Key Points
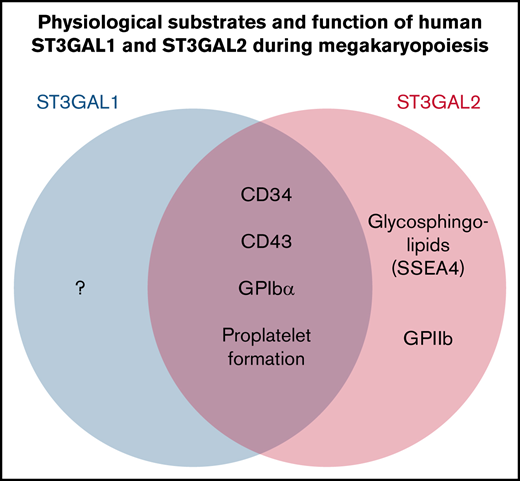
ST3GAL1 and ST3GAL2 have both overlapping and unique substrate specificities in O-glycan sialylation during megakaryopoiesis.
O-glycan sialylation is dispensable for MK production but indispensable for MK proplatelet formation.
Abstract
Although the sialyltransferases ST3GAL1 and ST3GAL2 are known to transfer sialic acid to the galactose residue of type III disaccharides (Galβ1,3GalNAc) in vitro, sialylation of O-linked glycosylated proteins in living cells has been largely attributed to ST3GAL1. To examine the role of ST3GAL2 in O-sialylation, we examined its expression during differentiation of human-induced pluripotent stem cells (iPSCs) into hematopoietic progenitor cells (HPCs) and megakaryocytes (MKs). ST3GAL1 and ST3GAL2 each became highly expressed during the differentiation of iPSCs to HPCs but decreased markedly in their expression upon differentiation into MKs, suggesting coordination of expression during megakaryopoiesis. To further delineate their role in these processes, we generated ST3GAL1-, ST3GAL2-, and doubly deficient human iPSC lines. Binding of the peanut agglutinin lectin, which reports the presence of unsialylated Galβ1,3GalNAc glycan chains, was strongly increased in HPCs and MKs derived from double-knockout iPSCs and remained moderately increased in cells lacking either one of these sialyltransferases, demonstrating that both can serve as functional cellular O-glycan sialyltransferases. Interestingly, the HPC markers CD34 and CD43, as well as MK membrane glycoprotein (GP) GPIbα, were identified as major GP substrates for ST3GAL1 and ST3GAL2. In contrast, O-sialylation of GPIIb relied predominantly on the expression of ST3GAL2. Finally, although disruption of ST3GAL1 and ST3GAL2 had little impact on MK production, their absence resulted in dramatically impaired MK proplatelet formation. Taken together, these data establish heretofore unknown physiological roles for ST3GAL1 and ST3GAL2 in O-linked glycan sialylation in hemato- and megakaryocytopoiesis.
Introduction
Mucin type O-glycosylation is a ubiquitous posttranslational modification, with an estimated 80% of proteins that traffic through the endoplasmic reticulum-Golgi secretory pathway becoming O-glycosylated.1 The process starts with the addition of N-acetylgalactosamine (GalNAc) to either a serine or threonine residue of the protein backbone to form the so-called Tn antigen (GalNAcα-O-Ser/Thr). The Tn antigen is normally extended by addition of galactose (Gal) by core1 β1,3-galactosyltransferase-1 (C1GALT1) and its molecular chaperone COSMC to form the core 1 structure (Galβ1,3GalNAc), also known as the T antigen. The T antigen is often then sialylated to form Sialyl-T or further converted to a core 2 structure by addition of N-acetylglucosamine (GlcNAc) by β1,6-N-acetylglucosaminyltransferases (C2GNT).2
Recent high-resolution mass spectrometry has identified with great precision a number of O-linked glycosylation sites in human platelet membrane glycoproteins3 ; however, the importance of protein O-glycosylation to platelet biology remains largely unstudied. Multiple knockout (KO) mouse models targeting either C1GALT14,5 or COSMC6 to abolish core 1 synthesis all exhibit severe thrombocytopenia in conjunction with a variety of platelet abnormalities, including reduced platelet surface glycoprotein receptor expression, impaired glycoprotein Ib-IX-V and αIIbβ3 activation, impaired proplatelet formation, and increased platelet clearance, strongly suggesting that proper protein O-glycosylation is required for platelet biogenesis and function, as well as platelet lifespan and clearance.
Among the 6 members of the α2,3-sialyltransferase (ST3GAL) family in mammalians, ST3GAL1 and ST3GAL2 are known to catalyze the addition of sialic acid to the terminal galactose of type III lactosamine (Galβ1,3GalNAc), which is found on O-glycosylated proteins and glycolipids. Despite seemingly close substrate specificity, in vitro studies have shown that ST3GAL1 prefers glycoprotein acceptors, whereas glycolipids serve as the predominant substrates for ST3GAL2.7-9 Whether this represents their differential physiological functions in cells remains unresolved.
To date, only a handful of substrates for ST3GAL1 and ST3GAL2 have been identified in living cells. CD8, CD43, and CD45 were identified as glycoprotein substrates for ST3GAL1 in mouse T cells that regulate T-cell homeostasis.10 ST3GAL1, responsible for sialylation of the core 1 structure on CD45 in human B cells, was reported to be required for regulating B-cell differentiation.11 In addition, several protein substrates for ST3GAL1 (eg, CD55, vasorin, GFRA1, and AXL) have been identified in human tumor cells that promote tumor growth and invasion.12-14 These findings support the concept that glycoproteins are common substrates for ST3GAL1 in cells. In contrast, ST3GAL2 has been shown to be responsible for adding terminal sialic acid residues to 2 gangliosides, GM1 and GD1b, to synthesize GD1a and GT1b, respectively, in mouse brain and adipose tissue.15,16 ST3GAL2 KO mice show a 50% reduction in ganglioside terminal sialylation but normal protein sialylation in brain, supporting the notion that ST3GAL2 acts primarily or exclusively on glycosphingolipid acceptors.17 Stage-specific embryonic antigen-4 (SSEA4), a well-known surface marker for pluripotent stem cells, belongs to the globo-series of glycosphingolipids and is also frequently overexpressed in cancer cells. The synthesis of SSEA4 from its precursor SSEA3, through addition of α2,3-linked sialic acid, has been found to be catalyzed by ST3GAL2 in several human tumor cells.18-21 Currently, however, no protein substrates for ST3GAL2 have been reported in either mouse or human cells.
