1, indicating very strong synergism (Figure 5C). In addition, combination treatment induced the downregulation of C-MYC, which is downstream of Notch and p-RB, a key cell cycle regulator in 2 independent T-ALL cell lines (RPMI-8402 and DND41; Figure 5D). Similarly, we observed a very strong synergism combining Notch inhibitors with a JAK1/2 inhibitor or a PI3Kδ inhibitor (Figure 5C). These findings suggest that Notch inhibition in combination with FDA-approved compounds targeting CDK4/6, IL-7R signaling, or the PI3K/AKT pathway should be more efficacious compared with single-agent treatment.
In vitro synergy between Notch inhibitors and multiple targeted therapies identified from the CRISPR screen. (A) Cell viability assay of T-ALL cells in response to PD-0332991, CB-103, or GSI. (B) Isobologram plots of T-ALL cells in response to a serial dilution of combination treatment of PD-0332991 with CB-103 (left) or GSI (right), starting at 3 ratios of their corresponding 50% inhibitory concentration. Purple, ratio (Notch inhibitor: PD-0332991) = 1:1; green, ratio = 1:2.5; gray, ratio = 1:0.5. x-axis shows the concentration of CB-103. The values shown are mean ± SD (n = 4 biologically independent samples, 2 independent experiments performed). (C) Table summarizing combination index (CI) of Notch inhibitors together with PD-0332991, Ruxolitinib, or CAL-101. (D) Total protein levels of NICD and c-MYC as well as phosphorylation level of p-RB in DND-41 cells (left) and RPMI-8402 cells (right). Cells were treated with DMSO or corresponding single drugs or drug combinations for 24 hours. Actin was used as the loading control.
In vitro synergy between Notch inhibitors and multiple targeted therapies identified from the CRISPR screen. (A) Cell viability assay of T-ALL cells in response to PD-0332991, CB-103, or GSI. (B) Isobologram plots of T-ALL cells in response to a serial dilution of combination treatment of PD-0332991 with CB-103 (left) or GSI (right), starting at 3 ratios of their corresponding 50% inhibitory concentration. Purple, ratio (Notch inhibitor: PD-0332991) = 1:1; green, ratio = 1:2.5; gray, ratio = 1:0.5. x-axis shows the concentration of CB-103. The values shown are mean ± SD (n = 4 biologically independent samples, 2 independent experiments performed). (C) Table summarizing combination index (CI) of Notch inhibitors together with PD-0332991, Ruxolitinib, or CAL-101. (D) Total protein levels of NICD and c-MYC as well as phosphorylation level of p-RB in DND-41 cells (left) and RPMI-8402 cells (right). Cells were treated with DMSO or corresponding single drugs or drug combinations for 24 hours. Actin was used as the loading control.
These promising in vitro results prompted us to assess their efficacy in xenotransplantation assays. RPMI-8402 T-ALL cells expressing a luciferase reporter were transplanted into NSG mice to monitor tumor growth and progression of disease over time. Animals with established tumor results were treated with either single-agent compounds (vehicle, CB-103, GSI, or PD-0332991) or combination therapy (CB-103 or GSI plus PD-0332991) for 2 weeks (Figure 6A). The kinetics of tumor progression showed a moderate and statistically significant reduction in tumor burden for both single-agent treatments of CB-103 or GSI compared with that for the vehicle (Figure 6B-C). Single-agent treatment of PD-0332991 revealed a robust reduction in tumor burden. However, the strongest reduction in tumor burden was observed when mice were treated with combination of PD-0332991 and either CB-103 or GSI (Figure 6C). To test whether combination treatment led to an increase in the overall survival of experimental animals, treatment was ceased after 2 weeks, and tumor relapse and survival rates were monitored. Despite the short treatment window, the dual-agent treatment of GSI plus PD-033291 translated into significant prolonged overall survival compared with other treatment regiments (Figure 6D).
