Key Points
Human iMφs show strong antimicrobial activity against S aureus in vitro and in vivo.
iMφs show accelerated and pronounced activation of a pro-inflammatory transcriptomic profile compared with monocyte-derived macrophages.
Abstract
Primary or secondary immunodeficiencies are characterized by disruption of cellular and humoral immunity. Respiratory infections are a major cause of morbidity and mortality among immunodeficient or immunocompromised patients, with Staphylococcus aureus being a common offending organism. We propose here an adoptive macrophage transfer approach aiming to enhance impaired pulmonary immunity against S aureus. Our studies, using human-induced pluripotent stem cell-derived macrophages (iMφs), demonstrate efficient antimicrobial potential against methicillin-sensitive and methicillin-resistant clinical isolates of S aureus. Using an S aureus airway infection model in immunodeficient mice, we demonstrate that the adoptive transfer of iMφs is able to reduce the bacterial load more than 10-fold within 20 hours. This effect was associated with reduced granulocyte infiltration and less damage in lung tissue of transplanted animals. Whole transcriptome analysis of iMφs compared with monocyte-derived macrophages indicates a more profound upregulation of inflammatory genes early after infection and faster normalization 24 hours postinfection. Our data demonstrate high therapeutic efficacy of iMφ-based immunotherapy against S aureus infections and offer an alternative treatment strategy for immunodeficient or immunocompromised patients.
Introduction
Patients with severe combined immunodeficiency often suffer from life-threatening bacterial, viral, and fungal infections because of profound defects in the development of lymphocytes, broadly affecting both cellular and humoral immunity.1 Other primary immunodeficiencies such as Wiskott-Aldrich syndrome, chronic granulomatous disease, STAT1 gain-of-function or hyperimmunoglobulin E syndrome are in particular characterized by impaired innate immunity with myeloid cell dysfunctions, predisposing patients to severe bacterial infections.2-4 Moreover, profound myeloid cell dysfunction can also be observed upon immunosuppressive treatment or in states of immune paralysis (eg, from severe influenza).
Among all pathogens, the gram-positive bacterium Staphylococcus aureus (S aureus), particularly methicillin-resistant S aureus (MRSA), is considered one of the major threats to immunocompromised patients,5-7 with the lungs being one of the most commonly affected organs. Despite the development of novel antibiotics, enhancing pulmonary immunity may be a promising treatment strategy to overcome acute respiratory infections in patients with severe combined immunodeficiency or other primary or secondary immunocompromised states.
The respiratory tract possesses a multistep immunity, wherein physical barriers such as mucosa and ciliary epithelial cells, in cooperation with innate and adaptive immune cells, protect the respiratory tract. Among all immune cells, macrophages are the most prevalent and constitute more than 95% of the cell population in bronchoalveolar space.8 Alveolar macrophages reside in the alveoli lumen and serve as essential regulators of lung tissue homeostasis and the first line of cellular immunity against respiratory pathogens.9 To this end, alveolar macrophages survey the lungs and orchestrate the engulfment of inhaled bacteria that evaded the respiratory tract's mechanical defenses.10 Following phagocytosis, macrophages eradicate bacteria within the phagosome by subsequent acidification and production of reactive oxygen and nitrogen species, antimicrobial proteins, and peptides. Activation of those microbicidal features promotes the secretion of pro-inflammatory cytokines and chemokines, antigen presentation, and activation of adaptive immune cells.11,12 Moreover, alveolar macrophages have the responsibility to balance and resolve the inflammatory response via clearing apoptotic neutrophils.13
Although antibiotics are the standard therapy against bacterial infections, the emergence of antibiotic-resistant strains and the prominent role of macrophages in pulmonary immunity suggest the adoptive transfer of macrophages as a promising therapeutic option to treat severe pulmonary bacterial infections. The bridge toward the immunotherapeutic application of macrophages to combat bacterial infections has been introduced by the intrapulmonary administration of human-induced pluripotent stem cell (iPSC)-derived macrophages in the context of gram-negative Pseudomonas aeruginosa-mediated pneumonia,14 which is typically seen in patients with cystic fibrosis. However, the overall feasibility of a macrophage-based immunotherapy in the context of primary or secondary immunodeficiencies and thereof associated gram-positive S aureus infection has not been investigated. Notably, macrophages sense gram-positive and gram-negative bacteria by different Toll-like receptors (TLRs), TLR2 and TLR4, respectively, which leads to differences in how these cells activate NF-κB and their subsequent bactericidal action.15
Thus, given the unmet clinical need and the high morbidity and mortality of S aureus in immunodeficient or immunocompromised patients, we aimed to elucidate the applicability of macrophages, which are derived from human iPSCs in the context of pulmonary S aureus infections in immunodeficient states.