Much of our understanding about the in vivo function of ST3GAL1 and 2 has been derived from genetic manipulation of their genes in mice. ST3GAL1-deficient but not ST3GAL2-deficient mice have thrombocytopenia, suggesting a unique role of ST3GAL1 in platelet biogenesis and/or clearance.22 The function of genes in mouse models, however, sometimes does not fully recapitulate their biological function in human cells. For example, humans and mice show different tissue distributions of ST3GAL1 and ST3GAL2; notably, ST3GAL2 is present in human, but undetectable in mouse, bone marrow.23,24 The distribution of these 2 enzymes in the hematopoietic lineage has not been well investigated. Both human and mouse platelets express ST3GAL1, but whether they also express ST3GAL2 is not known.25,26 Second, although glycoproteins do not seem to be suitable substrates for mouse ST3GAL2, they have been shown to be good substrates for human ST3GAL2 in in vitro assays, suggesting the potential for human ST3GAL2 to be involved in protein O-linked glycan sialylation in vivo.23
The development of human-induced pluripotent stem cell (iPSC)-based systems, combined with the availability of efficient genome-editing technologies, has provided new opportunities for examining the function of genes in human cells. In this study, we generated ST3GAL1, ST3GAL2, and doubly deficient KO iPSC lines and performed comparative analysis of hematopoietic and megakaryocytic differentiation starting from iPSCs. We found that synthesis of the glycolipid marker SSEA4 is exclusively reliant on ST3GAL2 in iPSCs. In contrast, ST3GAL1 and ST3GAL2 exhibit partially overlapping substrate specificities in hematopoietic progenitor cells (HPCs) and megakaryocytes (MKs). Surprisingly, ST3GAL2, but not ST3GAL1, appears to play a dominant role in sialylating GPIIb. Finally, although disrupting ST3GAL1 and ST3GAL2 has no significant impact on MK production, their absence results in dramatically impaired MK proplatelet formation. Taken together, these data establish heretofore unknown physiological roles for ST3GAL2 in O-linked glycan sialylation in hemato- and megakaryocytopoiesis and highlight a previously unappreciated role for O-linked glycan sialylation in the process of terminal platelet generation.
Materials and methods
gRNA plasmid constructs
Guide RNAs (gRNAs) were designed using the CRISPR Design Tool (https://benchling.com/crispr) to minimize off-target effects and selected to precede a 5′-NGG protospacer-adjacent motif. gRNAs used in this study were: ST3GAL1-gRNA1: 5′-GAACTACTCCCACACCATGG-3′, ST3GAL1-gRNA2: 5′-GGGGTCTGGTAATGAGAGTG-3′, ST3GAL2-gRNA1: 5′-CGGAG-AGGAACCACACCCGC-3′, ST3GAL2-gRNA2: 5′-GTGAGGAGTACAGCCATGGG-3′. Oligos were annealed and cloned into the BbsI site of the clustered regularly interspaced short palindromic repeats (CRISPR)-associated protein 9 (Cas9) expression plasmids PX459 V2.0 (Addgene, Cambridge, MA).
Flow cytometric analysis
Cultured cells were incubated with fluorescein isothiocyanate–conjugated peanut agglutinin (PNA), Ricinus communis agglutinin (RCA-I), Sambucus nigra agglutinin, or biotin-conjugated Maackia amurensis agglutinin (MAL-II) (Vector Laboratories Inc., Burlingame, CA) or fluorescently labeled antibodies for 20 minutes at room temperature. The antibodies used were Alexa Fluor 488-conjugated anti-SSEA3, Alexa Fluor 647-conjugated anti-SSEA4, PE-conjugated anti-TRA-1-60-R, PE-conjugated anti-CD43 (10G7), APC-conjugated anti-CD34 (581), APC-conjugated anti-CD42b, PE-conjugated anti-CD71, APC-conjugated anti-CD235ab (Biolegend, San Diego, CA), and PE-conjugated anti-CD41a (BD Biosciences, San Jose, CA). Flow cytometry was performed using a BD LSRII flow cytometer. Flow cytometry data were analyzed using FlowJo software (Tree Star Inc., Ashland, OR).
Immunoprecipitation
iPSCs, HPCs, MKs, and erythroblasts (EBs) were lysed in 20 mM tris(hydroxymethyl)aminomethane (pH7.4), 150 mM NaCl, 1% Triton X-100, 0.1 mM CaCl2, and protease inhibitor cocktail (Thermo Fisher Scientific, Waltham, MA). Whole-cell lysates were obtained after centrifugation at 17 000g for 15 minutes at 4°C. PNA-agarose beads (Vector Laboratories Inc.) were preblocked with 3% bovine serum albumin in phosphate-buffered saline, washed, and mixed with HPC lysates generated as described above. After overnight incubation at 4°C, PNA-reactive glycoproteins were eluted with reducing sodium dodecyl sulfate (SDS) sample buffer. For immunoprecipitation, whole-cell lysates were precleared with protein G Sepharose and then incubated with the anti-GPIIIa monoclonal antibody (mAb) AP3 or anti-GPIbα mAb AP1 overnight at 4°C. Immune complexes were collected on protein G Sepharose beads, then treated with neuraminidase (New England Biolabs Inc., Ipswich, MA) for 1 hour at 37°C and eluted with reducing SDS sample buffer.
