Key Points
VWF and FVIII have coevolved throughout mammalian diversification to maintain balance between primary and secondary hemostasis.
Ancestral VWF molecules exhibit unique biochemical characteristics that may be beneficial to pharmaceutical development.
Abstract
Ancestral sequence reconstruction provides a unique platform for investigating the molecular evolution of single gene products and recently has shown success in engineering advanced biological therapeutics. To date, the coevolution of proteins within complexes and protein–protein interactions is mostly investigated in silico via proteomics and/or within single-celled systems. Herein, ancestral sequence reconstruction is used to investigate the molecular evolution of 2 proteins linked not only by stabilizing association in circulation but also by their independent roles within the primary and secondary hemostatic systems of mammals. Using sequence analysis and biochemical characterization of recombinant ancestral von Willebrand factor (VWF) and coagulation factor VIII (FVIII), we investigated the evolution of the essential macromolecular FVIII/VWF complex. Our data support the hypothesis that these coagulation proteins coevolved throughout mammalian diversification, maintaining strong binding affinities while modulating independent and distinct hemostatic activities in diverse lineages.
Introduction
The bleeding disorders von Willebrand disease (VWD) and hemophilia A arise from qualitative or quantitative deficiencies in either von Willebrand factor (VWF) or coagulation factor VIII (FVIII), respectively. Each protein is the product of a distinct gene located on different chromosomes; however, their roles in hemostasis are coordinated and concomitant. VWF has critical functions in both primary and secondary hemostasis, and FVIII is an essential cofactor for the serine protease factor IX. VWF and FVIII circulate in a tight noncovalent complex. Association with VWF protects FVIII from proteolytic degradation, prolongs the plasma half-life of the cofactor, and localizes FVIII activation at the site of injury. Impairment of this association due to mutations in the binding site of either VWF or FVIII results in the bleeding disorders VWD type 2N and nonsevere hemophilia A. Understanding the FVIII-VWF interaction is paramount to the development of novel therapies for hemophilia A and VWD; however, many of the basic biological questions of their association have remained unanswered throughout the past 35 years of investigation.
Evolutionary studies of vertebrate coagulation have detected the earliest components of the coagulation system in lampreys and hagfish.1-4 These primitive systems generate fibrin exclusively through an extrinsic pathway of factor VIIa/tissue factor–mediated formation of the prothrombinase complex. In bony fishes, FVIII and factor IX emerged after genome duplication and subsequent diversification of the homologous coagulation proteins factor V and factor X, respectively. The emergence of additional coagulation factors allowed for increased thrombin amplification and fibrin generation and supported the development of increasingly complex vasculature systems. It remains unclear how FVIII’s dependence on VWF emerged despite occurring in every species investigated.
Known species-specific differences in FVIII biosynthesis, biochemistry, and immunology can be exploited for therapeutic engineering.5-13 These unique FVIII characteristics are the result of incremental amino acid replacements that can be traced throughout evolution by ancestral sequence reconstruction (ASR).14 Although species-specific differences in VWF biology have been observed regarding platelet activation and ristocetin binding,15-17 there has been limited investigation of VWF orthologs or the FVIII-VWF interaction across species. The current study characterizes the molecular evolution of mammalian VWF and investigates the hypothesis that FVIII and VWF coevolved as critically interdependent partners. Using ancestral VWF (AnVWF) sequences spanning several mammalian lineages, the data show that VWF and FVIII evolved coordinately. To the best of our knowledge, this report provides the first phylogenetic and biochemical evidence of molecular coevolution within the hemostatic system and the first investigation of protein complex evolution within higher vertebrates.
Methods
AnVWF sequence inference
AnVWF sequence reconstruction was performed as described previously18,19 by using 59 extant VWF sequences, MUSCLE, and MrBayes programs. Ancestral sequences were inferred by using PAML version 4.1.
AnVWF and ancestral FVIII DNA synthesis
Ancestral amino acid sequences An101-, An88-, An84-, An70-, and An63-VWF were codon optimized (co) for human host cell expression and synthesized by using GenScript and subcloned into the pCI-Neo vector. In addition, a C-terminal glycine hinge (GGRGG), TEV protease motif (ENLYFQG), and 3x FLAG tag (DYKDDDDK)3 with the nucleic acid sequence GGCGGCAGAGGAGGAGAGAACCTGTACTTCCAGGGCGACTATAAGGACGATGACGATAAGGATTACAAAGATGATGACGATAAGGATTATAAAGACGATGACGATAAGTGA were added to each ancestral sequence. Ancestral FVIII complementary DNAs were generated and purified as previously described.7
VWF enzyme-linked immunosorbent assay
Enzyme-linked immunosorbent assay (ELISA) was performed by using polyclonal antibodies A0082 and P0226 (Dako). Absorbance was interpolated to a standard curve of World Health Organization (WHO)-defined human standard plasma. VWF samples were sufficiently diluted to ensure antibody excess, and parallel absorbance standard curves were obtained for all constructs.
AnVWF expression and purification
Transient VWF plasmid transfections were performed in HEK293T and HEK116 cells. Polyclonal stable cell lines were generated in HEK293 cells. Monoclonal producer cell lines were generated from HEK116 cells using Geneticin selection. Protein was purified from serum-free Opti-MEM media (Thermo Fisher Scientific). Briefly, AnVWF was diluted 1:1 with 20 mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid (HEPES), 5 mM CaCl2, and 0.01% Tween 80 at pH 7.2 before heparin-sepharose affinity chromatography. Columns were equilibrated and washed with 20 mM HEPES, 5 mM CaCl2, 50 mM NaCl, and 0.01% Tween 80 at pH 7.2. AnVWF was eluted with 650 mM NaCl. Fractions were concentrated in Amicon 50K NMWL filters (MilliporeSigma) and stored for 48 to 72 hours at 4°C. Samples were centrifuged, and VWF containing supernatant was stored at −80°C. Final concentrations of AnVWF were determined by ELISA.