Combination of Notch inhibitors and multiple targeted therapies leads to decreased tumor burden and prolonged survival in human T-ALL cell line xenograft model. (A) Schematic representation of human T-ALL cell line xenograft model and drug treatment study. (B) Representative bioluminescence imaging at days indicated after treatment of each group. (C) Quantification of tumor burden measured by bioluminescent signals at days indicated after treatment of each group, testing Notch inhibitors alone or in combination with PD-0332991. Y-axis shows log10FC of signals on day 11 or day 18 after treatment comparing with initiation of treatment. Data are shown in box and whisker plots showing all data points. One-way ANOVA was performed. (D) Kaplan-Meier survival analysis of NSG mice within each treatment group testing Notch inhibitors, PD-0332991, or in combination. (E) Kaplan-Meier survival analysis of NSG mice within each treatment group, testing CB-103, MK-2206, or in combination. (F) Kaplan-Meier survival analysis of NSG mice within each treatment group, testing CB-103, venetoclax, or in combination. Experiments shown here were performed with the T-ALL cell line RPMI-8402. Log-rank (Mantel-Cox) test; P value as indicated, ∗P < .0332; ∗∗P < .0021; ∗∗∗P < .0002; ∗∗∗∗P < .0001.
Combination of Notch inhibitors and multiple targeted therapies leads to decreased tumor burden and prolonged survival in human T-ALL cell line xenograft model. (A) Schematic representation of human T-ALL cell line xenograft model and drug treatment study. (B) Representative bioluminescence imaging at days indicated after treatment of each group. (C) Quantification of tumor burden measured by bioluminescent signals at days indicated after treatment of each group, testing Notch inhibitors alone or in combination with PD-0332991. Y-axis shows log10FC of signals on day 11 or day 18 after treatment comparing with initiation of treatment. Data are shown in box and whisker plots showing all data points. One-way ANOVA was performed. (D) Kaplan-Meier survival analysis of NSG mice within each treatment group testing Notch inhibitors, PD-0332991, or in combination. (E) Kaplan-Meier survival analysis of NSG mice within each treatment group, testing CB-103, MK-2206, or in combination. (F) Kaplan-Meier survival analysis of NSG mice within each treatment group, testing CB-103, venetoclax, or in combination. Experiments shown here were performed with the T-ALL cell line RPMI-8402. Log-rank (Mantel-Cox) test; P value as indicated, ∗P < .0332; ∗∗P < .0021; ∗∗∗P < .0002; ∗∗∗∗P < .0001.
The PI3K-AKT axis was identified as a main switch of downstream signaling events responsible for resistance to Notch inhibition in the CRISPR/Cas9 screen, RNA-seq, and proteomics data. Therefore, we also tested dual treatment of the AKT inhibitor (MK-2206) combined with CB-103 and observed significant prolongation of overall survival with combination compared with single-agent therapy (Figure 6E).
In light of increased BCL2 expression in our RNA-seq data and a recent report on complete clinical response of a patient with relapse refractory T-ALL treated with CB-103 in combination with other drugs including venetoclax,10 we proceeded to assess the efficacy of combining CB-103 and venetoclax in our model. Indeed, this combination treatment significantly extended overall survival compared with single-agent treatment in a comparable range as with CB-103 plus MK-2206 (Figure 6F). Overall, the CRISPR/Cas9 screen in T-ALL cells unveiled potentially novel avenues of combination therapies.