Proplatelet formation assay
Eight-well chamber slides were coated with 100 μg/mL fibrinogen overnight at 4°C. Enriched iPSC-derived MKs were cultured in 8-well chamber slides for 14 hours at 37°C and 5% CO2, then fixed with 2% paraformaldehyde for 20 minutes and permeabilized with Triton X-100 (0.5%) for 5 minutes. Cells were first stained with rabbit anti-GPIbα antibody (LifeSpan BioSciences, Inc., Seattle, WA). After wash, the cells were further stained with AF488-conjugated mouse anti-rabbit immunoglobulin G (Jackson ImmunoResearch Laboratories, West Grove, PA), TRITC-phalloidin (Sigma, Waltham, MA), and 4′,6-diamidino-2-phenylindole. The cell images were observed by using Nikon Eclipse Ti2 inverted microscope with a 60× objective lens. Cells with cytoplasmic processes longer than the diameter of the cytoplasm were defined as proplatelet-forming MKs.
Statistical analysis
One-way analysis of variance (ANOVA) with Dunnett’s test comparing with wild-type (WT) cells was performed with GraphPad Prism 9. All bar graphs with error bars represent means plus or minus standard error of the mean (SEM). A P value <.05 was assumed to represent statistical significance.
Additional methodology
See supplemental Methods for iPSC culture, differentiation, and western blot analysis.
Results
Expression levels of ST3GAL1 and ST3GAL2 are coordinately regulated during hematopoiesis
To examine the expression and function of ST3GAL1 and ST3GAL2 during human hematopoiesis, we sequentially differentiated SSEA3+/SSEA4+ iPSCs into CD34+/CD43+ HPCs, CD41+/CD42b+ MKs, and CD71+/CD235+ EBs using previously published protocols27,28 (depicted in Figure 1A-B). Western blot analysis (Figure 1C) revealed modest initial expression levels for both ST3GAL1 and ST3GAL2 in human iPSCs that, upon differentiation into HPCs, became markedly increased, followed by a dramatic decrease in expression as the cells further differentiated into MKs and EBs. Consistently, analysis of the published RNA sequencing data set from iPSC-derived HPCs and MKs (GSE119828) showed decreased mRNA expression of ST3GAL1 and ST3GAL2, as well as C1GALT1 and its chaperon COSMC that catalyze core 1 synthesis, in MKs compared with HPCs (supplemental Figure 1A). This interesting pattern suggests ST3GAL1 and ST3GAL2 are coordinately regulated to carry out O-glycan sialylation during different stages of hematopoiesis from iPSCs. We further analyzed the published single-cell RNA sequencing data isolated from human bone marrow (GSE120221). Surprisingly, bone marrow hematopoietic stem cells mainly express ST3GAL1 but not ST3GAL2, whereas bone marrow MKs mainly express ST3GAL2 but not ST3GAL1 (supplemental Figure 1B-E). This may represent the difference between in vitro and in vivo hematopoiesis and further emphasizes an important role of ST3GAL2 for O-glycan sialylation in MKs.
Expression of ST3GAL1 and ST3GAL2 during MK and EB differentiation from human iPSCs. (A) Schematic for generating MKs and EBs from iPSCs. (B) Flow cytometric analysis showed production of CD34+/CD43+ HPCs, CD41+/CD42b+ MKs, and CD71+/CD235ab+ EBs from SSEA3+/SSEA4+ iPSCs. (C) Western blot showed the expression of ST3GAL1 and ST3GAL2 at different differentiation stages.
Expression of ST3GAL1 and ST3GAL2 during MK and EB differentiation from human iPSCs. (A) Schematic for generating MKs and EBs from iPSCs. (B) Flow cytometric analysis showed production of CD34+/CD43+ HPCs, CD41+/CD42b+ MKs, and CD71+/CD235ab+ EBs from SSEA3+/SSEA4+ iPSCs. (C) Western blot showed the expression of ST3GAL1 and ST3GAL2 at different differentiation stages.
Genotypic and phenotypic characterization of iPSC lines lacking ST3GAL1, ST3GAL2, or both sialyltransferases
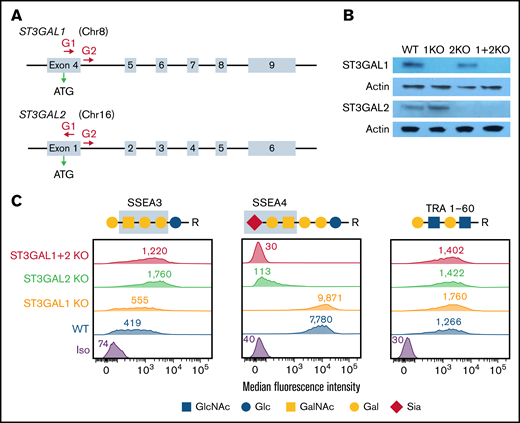
To characterize the actions of ST3GAL1 and ST3GAL2 during stem cell differentiation, we generated KO iPSC lines lacking ST3GAL1, ST3GAL2, or both, using standard CRISPR/Cas9 gene-editing technology. The gRNAs used to target specific exons within the ST3GAL1 and ST3GAL2 genes to achieve predictable deletions from the genome are shown in Figure 2A. Individual colonies were analyzed by polymerase chain reaction to verify biallelic deletion of the targeted gene in the genome (supplemental Figure 2). Western blot further confirmed the loss of ST3GAL1 and/or ST3GAL2 expression in the corresponding iPSCs (Figure 2B). Disruption of ST3GAL1 and/or ST3GAL2 did not affect the morphology or proliferation of iPSCs (data not shown).