RNA isolation and reverse transcription polymerase chain reaction
RNA was purified by using the RNeasy kit (Qiagen) according to manufacturer’s instructions. Cell counts, RNA collections, and VWF determinations were performed simultaneously. Transcript analysis was conducted as previously described14 using universal VWF primers within the D3 domain (forward 5′-GAGAACGGCTACGAGTGCG, reverse 5′-CTGGAGGACAGTGTGCGTG). Transcripts per cell were calculated by using an estimated 25 pg RNA per HEK116 cell.
Multimer analysis and ADAMTS13 cleavage
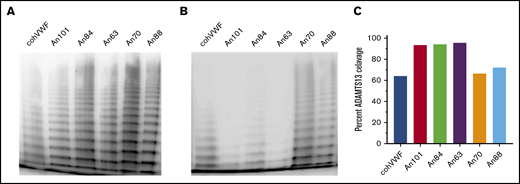
Multimer analysis was performed by using 60 ng VWF as previously reported.20-22 Multimers were visualized by chemiluminescent imaging of antibody P0226 (Dako). AnVWF digestion using recombinant human ADAMTS13 was performed as previously described.23,24 Densitometry was analyzed by using ImageJ software (National Institutes of Health) and calculated as percentage of multimers remaining after ADAMTS13 cleavage.
In vivo AnVWF production and FVIII rescue
All animal procedures were in accordance with the Canadian Council on Animal Care guidelines and approved by the Queen’s University Animal Care Committee. Linear DNA, 10 μg, in TransIT-EE buffer (Mirus Bio) was hydrodynamically injected into 8- to 12-week-old VWF−/− female mice. Retro-orbital samples were collected at 24 hours’ postinjection. VWF antigen was determined by ELISA and FVIII activity by chromogenic SP4 assay using a WHO-defined human standard plasma. Molar concentrations of FVIII were determined by using a specific-activity of 4000 IU/mg for murine FVIII.5
Clearance
AnVWF (20 μg) was injected via tail vein in 8- to 12-week-old male VWF−/− mice. Retro-orbital sampling was performed from alternate eyes at 2 time points. VWF determination was performed according to ELISA as described earlier.
VWF:GP1bM and VWF:RCo activity assays
The Innovance VWF:Ac GP1bM assay was performed on a BCS-XP coagulometer (Siemens) according to the manufacturer’s instructions and compared with WHO-defined human standard plasma. Specific-activity was defined as the ratio of GP1bM activity to VWF antigen concentration in international units per milligram. Ristocetin cofactor (VWF:RCo) activity was measured via platelet aggregometry using washed normal platelets and compared with a WHO-defined standard plasma, as previously described.25
Surface plasmon resonance
Studies were performed at 25°C using a Biacore T200 (GE Healthcare) and CM5 sensor chips (GE Healthcare) equilibrated with immobilization running buffer (20 mM HEPES, 150 mM NaCl, 0.02% Tween 20, pH 7.4). Recombinant anti-Flag antibody (M2, #F1804; Sigma) was immobilized using the Amine Coupling Kit (GE Healthcare) according to the manufacturer’s instructions to a range of ∼2000 to ∼7000 resonance units (RU). VWF-Flag variants were diluted to 50 to 100 µg/mL and injected at 2 µL/min. VWF immobilization level was between ∼200 and ∼500 RU. Flow cell 1 was immobilized with the anti-Flag antibody alone as a reference for systematic noise and instrument drift.
Surface plasmon resonance measurements were performed at 30 µL/min using single-cycle kinetics with 5 sequential 120-second injections at increasing concentrations of each FVIII variant (0.55-45 nM)26 followed by a final 300-second dissociation. The surface was regenerated with 20 mM HEPES, 600 mM NaCl, 350 mM CaCl2, and 0.02% Tween 20 at pH 7.4 for 2 minutes. Measurements included zero-concentration cycles of analyte before sampling for subtraction.27 All FVIII samples were measured in triplicates per run, and each VWF variant was immobilized to 3 different chips at different flow cell positions. Data were analyzed by using Biacore T200 Evaluation Software 3.0 (GE Healthcare). Sensorgrams were fitted globally by using the steady-state model after double reference subtraction.
Results
Phylogenetic tree and sequence reconstruction
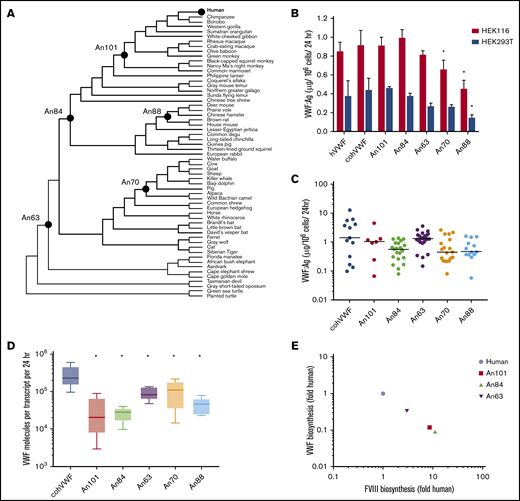
ASR has enabled the engineering of complex proteins when rational design and directed evolution methods have been prohibitive. Using 59 extant VWF sequences (numbered 1-59), we performed ASR as described previously18 to generate 53 ancestral sequences (numbered 60-112) (Figure 1A; supplemental Figure 1A-B). In addition, extant sequences for coagulation factors V, VII, VIII, IX, and X were applied to ASR analysis. Consistent with analyses based on the mirrortree method,28 independent reconstruction of these protein sequences resulted in identical phylogenetic trees, providing a singular representation for the molecular evolution of multiple procoagulant proteins.29 These phylogenetic data suggest that the components of the coagulation cascade recapitulate species evolution and coevolved throughout mammalian diversification.