Discussion
Next-generation sequencing analyses of primary T-ALL samples and cell lines has identified NOTCH1 as being among the most frequently mutated genes throughout different T-ALL subgroups.5,6 This, together with the identification of gain-of-function mutations in other tumor entities,8 highlights the Notch pathway as a therapeutic target for precision medicine. However, a major issue with personalized medicine is the establishment of resistance causing relapse. Thus, we performed an unbiased genome-wide LoF CRISPR screen in a NOTCH1-driven T-ALL cell line (DND-41). The moderate response of this cell line to 2 pharmacological Notch inhibitors presents an opportunity to identify candidate genes that cover both aspects of the response, including resistance to and synergism with Notch inhibition. We identified and validated that loss or downregulation of PIK3R1 in several human T-ALL cell lines is responsible for resistance to both GSI- and CB-103–mediated Notch inhibition, implicating a generic resistance mechanism. Aberrant activation of the PI3K pathway has been demonstrated to contribute to various cancer types including T-ALL.20-23 Of patients with T-ALL, between 23% and 27% harbor mutations in PI3K pathway genes,5,24,25 raising the question whether all patients with T-ALL with activating NOTCH1 mutations and aberrations within the PIK3 signaling cascade might be resistant to Notch inhibitors. A previous report linked PTEN, which negatively regulates PI3K/AKT signaling, in human T-ALL cell lines to GSI resistance.26 However, this conclusion is not consistent with observations that GSI sensitivity was comparable in Notch1-driven T-ALL cells obtained from wild-type and Pten-deficient mice.27 Similarly, multiple human T-ALL cell lines carrying mutant PTEN alleles are sensitive to GSI.25 Thus, PTEN deficiency may not be linked to resistance to pharmacological Notch inhibition a priori but might be dependent on the time point amid the T-ALL transformation process. Hence, the loss of PIK3R1 or PTEN during drug-mediated selection in fully established T-ALL may lead to rapid and high activation of the AKT pathway, resulting in continuous proliferation, survival and, thus, resistance to pharmacological Notch inhibitors, as observed and validated in our genetic CRISPR-based screen. The hypothesis is supported by findings using a mouse model of NOTCH1-induced T-ALL with subsequent loss of the Pten gene once T-ALL has been established. In this model loss of Pten indeed resulted in the development of GSI resistance, unlike the Pten KO models, in which Pten was already lost at the onset of Notch-mediated disease.27,28 Therefore, the prediction would be that patients with T-ALL with LoF mutations of PIK3R1 or PTEN or activating mutations in PI3K catalytic subunits at disease onset still respond to Notch inhibition. Nonetheless, individuals that acquire such mutations during treatment or at late-stage disease are more likely to be resistant to pharmacological Notch inhibitors because of elevated activation of AKT signaling.
Pharmacological inhibition of Notch signaling in Notch-driven T-ALL results in cell cycle arrest and apoptosis, mostly through downregulation of C-MYC expression.9,29 Interestingly, although MYC transcript levels are downregulated similarly in CB-103–treated PIK3R1 KO and NT control cells (supplemental Figure 3E), MYC protein levels of CB-103–treated PIK3R1 KO cells were higher than that in NT control cells but lower than that in vehicle-treated cells (Figure 4E), suggesting that increased AKT signaling maintains C-MYC protein levels, at least in part, through posttranscriptional mechanisms, as previously reported for other cancers.30,31
To the best of our knowledge, our study provides for the first time, a comprehensive analysis of disrupted phosphorylation modification on splicing factors (supplemental Figure 6A-B) upon altered PI3K signaling in response to Notch inhibition. This was correlated with differential expression levels of transcript isoforms of genes enriched in cell cycle and antiapoptotic pathways as well as in regulation of RNA splicing (supplemental Figure 6C-D). Recently, oncogenic PI3K signaling was shown to induce expression of alternatively spliced isoforms linked to proliferation and metabolism in breast cancer.18 The PI3K-AKT pathway has been shown to regulate several proteins of the splicing machinery.32 Interestingly, genetic alterations in RNA processing factors were identified in 11% of pediatric T-ALL cases.6 Inhibitors against SF3B1, a key U2 spliceosome component, which is also differentially phosphorylated (supplemental Figure 6A-B), have been shown to inhibit growth of T-ALL and other leukemias.33 Whether treating PIK3R1 KO T-ALL cells with a SF3B1-inhibitor would resensitize them to pharmacological Notch inhibitors or simply kill T-ALL cells remains to be addressed. Also, the question remains how the altered phosphorylation profile of splicing factors could cause transcript isoform alterations and specifically contribute to the resistance phenotype. One possible explanation is that the subcellular localization and activity of splicing factors might be phosphorylation dependent and contribute to the expression of particular splice variants involved in key oncogenic pathways. Whether alternative splicing profiles can predict response to Notch inhibition in T-ALL and other cancer contexts requires future exploration.