Generation and characterization of ST3GAL1 and/or ST3GAL2 KO iPSC lines. (A) Schematic illustration of the ST3GAL1 and ST3GAL2 locus showing the location of the gRNA binding sites (red arrows) to guide Cas9 to its cleavage site. ATG start codon for gene translation is marked by the green arrow. (B) Western blot demonstrating the loss of expression of ST3GAL1 and/or ST3GAL2 in corresponding KO iPSC lines. (C) Flowcytometric analysis demonstrating the expression of SSEA3, SSEA4, and TRA 1-60 surface markers on different iPSC lines. Blue boxes highlight the carbohydrate epitopes recognized by anti-SSEA3 and anti-SSEA4 antibodies.
Generation and characterization of ST3GAL1 and/or ST3GAL2 KO iPSC lines. (A) Schematic illustration of the ST3GAL1 and ST3GAL2 locus showing the location of the gRNA binding sites (red arrows) to guide Cas9 to its cleavage site. ATG start codon for gene translation is marked by the green arrow. (B) Western blot demonstrating the loss of expression of ST3GAL1 and/or ST3GAL2 in corresponding KO iPSC lines. (C) Flowcytometric analysis demonstrating the expression of SSEA3, SSEA4, and TRA 1-60 surface markers on different iPSC lines. Blue boxes highlight the carbohydrate epitopes recognized by anti-SSEA3 and anti-SSEA4 antibodies.
Pluripotency markers on the stem cell surface are often carbohydrate antigens. Interestingly, disruption of ST3GAL2, but not ST3GAL1, abolished the expression of the pluripotency marker SSEA4 and accordingly increased the expression of its precursor SSEA3, indicating ST3GAL2 is responsible for the synthesis of SSEA4 in iPSCs (Figure 2C). Despite their common use as pluripotency markers, SSEA3 and SSEA4 are not essential for maintaining pluripotency of human embryonic stem cell.29 Therefore, the loss of SSEA4 expression in ST3GAL2 KO and ST3GAL1 + 2 KO iPSCs does not represent impaired pluripotency of the cells. Indeed, ablation of ST3GAL1 and ST3GAL2 had no effect on the expression of another carbohydrate pluripotency marker, TRA 1-60 (Figure 2C). PNA, a lectin that specifically binds to unsialylated Galβ1,3GalNAc structure, and RCA-I, a β-Gal-binding lectin, showed increased binding to ST3GAL1 + 2 KO iPSCs (supplemental Figure 3A). Accordingly, the binding to MAL-II, which recognizes α2,3 sialic acid linkages, was decreased in ST3GAL1 + 2 KO iPSCs (supplemental Figure 3A). As expected, the α2,6 sialic acid linkages were not significantly affected by disruption of ST3GAL1 and 2, as indicated by similar binding of Sambucus nigra agglutitin between WT and double-KO cells (supplemental Figure 3A).
ST3GAL1 and ST3GAL2 are each able to modify multiple glycoprotein substrates in HPCs
Hematopoietic differentiation from ST3GAL1 and ST3GAL2 single- and double-KO cell lines all showed normal expression of the diagnostic mesodermal makers KDR and CD31 on Day 6 (supplemental Figure 4) and gave rise to CD34+ HPCs (Figure 3A-B), demonstrating that disruption of ST3GAL1 and ST3GAL2 does not significantly impair hematopoietic differentiation. Moreover, western blot showed that disruption of ST3GAL1 or ST3GAL2 did not cause dramatic compensatory upregulation of the other enzyme in HPCs (supplemental Figure 5A).
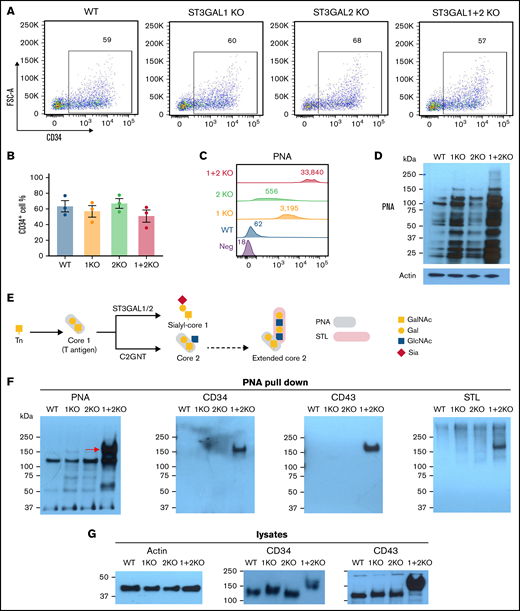
ST3GAL1 and ST3GAL2 modify multiple glycoproteins in HPCs and outcompete the formation of core 2 O-glycans on CD34 and CD43. (A) Flowcytometric analysis showed production of CD34+ HPCs from different iPSC lines. (B) Quantitated yield of CD34+ HPCs from different iPSC lines. Values represent the means plus or minus SEM from 3 independent experiments. No significance was found by 1-way ANOVA with Dunnett’s test compared with WT cells. (C) Flowcytometric analysis of PNA binding to iPSC-derived HPCs. Numbers indicate median fluorescence intensities. (D) PNA blot revealed multiple glycoprotein substrates for ST3GAL1 and ST3GAL2 in lysates from HPCs. (E) Schematic of O-glycan biosynthesis and plant lectin binding epitopes. (F) Pull-down experiment using PNA-agarose beads from lysates of iPSC-derived HPCs, followed by SDS-PAGE and immunoblot with either PNA, STL lectins or sheep anti-CD34 or goat anti-CD43 polyclonal antibodies. (G) Western blot analysis of CD34 and CD43 expression in whole-cell lysates from iPSC-derived HPCs. SDS-PAGE, SDS-polyacrylamide gel electrophoresis.