AnVWF biosynthetic efficiency correlates with ancestral FVIII. (A) Phylogenetic tree depicting ancestral sequence reconstruction of 59 extant VWF sequences. VWF sequences selected for de novo synthesis are labeled as closed circles. (B) Production rates of AnVWF protein following transient transfection of HEK116 (red bars) and HEK293T (blue bars) cells. Error bars represent SD (n = 4). *P < .05 (Dunnett’s one-way ANOVA). (C) Stable, monoclonal producer cell lines were produced from HEK116 cells, and VWF biosynthesis was measured via ELISA. Horizontal bars represent median values; n = 13, 7, 23, 29, 19, and 14 for cohVWF, An101, An84, An63, An70, and An88, respectively. No significant differences were detected compared with cohVWF (Dunn’s one-way ANOVA). (D) Transcript analysis of monoclonal producer cell lines at steady state was performed by one-step reverse transcription polymerase chain reaction. VWF antigen was normalized to transcript levels to determine biosynthetic efficiency. Box plot reveals range, 25th and 75th percentiles, and median values; n = 6 for An70 and An88 and n = 5 for others. All AnVWF constructs displayed significantly reduced (*P < .05) biosynthetic efficiency compared with cohVWF (Dunnett’s one-way ANOVA). (E) Correlation of VWF and FVIII biosynthetic efficiency was compared as fold change relative to human. FVIII biosynthetic efficiency was reported previously.14 Linear regression equation (not shown) is y = −1.002x + 6.4 × 10−5 with r2 = 1.
AnVWF biosynthetic efficiency correlates with ancestral FVIII. (A) Phylogenetic tree depicting ancestral sequence reconstruction of 59 extant VWF sequences. VWF sequences selected for de novo synthesis are labeled as closed circles. (B) Production rates of AnVWF protein following transient transfection of HEK116 (red bars) and HEK293T (blue bars) cells. Error bars represent SD (n = 4). *P < .05 (Dunnett’s one-way ANOVA). (C) Stable, monoclonal producer cell lines were produced from HEK116 cells, and VWF biosynthesis was measured via ELISA. Horizontal bars represent median values; n = 13, 7, 23, 29, 19, and 14 for cohVWF, An101, An84, An63, An70, and An88, respectively. No significant differences were detected compared with cohVWF (Dunn’s one-way ANOVA). (D) Transcript analysis of monoclonal producer cell lines at steady state was performed by one-step reverse transcription polymerase chain reaction. VWF antigen was normalized to transcript levels to determine biosynthetic efficiency. Box plot reveals range, 25th and 75th percentiles, and median values; n = 6 for An70 and An88 and n = 5 for others. All AnVWF constructs displayed significantly reduced (*P < .05) biosynthetic efficiency compared with cohVWF (Dunnett’s one-way ANOVA). (E) Correlation of VWF and FVIII biosynthetic efficiency was compared as fold change relative to human. FVIII biosynthetic efficiency was reported previously.14 Linear regression equation (not shown) is y = −1.002x + 6.4 × 10−5 with r2 = 1.
We synthesized 5 AnVWF complementary DNAs for biochemical characterization, chosen based on their evolutionary position between the common mammalian ancestor and the most extensively characterized extant VWF molecules. These AnVWF sequences, termed An101-, An84-, An63-, An70-, and An88-VWF, range in total amino acid replacements from 81 to 364, representing 97% to 87% identity to human VWF (Table 1). All cysteine residues are conserved in ancestral sequences.
Ancestral VWF sequence identity and classification
| VWF . | Amino acid replacements . | % Human identity . | Predicted age (mya) . | Classification . |
|---|---|---|---|---|
| cohVWF | 0 | 100 | — | Primate |
| An101 | 81 | 97.1 | 43.2 | Primate: Simiiformes |
| An84 | 206 | 92.7 | 89.8 | Euarchontoglires |
| An63 | 245 | 91.3 | 105.5 | Eutheria |
| An70 | 307 | 89.1 | 64.2 | Ungulate: Artiodactyla |
| An88 | 364 | 87.1 | 32.7 | Rodentia: Muroidea |
| VWF . | Amino acid replacements . | % Human identity . | Predicted age (mya) . | Classification . |
|---|---|---|---|---|
| cohVWF | 0 | 100 | — | Primate |
| An101 | 81 | 97.1 | 43.2 | Primate: Simiiformes |
| An84 | 206 | 92.7 | 89.8 | Euarchontoglires |
| An63 | 245 | 91.3 | 105.5 | Eutheria |
| An70 | 307 | 89.1 | 64.2 | Ungulate: Artiodactyla |
| An88 | 364 | 87.1 | 32.7 | Rodentia: Muroidea |
mya, million years ago.
Biosynthesis, multimerization, and ADAMTS13 cleavage
Using human cell lines clinically employed in heterologous recombinant production of VWF and FVIII, we investigated the biosynthetic production of AnVWF. VWF antigen levels were determined via ELISA by using cross-reactive polyclonal anti-human VWF antibodies previously validated for measuring VWF orthologs, including murine VWF.30 Parallel standard curves were obtained for all ancestral constructs, allowing concentration determinations from standard human plasma (supplemental Figure 2A-B). Transient transfection of either HEK116 or HEK293T cells resulted in equivalent production of the codon-optimized human (coh) VWF compared with wild type (Figure 1B). Compared with cohVWF, An70- and An88-VWF exhibited significantly reduced production in one or both cell lines (P = .0284 and .0003, respectively, for HEK116 cells; P = .0024 for An88-VWF in 293T cells; one-way analysis of variance [ANOVA]). Analysis of stable monoclonal cell lines at steady-state, however, revealed no significant difference in VWF production despite lower median values (Dunn’s one-way ANOVA) (Figure 1C). To determine if observed VWF production is transcript dependent, messenger RNA analysis was performed at the time of antigen determination. Biosynthetic efficiencies of all AnVWF molecules were significantly reduced compared with cohVWF, suggesting that translation and/or secretion are rate-limiting (Dunnett’s ANOVA) (Figure 1D).
Comparing biosynthesis of AnVWF vs previously reported ancestral FVIII sequences,14 we observed an inverse correlation throughout primate evolution (Figure 1E), suggesting a conserved balance of protein production. These changes are not sequential, exhibiting temporally random but reciprocal changes in the partner protein’s biosynthesis.