The complementary part of the study was to identify potential candidate genes or pathways for combination therapies with pharmacological Notch inhibitors. Our CRISPR screen led to the identification of PIK3CD, IL-7R/JAK1, and CDK6:CCND3 as potential targets (Figure 1B-C). It is interesting that sgRNAs against NOTCH1 score as negatively depleted, pointing to possibly incomplete pharmacological inhibition of the pathway. Unfavored loss of PI3KCD (encoding catalytic PI3K subunit P110δ) was notable because its activity is modulated by the negative regulatory subunit p85α, which we identified as a resistance-associated protein to Notch inhibition. Furthermore, 9% of patients with T-ALL34 harbor activating IL-7R mutations, which causes constitutive JAK1 signaling in T-ALL.35 Interestingly, both IL-7R and the downstream mediator JAK1 were identified to be preferentially lost in the presence of Notch inhibition. A previous report linked the synergistic action of GSI with JAK1 inhibitors in vitro in T-ALL,36 further corroborating our potential targets. We also focused on the CDK6::cyclinD3 complex, as it is reportedly essential for the initiation and maintenance of T-ALL37-39 and may, thus, be justified as a promising candidate to be targeted together with chemotherapy in T-ALL.40
We explored a panel of FDA-approved inhibitors and tested them first for their ability to function synergistically with CB-103 or GSI in vitro. Interestingly, PD-0332991 (inhibiting CDK4/6), ruxolitinib (inhibiting JAK1/2), and CAL-101 (inhibiting PIK3CD) appear to function synergistically with both CB-103 or GSI (Figure 5C). We further assessed both CB-103 and GSI in combination with PD-0332991, MK-2206, and venetoclax in xenotransplantation assays. All different combination treatments showed significant prolonged survival when compared with single-agent treatment. Although, all our results were obtained with established patient-derived Notch-driven T-ALL cell lines, which might be considered as a limitation of the study, the best combination in terms of overall survival was obtained by simultaneous inhibition of CDK4/6 and Notch signaling, indicating that such combination therapies might be worthwhile to be considered in future clinical combination trials.
Acknowledgments
The authors acknowledge Ute Koch for critical reading, reviewing and editing of the manuscript, the staff from the Flow Cytometry Core Facility, Gene Expression Core Facility, and Center of Phenogenomics at École Polytechnique Fédérale de Lausanne for excellent technical support. The authors thank Yueyun Zhang for her technical help. The visual abstract was created with BioRender.com.
This work is supported by SNSF 310030_188505 and 31003A_165966 (F.R.).
Authorship
Contribution: L.C. was responsible for designing and performing experiments, analyzing data, interpreting results, writing original draft, and reviewing and editing the draft manuscript; G.A.R.B., N.F., and Y.L. were responsible for analyzing bioinformatics data and reviewing and editing the draft manuscript; F.A., R.H., and M.P. were responsible for analyzing proteomics data and reviewing and editing the draft manuscript; and F.R. was responsible for supervision, project administration, funding acquisition, writing original draft, and reviewing and editing the draft manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Freddy Radtke, École Polytechnique Fédérale de Lausanne, Swiss Institute for Experimental Cancer Research, Station 19, CH-1015 Lausanne, Switzerland; e-mail: freddy.radtke@epfl.ch.
References
Author notes
CRISPR screen data and RNA-seq data were deposited to Gene Expression Omnibus database (accession numbers GSE221576 and GSE221577, respectively).
Proteomics data were deposited to the ProteomeXchange Consortium via the PRIDE partner repository (accession number PXD038908).
Data are available on request from the corresponding author, Freddy Radtke (freddy.radtke@epfl.ch).
The full-text version of this article contains a data supplement.