ST3GAL1 and ST3GAL2 modify multiple glycoproteins in HPCs and outcompete the formation of core 2 O-glycans on CD34 and CD43. (A) Flowcytometric analysis showed production of CD34+ HPCs from different iPSC lines. (B) Quantitated yield of CD34+ HPCs from different iPSC lines. Values represent the means plus or minus SEM from 3 independent experiments. No significance was found by 1-way ANOVA with Dunnett’s test compared with WT cells. (C) Flowcytometric analysis of PNA binding to iPSC-derived HPCs. Numbers indicate median fluorescence intensities. (D) PNA blot revealed multiple glycoprotein substrates for ST3GAL1 and ST3GAL2 in lysates from HPCs. (E) Schematic of O-glycan biosynthesis and plant lectin binding epitopes. (F) Pull-down experiment using PNA-agarose beads from lysates of iPSC-derived HPCs, followed by SDS-PAGE and immunoblot with either PNA, STL lectins or sheep anti-CD34 or goat anti-CD43 polyclonal antibodies. (G) Western blot analysis of CD34 and CD43 expression in whole-cell lysates from iPSC-derived HPCs. SDS-PAGE, SDS-polyacrylamide gel electrophoresis.
PNA binds to nonsialylated core 1 and core 1–derived glycans (depicted schematically in Figure 3E). Flow cytometric analysis showed negligible surface PNA binding to WT HPCs, reporting the absence of an exposed core 1 structure on the cell surface (Figure 3C). Both ST3GAL1 KO and ST3GAL2 KO HPCs, however, showed significant surface PNA binding compared with WT HPCs, whereas ST3GAL1 + 2 KO HPCs exhibited dramatically increased levels of PNA binding compared with single-KO cells (Figure 3C; supplemental Figure 5B), demonstrating the absence of terminal O-linked sialic acid residues. To rule out the possibility that the phenotype is caused by an off-target effect of CRISPR, we established additional clones of each cell type and confirmed consistent phenotype in these clones (supplemental Figure 5C). Consistently, disruption of ST3GAL1 and 2 caused increased RCA-I binding and decreased MAL-II binding in HPCs (supplemental Figure 3B), further supporting the loss of α2,3-linked sialic acids on the cell surface. Because surface PNA can also bind to unsialylated glycosphingolipids in the plasma membrane, we next analyzed PNA binding specifically to glycoprotein substrates. As shown in Figure 3D, PNA western blot analysis revealed numerous glycoprotein targets in ST3GAL1 and ST3GAL2 single-KO cells and an even larger number of unsialylated glycoproteins in the double-KO cells (Figure 3D; supplemental Figure 5D). These data, therefore, demonstrate that ST3GAL1 and ST3GAL2 are each able to perform sialylation of O-linked glycans in HPCs, with many, but not completely, overlapping glycoprotein targets.
CD34 and CD43 are overlapping glycoprotein substrates for ST3GAL1 and ST3GAL2
To identify specific glycoprotein substrates for ST3GAL1 and ST3GAL2, PNA-agarose beads were used to pull-down glycoproteins with exposed core 1 or core 2 O-linked glycans from HPC detergent lysates. As shown in Figure 3F, PNA immunoblots revealed several candidate bands, including a prominent ∼160 kDa band present exclusively in ST3GAL1 + 2 KO cells. Because the HPC markers CD34 and CD43 have been reported to be extensively O-glycosylated and sialylated,30,31 we postulated that they might serve as glycoprotein substrates for ST3GAL1 and/or ST3GAL2. Indeed, blotting with CD34 or CD43 polyclonal antibodies positively identified these 2 glycoproteins as targets for these 2 sialyltransferases (Figure 3F). Interestingly, neither CD34 nor CD43 became PNA+ unless both sialyltransferases were missing, demonstrating that both ST3GAL1 and ST3GAL2 are each capable of independently sialylating these glycoproteins and therefore exhibit overlapping glycoprotein substrate specificities. Interestingly, the electrophoretic mobility of CD34 and CD43 in ST3GAL1 + 2 KO cells was noticeably decreased (Figure 3G), consistent with the absence of core 1 sialylation. As shown schematically in Figure 3E, ST3GAL1 and C2GNT normally compete for the same core 1 substrate. In the absence of ST3GAL1 and ST3GAL2, therefore, one might expect an increase in core 2 production catalyzed by C2GNT.10 To examine whether these slower mobility species might be due to their carrying extended, unsialylated core 2 glycan, we probed the blots of the PNA pull-downs with Solanum tuberosum lectin (STL), a lectin that reacts with poly-LacNAc structure present on extended core 2 O-glycans (and also on N-glycans). As shown in the rightmost panel of Figure 3F, STL reacted with the 160 kDa band pulled down by PNA. This finding, together with the decreased mobility of CD34 and CD43 in double-KO cells, suggest the presence of core 2 glycans on CD34 and CD43 in double-KO cells. Taken together, these data demonstrate that ST3GAL1 and ST3GAL2 work redundantly for O-glycan sialylation on CD34 and CD43 in HPCs and that when present, their activity normally outcompetes C2GNT for core 1 substrates.