Multimer analysis confirmed the presence of high molecular weight (HMW) multimers equivalent to cohVWF (Figure 2A). The formation of HMW multimers in the oldest ancestral sequence, An63-VWF, suggests that all mammalian VWF sequences form HMW multimers. These data are in agreement with the observation of HMW multimers in zebrafish31 but not hagfish,1 suggesting trait emergence between 435 and 615 million years ago.
AnVWF shows conserved multimerization and human ADAMTS13 proteolysis. Purified recombinant AnVWF was analyzed for multimer formation using sodium dodecyl sulfate–agarose electrophoresis following incubation in the absence (A) or presence (B) of recombinant human ADAMTS13. (C) Percentage of VWF multimer cleavage was determined by densitometry before and after enzyme addition and normalized to background signal.
AnVWF shows conserved multimerization and human ADAMTS13 proteolysis. Purified recombinant AnVWF was analyzed for multimer formation using sodium dodecyl sulfate–agarose electrophoresis following incubation in the absence (A) or presence (B) of recombinant human ADAMTS13. (C) Percentage of VWF multimer cleavage was determined by densitometry before and after enzyme addition and normalized to background signal.
In circulation, the metalloprotease ADAMTS13 cleaves HMW multimers into smaller multimers and monomers. Throughout VWF evolution, however, ADAMTS13 proteolysis has undergone species-specific diversification abrogating cross-compatibility in some instances16 but not others.32 Interestingly, An70- and An88-VWF within the carnivore/ungulate and rodent lineages, respectively, exhibited ADAMTS13-mediated cleavage comparable to human VWF (Figure 2B-C), suggesting that substrate diversification occurred more recently. In contrast, early ancestral and primate lineage VWF molecules (An63-, An84-, and An101-VWF) all exhibited increased and near complete cleavage of HMW multimers (Figure 2C).
AnVWF specific–activity by VWF:GPIbM and VWF:RCo
VWF activates the GPIb/IX/V complex on platelets through the VWF-A1 domain. To measure VWF activity in vitro, ristocetin is added to uncoil the mechanosensory domain of VWF, independent of shear stress. However, ristocetin does not bind all mammalian VWF molecules. To test the activity of AnVWF molecules in a ristocetin-independent manner, the GP1bM assay was used. An70- and An88-VWF displayed 1.5- to 2-fold increased specific-activity over cohVWF. The VWF molecules An101-, An84-, and An63-VWF exhibit conserved but reduced platelet activation. These differences are likely due to amino acid replacements surrounding and within the A1 domain; however, there are no unique residue substitutions in the A1 domain of An101-VWF outside of the autoinhibitory modules (AIMs).33 Aggregation of washed human platelets in the presence of ristocetin revealed specific-activities similar to GPIbM determinations, with the exception of An88-VWF. Consistent with the observation that ristocetin does not uncoil mouse or rat VWF,17 An88-VWF within the rodent lineage displayed a 4.6-fold discrepancy in activity between these assays. Ristocetin does not uncoil canine or porcine VWF15,34 but is able to uncoil VWF from sheep and goats.17 Considering that ristocetin activation was observed with An70-VWF (preceding pig and sheep but not dog) (Figure 1A), our data suggest that the ristocetin binding sequence may have been lost in two or more evolutionary branches within the past 78 million years.
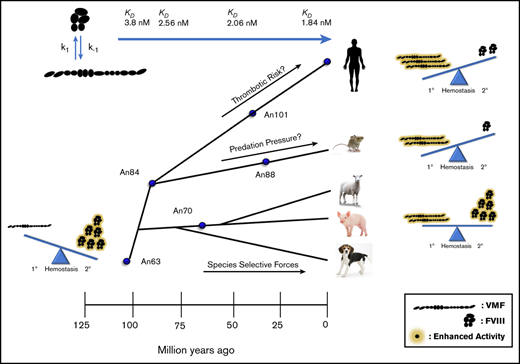
VWF and FVIII are critical components of primary and secondary hemostasis, respectively. A recent, large epidemiologic study highlights that the predominant association between the coagulation proteins and increased risk of venous thrombotic disease is a quantitative variance of FVIII and VWF levels.35 The critical balance between hemostasis and thrombosis has likely driven a strong selective pressure on the biological activity of the coagulation proteins. Strikingly, FVIII and VWF specific-activities are inversely correlated throughout evolution (Figure 3). Although this analysis of AnVWF activity is restricted to human GPIb/platelets, our data suggest that primate evolution may have promoted primary hemostatic coverage and reduced the effect of secondary hemostatic amplification, perhaps as a modulator of thrombosis. However, a deeper analysis of associated genes within hemostasis is necessary to adequately evaluate the global effect on the hemostatic process.