ST3GAL1 and 2 have both overlapping and unique glycoprotein substrate specificities sialylating membrane glycoproteins of the megakaryocyte lineage
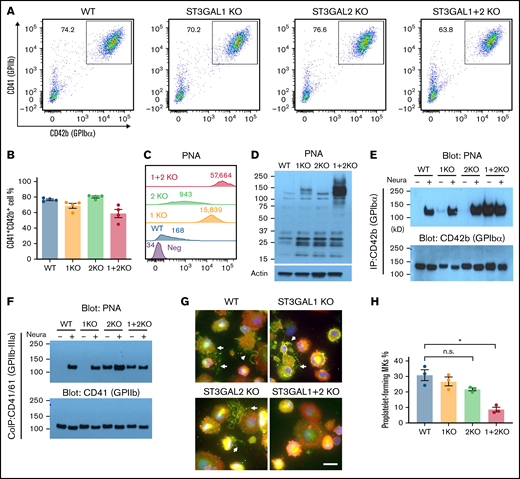
As shown in Figure 4A-B, ST3GAL1 KO, ST3GAL2 KO, and double-KO cell lines were fully capable of differentiating into CD41+/CD42b+ MKs, demonstrating that disruption of these 2 O-sialyltransferases has little impact on megakaryopoiesis. Moreover, the overall expression of MK surface glycoprotein GPIIb (CD41) and GPIbα (CD42b) were not affected by the absence of ST3GAL1 and ST3GAL2 (Figure 4A). In addition, DNA ploidy level of MKs from all the cell lines remained comparable (supplemental Figure 6A-B), indicating disruption of ST3GAL1 and ST3GAL2 does not affect MK maturation.
Loss of ST3GAL1 and ST3GAL2 disrupts O-glycan sialylation of major surface glycoproteins on MKs and impairs proplatelet formation. (A) Flow cytometric analysis showed production of CD41+/CD42b+ MKs from different iPSC lines. (B) Quantitated yield of CD41+/CD42b+ MKs from different iPSC lines. Values represent the means plus or minus SEM from 4 independent experiments. No significance was found by 1-way ANOVA with Dunnett’s test compared with WT cells. (C) Flowcytometric analysis of PNA binding to iPSC-derived MKs. Numbers indicate median fluorescence intensities. (D) PNA blot revealed a subset of glycoprotein substrates for ST3GAL1 and ST3GAL2 in lysates from MKs. (E) Immunoprecipitation with anti-GPIbα monoclonal antibody AP1 from lysates of iPSC-derived MKs, followed by SDS-PAGE and immunoblot with either PNA or rabbit anti-GPIbα polyclonal antibody. Immunoprecipitated GPIbα from different cell lines was treated with neuraminidase for complete exposure of PNA-reactive epitopes as positive controls. (F) Coimmunoprecipitation of GPIIb-IIIa complex with anti-GPIIIa monoclonal antibody AP3 from lysates of iPSC-derived MKs, followed by SDS-PAGE and immunoblot with either PNA or rabbit anti-GPIIb polyclonal antibody. (G) Immunofluorescence staining of iPSC-derived MKs cultured on fibrinogen-coated glass slides. The cells were labeled with phalloidin (red), anti-GPIbα antibody (green), and 4′,6-diamidino-2-phenylindole (blue). Scale bar, 20 μm. White arrows indicate proplatelet-forming MKs. (H) Quantitated percentage of proplatelet-forming MKs from each cell line. Values represent the means plus or minus SEM from 3 independent experiments. *P < .05 by 1-way ANOVA with Dunnett’s test compared with WT cells. n.s., not significant; SDS-PAGE, SDS-polyacrylamide gel electrophoresis.
Loss of ST3GAL1 and ST3GAL2 disrupts O-glycan sialylation of major surface glycoproteins on MKs and impairs proplatelet formation. (A) Flow cytometric analysis showed production of CD41+/CD42b+ MKs from different iPSC lines. (B) Quantitated yield of CD41+/CD42b+ MKs from different iPSC lines. Values represent the means plus or minus SEM from 4 independent experiments. No significance was found by 1-way ANOVA with Dunnett’s test compared with WT cells. (C) Flowcytometric analysis of PNA binding to iPSC-derived MKs. Numbers indicate median fluorescence intensities. (D) PNA blot revealed a subset of glycoprotein substrates for ST3GAL1 and ST3GAL2 in lysates from MKs. (E) Immunoprecipitation with anti-GPIbα monoclonal antibody AP1 from lysates of iPSC-derived MKs, followed by SDS-PAGE and immunoblot with either PNA or rabbit anti-GPIbα polyclonal antibody. Immunoprecipitated GPIbα from different cell lines was treated with neuraminidase for complete exposure of PNA-reactive epitopes as positive controls. (F) Coimmunoprecipitation of GPIIb-IIIa complex with anti-GPIIIa monoclonal antibody AP3 from lysates of iPSC-derived MKs, followed by SDS-PAGE and immunoblot with either PNA or rabbit anti-GPIIb polyclonal antibody. (G) Immunofluorescence staining of iPSC-derived MKs cultured on fibrinogen-coated glass slides. The cells were labeled with phalloidin (red), anti-GPIbα antibody (green), and 4′,6-diamidino-2-phenylindole (blue). Scale bar, 20 μm. White arrows indicate proplatelet-forming MKs. (H) Quantitated percentage of proplatelet-forming MKs from each cell line. Values represent the means plus or minus SEM from 3 independent experiments. *P < .05 by 1-way ANOVA with Dunnett’s test compared with WT cells. n.s., not significant; SDS-PAGE, SDS-polyacrylamide gel electrophoresis.
To further explore the function of ST3GAL1 and ST3GAL2 in MKs, we examined the binding of PNA to each of these MK cell lines. Like that observed in HPCs, both ST3GAL1 KO and ST3GAL2 KO MKs showed increased surface PNA binding compared with WT MKs, with PNA binding exceptionally strong on ST3GAL1 + 2 KO MKs (Figure 4C; supplemental Figure 6C-D). Consistent with these observations, western blot analysis revealed markedly increased PNA binding in ST3GAL1 + 2 double-KO cells (Figure 4D; supplemental Figure 6E). In addition, increased RCA-I binding and decreased MAL-II binding were found in ST3GAL1 + 2 KO MKs (supplemental Figure 3C), further supporting the loss of α2,3-linked sialic acids in these cells. Altogether, these data demonstrate that both ST3GAL1 and ST3GAL2 are able to sialylate O-linked glycans on a number of yet-to-be-identified proteins of the megakaryocyte lineage.
In this regard, GPIbα is known to contain a mucin-like region containing a large number of potential O-linked glycosylation sites. To determine whether any of these might be substrates for ST3GAL1 and/or ST3GAL2, we immunoprecipitated GPIbα from MK lysates and subjected it to western blot analysis using PNA to probe its sialylation status. As shown in Figure 4E, GPIbα from WT, ST3GAL1 KO, or ST3GAL2 KO MKs reacted weakly or not at all with PNA, indicating that the O-linked glycans of GPIbα are normally completely sialylated. In contrast, GPIbα derived from ST3GAL1 + 2 KO cells reacted strongly with PNA even without pretreatment with neuraminidase, demonstrating that both ST3GAL1 or ST3GAL2 are able by themselves to transfer sialic acid to the T antigens of GPIbα during biosynthesis.
Previous studies have shown that GPIIb, one of the most abundant glycoproteins on the platelet surface, is modified with core 1 O-linked glycans attached to serine residues 845 and 847 near the C-terminus of its extracellular domain.3 To identify the enzyme(s) responsible for transferring sialic acids to these T antigens on GPIIb, we coimmunoprecipitated GPIIb-IIIa complexes from MK lysates and subjected them to PNA western blot analysis. As shown in Figure 4F and supplemental Figure 6F, GPIIb from WT and ST3GAL1 KO cells supported PNA binding only after neuraminidase treatment, indicating that GPIIb from these 2 cell lines is fully sialylated. In contrast, GPIIb from ST3GAL2 KO cells showed strong PNA binding even without neuraminidase treatment, demonstrating that ST3GAL2, and not ST3GAL1, is the major enzyme responsible for T antigen sialylation on GPIIb and providing one of the first examples of a unique glycoprotein substrate for this sialyltransferase. Interestingly, loss of sialic acids on O-glycan of GPIIb does not significantly affect GPIIb-IIIa activation upon thrombin stimulation, as indicated by similar Pac-I binding (supplemental Figure 7A-B) and fibrinogen binding (supplemental Figure 7C-D) of ST3GAL1/2-deficient MKs to that of WT MKs.
Disruption of ST3GAL1 and ST3GAL2 impairs proplatelet formation
The gene Slc35a1 encodes the cytidine 5′-monophosphate (CMP)-sialic acid transporter that transports CMP-sialic acid from the cytoplasm into the Golgi apparatus, and Slc35a1-deficient mice show significantly reduced sialylation in MKs and impaired proplatelet formation, suggesting sialic acids on the MK surface play a role in proplatelet formation.32 Similarly, disruption of the gene encoding C1GALT1 in mouse bone marrow blocks core 1 O-glycan synthesis and also impairs proplatelet formation.4 To further examine whether ST3GAL1 or 2 might play a role in this process, we plated MKs on a fibrinogen-coated surface under conditions that promote proplatelet formation. As shown in Figure 4G and quantified in Figure 4H, whereas both WT, ST3GAL1 KO, and ST3GAL2 KO cells displayed long, extended, fragmented protrusions typical of proplatelet formation, ST3GAL1 + 2 KO cells exhibited severe impairment of proplatelet formation, demonstrating that each of these 2 sialyltransferases are able to add terminal sialic acid residues to O-linked glycans that play a critical role in the terminal stages of MK differentiation.
Overlapping substrate specificity of ST3GAL1 and ST3GAL2 in erythroblasts
To examine whether ST3GAL1 or ST3GAL2 might be required for differentiation down the erythroid lineage, iPSC lines lacking 1 or both sialyltransferases were differentiated into HPCs similar to those shown in Figure 3A and then further differentiated into transferrin receptor (CD71)-positive EBs. As shown in supplemental Figure 8A-B, deficiency of either or both ST3GAL1 and ST3GAL2 had no effect on erythropoiesis. Flow cytometric analysis of surface PNA binding revealed partial functional redundancy of ST3GAL1 and ST3GAL2 as loss of both sialyltransferases resulting in stronger PNA binding compared with loss of either one alone (supplemental Figure 8C). ST3GAL2, however, appears to play a more dominant role in sialylating O-glycans in mature EBs as its loss results in ∼3 times the level of PNA binding compared with cells lacking ST3GAL1.
Discussion
Though ST3GAL1 and ST3GAL2 are well-studied type III oligosaccharide (Galβ1,3GalNAc) sialyltransferases, their glycoprotein targets in cells, especially human cells, remain largely undefined. Based on in vitro analyses, ST3GAL2 has been thought to have a strong preference for glycolipid acceptors, whereas sialylation of O-linked glycoprotein targets has been largely attributed to ST3GAL1.33,34 One of the major findings of the present study, however, is that these 2 enzymes actually share a number of glycoprotein substrates, including CD34, CD43, and platelet GPIbα (Figures 3F and 4E). Redundancy in the substrate specificity of ST3GAL1 and 2 for GPIbα might be of particular interest as desialylation of the O-glycans on mouse GPIbα has recently been shown to drive platelet clearance.35 At least in human MKs, and presumably human platelets, our data would suggest that both ST3GAL1 and ST3GAL2 are each able to sialylate GPIbα (Figure 4E; also see discussion below). In contrast, ST3GAL2 appears to be primarily responsible for O-sialylating platelet membrane GPIIb (Figure 4F), thus establishing that ST3GAL2 can function not only to catalyze the addition of sialic acid to the terminal galactose of type III lactosamine on glycolipids but also act as a glycoprotein sialyltransferase. The functional diversification of these 2 enzymes suggests that the substrate specificity of O-sialyltransferases not only depends on the acceptor glycans but is also, at least partially, affected by the protein substrate scaffold.