AnVWF specific-activity is reciprocal to ancestral FVIII. (A) Activities of AnVWF molecules were determined by VWF:GPIbM (red bars) and VWF:RCo (blue bars) and normalized to antigen levels to generate specific-activities. VWF:RCo data are presented as the mean of 2 purified AnVWF lots. Specific-activities of VWF (open squares) and FVIII (closed circles) from the primate (B) and rodent (C) lineages were plotted in relation to their predicted age of ancestry (million years ago [mya]) as determined by TimeTree. Specific-activities of ancestral FVIII were determined by one-stage coagulation activity normalized to absorbance after extinction coefficient correction. Specific-activities of orthologous FVIII protein was determined previously.5,12
AnVWF specific-activity is reciprocal to ancestral FVIII. (A) Activities of AnVWF molecules were determined by VWF:GPIbM (red bars) and VWF:RCo (blue bars) and normalized to antigen levels to generate specific-activities. VWF:RCo data are presented as the mean of 2 purified AnVWF lots. Specific-activities of VWF (open squares) and FVIII (closed circles) from the primate (B) and rodent (C) lineages were plotted in relation to their predicted age of ancestry (million years ago [mya]) as determined by TimeTree. Specific-activities of ancestral FVIII were determined by one-stage coagulation activity normalized to absorbance after extinction coefficient correction. Specific-activities of orthologous FVIII protein was determined previously.5,12
Coordinated nonsynonymous mutation of VWF and FVIII
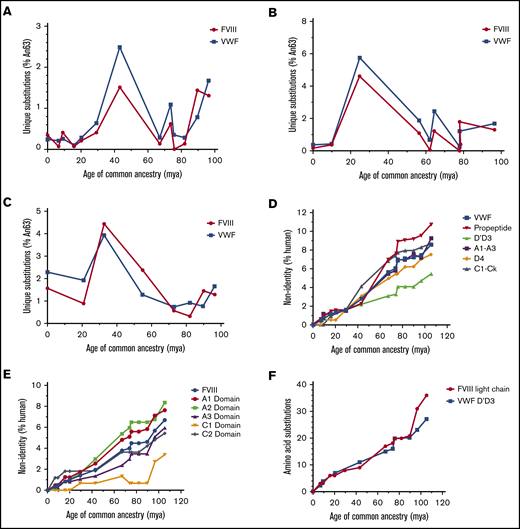
To identify the principal driver of coevolution, we analyzed the emergence of nonsynonymous mutations in ancestral VWF and ancestral FVIII sequences. Temporal analysis reveals a highly reciprocal pattern of mutation across all lineages (Figure 4A-C). The greatest number of substitutions in primate and ungulate VWF and FVIII was observed between 67 and 43 million years ago (Figure 4A). Rodent VWF and FVIII, however, have undergone continuous change over the past 100 million years (Figure 4C). Unlike the primate and ungulate lineages, which maintain a low percentage of new mutations during the past 10 million years (<0.4%), rodent FVIII and VWF exhibit increased substitutions of >1% and 2%, respectively, suggesting that different selective pressures and/or population sizes influence rodent coagulation factors.
Mammalian VWF and FVIII evolved under analogous selective pressure. Temporal emergence of unique amino acid substitutions diverging from the common ancestral sequence An63 are shown for primate (A), ungulate (B), and rodent (C) lineages. The number of unique residue differences is shown as the percentage of all amino acids. Total accumulation of mutations within specific subdomains of VWF (D) or FVIII (E) is shown as percent non-human relative to the size of each domain across primate evolution. Human VWF and FVIII define 0% nonidentity. (F) The number of amino acid substitutions within FVIII light chain or VWF D′D3 domains is shown for the primate lineage. mya, million years ago.
Mammalian VWF and FVIII evolved under analogous selective pressure. Temporal emergence of unique amino acid substitutions diverging from the common ancestral sequence An63 are shown for primate (A), ungulate (B), and rodent (C) lineages. The number of unique residue differences is shown as the percentage of all amino acids. Total accumulation of mutations within specific subdomains of VWF (D) or FVIII (E) is shown as percent non-human relative to the size of each domain across primate evolution. Human VWF and FVIII define 0% nonidentity. (F) The number of amino acid substitutions within FVIII light chain or VWF D′D3 domains is shown for the primate lineage. mya, million years ago.
Throughout primate evolution, the protein domains that experienced the greatest change are the VWF propeptide and the A1 and A2 domains of FVIII (Figure 4D-E). In the case of FVIII, this may provide an explanation for the differences in biosynthetic efficiencies and specific-activities previously observed across species. Conversely, the most conserved protein domains are the D′D3 domain of VWF and the C1 domain of FVIII, the latter of which has remained unchanged in primates for roughly 20 million years. The association of VWF and FVIII has been mapped to the D′D3 domain of VWF and the acidic a3 domain and C1/C2 domains of FVIII, respectively.36-38 Within rodent and ungulate lineages, the D′D3 VWF domains and the FVIII C1 domain are also the least disparate domains (supplemental Figure 3A-D). Notably, the rate of amino acid replacement throughout the primate VWF D′D3 domains parallels the FVIII light chain (Figure 4F).
FVIII-VWF affinity determination
We next investigated the evolution of the VWF-FVIII complex of each AnVWF molecule binding to each ancestral FVIII molecule (supplemental Figures 5 and 6; supplemental Table 1). Consistent with the high sequence conservation within the binding domains, each FVIII molecule bound VWF with low nanomolar to picomolar steady-state affinities (Table 2). We hypothesized that the greatest affinity would be observed between proteins of the same phylogenetic position (eg, human-human, An63-An63); however, this was only observed for the An88 FVIII-VWF pair. The strongest binding was observed between An70-VWF and human FVIII with a steady-state KD of 680 pM, and the weakest binding was observed between An63-VWF and human FVIII at 5.47 nM. Of the 36 protein interactions tested, An70-VWF exhibited 4 of the strongest affinity constants. An84- and An63-VWF together showed the 8 weakest affinity constants observed. An70- and An88-VWF exhibited the only significantly reduced KD values compared with the human-human KD (supplemental Table 2).
SPR steady-state KD determinations (nM)
| . | coh-VWF . | An101-VWF . | An84-VWF . | An63-VWF . | An70-VWF . | An88-VWF . |
|---|---|---|---|---|---|---|
| Human FVIII | 1.84 ± 0.19 | 2.35 ± 0.31 | 4.05 ± 0.14**** | 5.47 ± 0.41**** | 0.68 ± 0.15** | 1.14 ± 0.06 |
| An101-FVIII | 1.68 ± 0.25 | 2.06 ± 0.28 | 3.27 ± 0.30**** | 4.25 ± 0.30**** | 0.88 ± 0.23* | 1.13 ± 0.11 |
| An84-FVIII | 1.55 ± 0.09 | 1.87 ± 0.21 | 2.56 ± 0.23 | 2.90 ± 0.08 ** | 1.08 ± 0.30 | 1.02 ± 0.22 |
| An63-FVIII | 2.15 ± 0.28 | 2.62 ± 0.39 | 4.49 ± 0.60**** | 3.80 ± 0.29**** | 0.95 ± 0.10* | 1.71 ± 0.10 |
| An70-FVIII | 2.17 ± 0.29 | 2.67 ± 0.54 | 5.17 ± 0.33**** | 4.03 ± 0.39**** | 0.98 ± 0.02* | 1.84 ± 0.13 |
| An88-FVIII | 0.96 ± 0.08* | 1.01 ± 0.06 | 1.22 ± 0.27 | 1.17 ± 0.30 | 1.28 ± 0.29 | 0.77 ± 0.18** |
| . | coh-VWF . | An101-VWF . | An84-VWF . | An63-VWF . | An70-VWF . | An88-VWF . |
|---|---|---|---|---|---|---|
| Human FVIII | 1.84 ± 0.19 | 2.35 ± 0.31 | 4.05 ± 0.14**** | 5.47 ± 0.41**** | 0.68 ± 0.15** | 1.14 ± 0.06 |
| An101-FVIII | 1.68 ± 0.25 | 2.06 ± 0.28 | 3.27 ± 0.30**** | 4.25 ± 0.30**** | 0.88 ± 0.23* | 1.13 ± 0.11 |
| An84-FVIII | 1.55 ± 0.09 | 1.87 ± 0.21 | 2.56 ± 0.23 | 2.90 ± 0.08 ** | 1.08 ± 0.30 | 1.02 ± 0.22 |
| An63-FVIII | 2.15 ± 0.28 | 2.62 ± 0.39 | 4.49 ± 0.60**** | 3.80 ± 0.29**** | 0.95 ± 0.10* | 1.71 ± 0.10 |
| An70-FVIII | 2.17 ± 0.29 | 2.67 ± 0.54 | 5.17 ± 0.33**** | 4.03 ± 0.39**** | 0.98 ± 0.02* | 1.84 ± 0.13 |
| An88-FVIII | 0.96 ± 0.08* | 1.01 ± 0.06 | 1.22 ± 0.27 | 1.17 ± 0.30 | 1.28 ± 0.29 | 0.77 ± 0.18** |
SPR, surface plasmon resonance.
P < .05, **P < .005, ****P < .0001.
In addition to steady-state determinations, we performed kinetic affinity measurements but observed poor fits to a 1:1 binding model for An63-, An84-, or An101-VWF when complexed with the majority of FVIII molecules tested (supplemental Table 1; supplemental Figures 5 and 6). An88-FVIII exhibited strong 1:1 binding fits with 5 of the 6 VWF molecules tested, and both An70-VWF and An88-FVIII displayed binding characteristics that are predicted to be pharmaceutically advantageous.
AnVWF rescues FVIII in VWF−/− mice
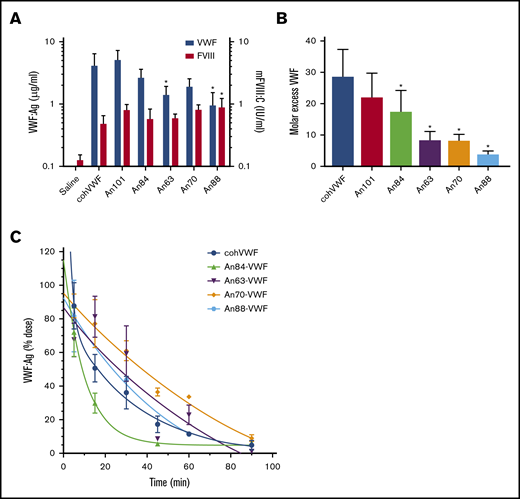
To determine if liver-derived AnVWF can functionally restore circulating levels of endogenous mouse FVIII (mFVIII) in vivo, linear DNA containing AnVWF sequences was hydrodynamically injected into VWF-deficient mice. VWF antigen levels ranged from 10% to 52% normal levels (Figure 5A). Production of An63- and An88-VWF were significantly lower than cohVWF production (P = .0357 and .013; Dunnett’s ANOVA). Endogenous mFVIII activity levels were subsequently increased from 13% in mice receiving saline, to a range of 49% to 90% normal. Mice injected with An88-VWF plasmid had significantly increased rescue of mFVIII despite significantly lower VWF concentrations (P = .0412; Dunnett’s ANOVA).
AnVWF rescues murine FVIII and exhibits reduced clearance in vivo. (A) VWF antigen (blue bars) and FVIII (red bars) activity was determined by ELISA and chromogenic assay, respectively, following hydrodynamic infusion of AnVWF plasmid DNA (n = 4) or saline (n = 9). Mice administered An88-VWF plasmid possessed significantly increased (*) FVIII activity (P = .0412; Dunnett’s ANOVA) despite reduced VWF present in circulation (P = .013; Dunnett’s ANOVA). (B) Molar ratios of VWF and FVIII were calculated. Molar excess of An84-, An63-, An70-, and An88-VWF was significantly reduced compared with cohVWF (*P < .05; Dunnett’s ANOVA). (C) Male VWF-deficient mice were administered 20 μg recombinant AnVWF. VWF antigen levels were determined by ELISA normalized to the initial dose via 2-phase decay, n = 3 per time point.
AnVWF rescues murine FVIII and exhibits reduced clearance in vivo. (A) VWF antigen (blue bars) and FVIII (red bars) activity was determined by ELISA and chromogenic assay, respectively, following hydrodynamic infusion of AnVWF plasmid DNA (n = 4) or saline (n = 9). Mice administered An88-VWF plasmid possessed significantly increased (*) FVIII activity (P = .0412; Dunnett’s ANOVA) despite reduced VWF present in circulation (P = .013; Dunnett’s ANOVA). (B) Molar ratios of VWF and FVIII were calculated. Molar excess of An84-, An63-, An70-, and An88-VWF was significantly reduced compared with cohVWF (*P < .05; Dunnett’s ANOVA). (C) Male VWF-deficient mice were administered 20 μg recombinant AnVWF. VWF antigen levels were determined by ELISA normalized to the initial dose via 2-phase decay, n = 3 per time point.