Recent studies have elucidated several complex mechanisms that regulate platelet clearance, such as phosphatidylserine upregulation and proapoptotic events, antibody-mediated platelet clearance, and glycan-mediated platelet clearance.36 Several lines of evidence suggest that sialylation of GPIbα is critically important for protecting platelets from clearance by the Ashwell-Morell receptor on hepatocytes and integrin αMβ2 on macrophages, respectively.37 The specific sialyltransferases involved, and their glycan substrates on GPIbα, however, are incompletely defined and remain the subject of intense investigation. There are 6 members of the ST3GAL family, each having different substrate specificities. ST3GAL1 and 2 use the Galβ1,3GalNAc structure (type III lactosamine) as their acceptor substrate, which corresponds to the core 1 structure on O-glycans, whereas ST3GAL3 and ST3GAL4 use both Galβ1,3GlcNAc (type I) and Galβ1,4GlcNAc (type II) structures as acceptor substrates. ST3GAL5 functions as a GM3 ganglioside synthase, whereas ST3GAL6 utilizes the Galβ1,4GlcNAc (type II) structure as an acceptor substrate.38 This study focused on ST3GAL1 and ST3GAL2 because these are the major sialyltransferases for the core 1 structures on O-glycosylated glycoproteins. The complex nature of the O-linked glycans attached to GPIbα was first examined nearly 40 years ago (shown schematically in supplemental Figure 9).39,40 More recently, King et al, using sophisticated mass spectrometry analysis, were able to identify 7 O-linked glycosylation sites within the mucin-like macroglycopeptide stem region of GPIbα.3 Due to technical limitations, it was not possible to simultaneously identify both the glycan structure and the precise sites to which they are attached for complex O-glycans; however, the authors were able to predict the presence of branched core 2 structure on GPIbα within the so-called mechanosensory domain (MSD), a juxtamembrane portion of the receptor that responds to tensile forces.41 Desialylation of all of the sialic acid residues within the mucin-rich core by an α2,3-neuraminidase has been shown to render the MSD susceptible to cleavage by ADAM17, which is reported by increased binding to the monoclonal antibody 5G6,42,43 leading to shedding of a large portion of the extracellular domain of the receptor and rapid platelet clearance.35 Similarly, ST3GAL4-deficient mouse platelets also become rapidly removed from circulation of WT recipient mice,44 suggesting that this sialyltransferase is one of those that plays a role in maintaining the sialylation state of GPIbα, a notion supported by the observation that removal of the extracellular domain of GPIbα rescues the clearance of ST3GAL4-deficient platelets from circulation. Interestingly, mAb 5G6 does not show increased binding to ST3GAL1 + 2-deficient megakaryocytes compared with WT cells, nor did we observe a decrease in the binding of mAb HIP1, an antibody that targets the N terminus of GPIbα (data not shown) in these cells, suggesting that exposure of the desialylated Galβ1,3GalNAc branch of core 2, though rendering GPIbα PNA+ (Figure 4E), is insufficient to disrupt the MSD and expose the ADAM17 cleavage site, perhaps because the Galβ1,4GlcNAc branch remains sialylated by ST3GAL4 and limits exposure of the terminal galactose of core 2 (supplemental Figure 9). Taken together, these data are consistent with the notion that the core 1 branch of the core 2 glycan becomes sialylated by ST3GAL1 or 2, whereas the longer Galβ1,4GlcNAcβ1,6GalNAc branch is a substrate for ST3GAL4 (or potentially ST3GAL3 and ST3GAL6).
Due to their anionic charge and their peripheral position in glycans at the cell surface, sialic acids are crucial players modulating cell function and regulating cell communications. Both mouse and human platelets contain high levels of O-glycans, with more sialic acids estimated on platelet O-glycans than on N-glycans.5 Genetic disruption of CMP-sialic acid transporter, which broadly impairs sialylation of both N-glycan and O-glycan, causes impaired proplatelet formation and thrombocytopenia.32 Consistent with those observations, we found that although ablation of ST3GAL1 + 2 does not significantly affect MK production, it disrupts MK proplatelet formation, consistent with the notion that sialic acids on O-glycans play an important role in terminal megakaryocyte differentiation and platelet production. The mechanism by which O-glycan sialylation contributes to proplatelet formation is unclear but is likely related to the function of still-to-be-identified receptors present on the surface of megakaryocytes.
In conclusion, we have discovered a number of novel and physiologically important roles for human ST3GAL2 for sialylation of O-glycosylated proteins that have been overlooked in the past. Illustration of the functional redundancy and diversification of ST3GAL1 and ST3GAL2 provides important new elements in the glycobiology of hematopoietic cells. Further characterization should help to identify the sialylglycoconjugates produced by them to fully understand their biological significance in the blood and vascular cells in which they are expressed.
Acknowledgment
This work was supported by grant R35 HL139937 (P.J.N.) from the National Institutes of Health, National Heart Lung and Blood Institute.
Authorship
Contribution: N.Z. designed and conducted experiments, analyzed data, and wrote the manuscript; P.J.N. designed experiments, analyzed data, and wrote the manuscript; and S.L. and W.C. analyzed single-cell RNA sequencing data.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Peter J. Newman, Blood Research Institute, Versiti, 8727 W. Watertown Plank Rd., Milwaukee, WI 53226; e-mail: pjnewman@versiti.org; and Nanyan Zhang, Blood Research Institute, Versiti, 8727 W. Watertown Plank Rd., Milwaukee, WI 53226; e-mail: nzhang@versiti.org.
References
Author notes
Requests for data sharing may be submitted to Nanyan Zhang (nzhang@versiti.org).
The full-text version of this article contains a data supplement.