At these nonsaturating VWF concentrations, the molar ratio of cohVWF:mFVIII was calculated at roughly 29:1. An88-VWF is a rodent lineage VWF sequence and shares 94% amino acid identity to mouse VWF compared with 83% for human VWF. The concentrations of An88-VWF to mFVIII approached but did not reach equimolar levels (Figure 5B). Considering the reduced biosynthesis and specific-activity of murine FVIII and enhanced half-life of murine FVIIIa,5 the strong affinity of An88-VWF for An88-FVIII and 1:1 binding fit support the hypothesis that these proteins coevolved to most efficiently maintain FVIII levels in circulation. These data support further investigation of An88-FVIII in other models of VWF deficiency to assess if the FVIII/VWF stoichiometry is consistently more favorable.
Reduced clearance of recombinant AnVWF
Clearance of VWF is mediated by the asialoglycoprotein receptor (ASGPR), lipoprotein receptor–related protein-1 (LRP1), and siglec-5 on the surface of macrophages and hepatocytes, as well as CLEC4M and stabilin-2 on sinusoidal endothelial cells. Although clearance of VWF is largely mediated by N- and O-linked glycosylation, amino acid replacements such as the R1205H Vicenza variant also affect clearance rates. These observed replacements occur throughout the D, A, and C domains of VWF, with the greatest concentration occurring in the D′D3 and A1 domains. Pharmacokinetic analysis of cohVWF and An84-, An63-, An70-, and An88-VWF in VWF-deficient mice found terminal half-lives of 23.3, 7.0, 59.8, 70.4, and 35.4 minutes, respectively (Figure 5C). Therefore, An63- and An70-VWF show a 2.6- and 3.0-fold prolonged half-life after intravenous injection.
Discussion
ASR has enabled studies of molecular evolution of numerous gene products, including hormone receptors,39,40 metabolic enzymes,41 oxidases,42 and coagulation proteins,14,29 as well as phenylalanine/tyrosine ammonia-lyases, for pharmaceutical development.43 Investigations of resurrected proteins have revealed insights into mechanisms of evolution and epistasis, emerging complexity within enzyme systems, and global trends of diminishing thermostability and promiscuity of enzymes.43,44 Furthermore, this expanding field enables identification of sequence determinants responsible for novel biochemical functions for therapeutic protein engineering due to the limited number of replacements between branch points. In addition to direct identification of novel VWF sequences with therapeutically advantageous properties, this is the first known report of coevolution of interdependent mammalian therapeutic proteins supported by both phylogenetic analysis and biochemical characterization.
Investigating the molecular evolution of VWF proteins revealed an increase in biosynthetic efficiency coupled with increased specific-activity. These data suggest that primate evolution may have promoted primary hemostatic coverage and reduced the effect of secondary hemostatic amplification, at least from the perspective of platelet activation, as a potential modulator of thrombosis. Within all lineages, the temporal emergence of novel substitutions in either protein is mirrored by substitutions in the other, with most mutations occurring outside of the VWF-FVIII binding sites. The increased emergence of substitutions coincides with the Cretaceous–Paleogene extinction ∼66 million years ago. Although primate species, in contrast to ungulates, are not suggested to have greatly diversified during the Cretaceous–Paleogene extinction,45,46 our evidence suggests that primate hemostatic proteins underwent increased diversification at this time, possibly supporting primate development during the following Eocene and Oligocene epochs.
In our investigation of the VWF-FVIII complex, we observed a heavily conserved picomolar to nanomolar affinity across all FVIII and VWF combinations. This finding is consistent with the observations that the D′D3 domain of VWF and the C1 domain of FVIII are the most conserved domains across all lineages (supplemental Figure 3A-D), suggesting a strong negative selection on mutations abrogating their association. Interestingly, early VWF molecules An63, An84, and An101 exhibited poor 1:1 kinetic binding to FVIII. Instead, these VWF molecules more appropriately fit a bivalent model of binding, perhaps suggesting changes in the FVIII-VWF multimeric complex and stoichiometry. Although each monomer of VWF contains a binding site for FVIII, the observed stoichiometry of VWF to FVIII in circulation is 50:1.47 In our study of FVIII rescue in VWF-deficient mice (Figure 5A-B), we observed significant decreases in the molar excess of AnVWF, supporting the presence of an additional binding site or altered stoichiometry.
The domains of greatest substitution, and likely under positive selection, are the propeptide and A domains of VWF and the A1 and A2 domains comprising the heavy chain of FVIII. Characterization of FVIII through ortholog scanning and alanine mutagenesis has identified sequences within the heavy chain responsible for biosynthesis, secretion, and engagement of the unfolded protein response.6,48-50 Similarly, gene therapy studies codelivering the heavy and light chain of FVIII in trans show a stoichiometric imbalance favoring light chain synthesis.51 Ultimately, the key biochemical differences of FVIII across species are mediated primarily by amino acid residues within the heavy chain domains.5,7,9,12
A closer look at the VWF amino acid substitutions within A1 domain and AIMs of these ancestral sequences reveals a finite number of substitutions, which may account for increased specific-activity. Substitutions unique to An70- or An88-VWF within the A1 domain, residues 1272-1458, include E1292D, G1330A, K1332R, Q1353E, I1380V, M1393L, I1410T and D1333A, A1381V, and V1439L, respectively. Interestingly, none of these mutations is associated with VWD type 2B or any other subtype. Patients with type 2M and 2A were found to have D1302G or R1308H mutations52 ; however, these substitutions are observed in An63-, An84-, An70-, and An88-VWF sequences. Similarly, V1314F or V1314D are identified in type 2A and 2B diagnoses,53 although An63-, An84-, An70-, and An88-VWF have an isoleucine substitution at this residue. These data highlight the importance of epistasis considerations in protein engineering and pathology. Within the N-terminal AIM, unique substitutions Q1238R, V1244E, A1250G, and Y1271F or P1254S and D1269N are observed in An70- or An88-VWF, respectively. Additional substitutions D1249E, L1257P, I1262T, and S1263P are observed in all ancestral sequences. The C-terminal AIM contains unique substitutions A1464V, P1470R, and D1472L in An70 and M1473V found in both An70- and An88-VWF sequences. Of interest, D1472H, a variant that affects ristocetin-based activity measurements but not in vivo activity,54 is found in An101-VWF. At this residue, D1472Q is observed in An84-, An63-, and An88-VWF. Other residues of substitution within the CAIM include P1466A and L1469M/Q. None of these substitutions is reported in VWD pathologies but may influence the activity of in vitro VWF assays.
Within the D′ and D3 domains of VWF, several substitutions are observed that may affect the FVIII-VWF complex affinity. Within the D′ domain of An70- and An88-VWF, the molecules showing the strongest affinity for FVIII, these substitutions include M799V, M801T, R820K, and K833R, with only K833 residing within a structured β-sheet. Within the D3 domains, An70- and An88-VWF harbor I1001V and S1040V substitutions. As supported by our binding data, none of these mutations is associated with patient type 2N diagnoses.
As evidence of the pharmaceutical development potential of ASR, reconstruction of VWF sequences in this study resulted in proteins with enhanced and potentially advantageous biochemical characteristics. Specifically, An70-VWF exhibits increased platelet activation, increased affinity to FVIII, and a significantly prolonged half-life in mice, as well as a 3-fold improved binding affinity to human collagen (supplemental Figure 7). These characteristics are critical for pharmaceutical development of next-generation biologics and gene therapies for hemophilia A and VWD. However, despite high identity to human sequences, these proteins are feared to harbor immunogenic properties due to non-human epitopes.55-58 Currently, investigations of coagulant protein immunogenicity are limited by existing animal models; however, in silico predictions of B- and T-cell epitopes are increasing in power and accuracy. An analysis of T-cell epitopes within a human-porcine FVIII chimeric protein, termed ET3′, revealed no observable difference from the human FVIII epitope landscape,59 and a similar finding was observed for ancestral FVIII proteins for B-cell epitopes.14
Together, our study validates the utility of ASR for protein therapeutic engineering as well as direct therapeutic discovery. We provide evidence that FVIII and VWF coevolved throughout mammalian evolution and diversification under both positive and negative selection. These findings enable the development of novel protein complex therapeutics and gene therapies and provide a platform for the exploration of protein–protein interactions throughout evolution.
Requests for data sharing may be submitted to the corresponding author (Philip M. Zakas; e-mail: PZakas12@gmail.com). The multiple sequence alignment is electronically available upon request.
Acknowledgments
This work was partially supported by CIHR Foundation (grant FDN154285) (D.L.); National Institutes of Health, National Heart, Lung, and Blood Institute (grant HL137128) (C.B.D.); National Institutes of Health, National Institute of Arthritis, Musculoskeletal and Skin Diseases Research (grant R01AR069137) (E.A.G.); Department of Defense (grant MURI W911NF-16-1-0372) (E.A.G.); and Human Frontier Science Program (grant RGP0041) (E.A.G.).
Authorship
Contribution: P.M.Z., C.B.D., and D.L. conceived the study; P.M.Z., C.W.C., and C.B. performed the experiments; K.N. and C.D. performed in vivo procedures; A.B., M.B., B.S.-W., and C.K. performed binding studies; C.E.R. and E.A.G. performed the bioinformatics and molecular evolutionary analyses; P.M.Z., H.T.S, E.A.G., C.B.D., and D.L. wrote the manuscript; and all authors read and approved the manuscript submission.
Conflict-of-interest disclosure: P.M.Z., C.E.R., H.T.S., C.B.D., E.A.G., and D.L. are inventors on patent applications describing ancestral variants of coagulation factors. C.B.D. and H.T.S. are cofounders of Expression Therapeutics and own equity in the company. Expression Therapeutics licenses the intellectual property associated with ancestral coagulation factors. The terms of this arrangement have been reviewed and approved by Emory University and Georgia Institute of Technology in accordance with their conflicts of interest policies. The remaining authors declare no competing financial interests.
The current affiliation for P.M.Z. is Tessera Therapeutics, Cambridge, MA.
The current affiliation for E.A.G. is Department of Biology, Georgia State University, Atlanta, GA.
Correspondence: Philip M. Zakas, 345 Franklin St, Cambridge, MA 02139; e-mail: pzakas12@gmail.com.
References
Author notes
The full-text version of this article contains a data supplement.

![AnVWF specific-activity is reciprocal to ancestral FVIII. (A) Activities of AnVWF molecules were determined by VWF:GPIbM (red bars) and VWF:RCo (blue bars) and normalized to antigen levels to generate specific-activities. VWF:RCo data are presented as the mean of 2 purified AnVWF lots. Specific-activities of VWF (open squares) and FVIII (closed circles) from the primate (B) and rodent (C) lineages were plotted in relation to their predicted age of ancestry (million years ago [mya]) as determined by TimeTree. Specific-activities of ancestral FVIII were determined by one-stage coagulation activity normalized to absorbance after extinction coefficient correction. Specific-activities of orthologous FVIII protein was determined previously.5,12](https://ash.silverchair-cdn.com/ash/content_public/journal/bloodadvances/5/3/10.1182_bloodadvances.2020002971/2/m_advancesadv2020002971f3.png?Expires=1769824564&Signature=LwM2E-LRAMVoShV8Ul~G5TDuOA6Jd6VVIergY7ED9D~BJfal6fwEYoCafYA7QQfIUYNwOytH5dmPRdoU4~QwgTdcBfihnp2jKJjQ1YYP~18IHBYdZBEIkJEDAVIp5ip47HaNfqvG6XCRXPKqUngudCuTzCsl9qi99wN6-qzJgZ7s6U0tU03~p9ZSZlgZwmba1lx6BT2G3mpyNoaMnfxtorO5yb1WivAQRypPpNMIBDUTFuIvBJpRikED-~m4lP6b6jgbBp-1MkM6EBOUYfMY5mF0ckrK3E1vDhEay-lQhQs2s0V~4vExcheMfg24UUr9hI3WYzWh2co7yXQx~N33Hg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)