In our study, we made use of human iPSCs and generated macrophages (iMφs) to study the antimicrobial efficacy in vitro using both a methicillin-sensitive and a methicillin-resistant clinical isolate of S aureus. Therapeutic effect in vivo was investigated following adoptive transfer of iMφs in S aureus-mediated pulmonary infection models using immunodeficient mice.
The present study proves the immunotherapeutic application of iMφs in the context of S aureus-mediated pulmonary infections, which could have a profound clinical impact for a variety of immunocompromised patients and beyond.
Materials and methods
Study approval
Mice.
Human interleukin-3 (IL-3)/granulocyte-macrophage colony-stimulating factor (GM-CSF) knock-in (KI) mice (Rag2tm1.1Flv Csf2/Il3tm1.1(CSF2,IL3)Flv Il2rgtm1.1Flv/J) were obtained from the Jackson Laboratory and housed in the central animal facility of Hannover Medical School. Mice were maintained under specific pathogen-free conditions in individually ventilated cages with free access to food and water. All animal experiments were approved by the animal welfare committee of lower Saxony (“Niedersächsisches Landesamt für Verbraucherschutz und Lebensmittelsicherheit/LAVES”) and performed according to its guidelines.
Human materials.
Buffy coat blood from anonymous healthy donors was purchased from “Deutsches Rotes Kreuz, DRK.” Generation and the use of cells derived from buffy coat blood samples and human iPSCs were approved by the local ethical committee (approval numbers: 1427-2012, 2127-2014, 1303-2012).
Generation of macrophages from peripheral blood (PBMφ)
Monocytes were isolated from buffy coat blood by double gradient centrifugation. The first density gradient was performed using Ficoll solution (Sigma-Aldrich) to separate mononuclear cells (460g for 45 minutes). The iso-osmotic Percoll solution (Sigma-Aldrich) was used for the second-density gradient to isolate monocytes from peripheral blood mononuclear cells (800g for 20 minutes at room temperature). Isolated monocytes were cultured in suspension culture plates in RPMI 1640 medium (Thermo Fisher) supplemented with 10% fetal calf serum, 1.5 mM NEAA, 2 mM l-glutamine, 1 mM sodium pyruvate, 0.05 mM β-mercaptoethanol, 1% penicillin-streptomycin, 10 ng/mL hM-CSF, and 10 ng/mL hIL3 (all Peprotech). After 7 days, the cells were cultured in 10 ng/mL hM-CSF for a further 5-7 days.
Generation of iMφs
The generation of iMφs was performed in 6-well tissue culture plate for in vitro experiments or in a stirred-tank bioreactor system for in vivo experiments as previously described.14 In brief, the DASbox Mini bioreactor system (Eppendorf) with a 250-mL glass vessel was used in this study.
For hematopoietic differentiation, iPS cells were expanded in 6-well tissue culture plates and, after reaching about 80% confluency, embryoid body (EB) formation was performed. After 5 days, EBs were selected by sedimentation. EBs were transferred to a 6-well suspension plate or the bioreactor vessel containing 3 or 120 mL of differentiation media (X-VIVO 15 [Lonza], 1 mM l-glutamine, 0.05 mM β-mercaptoethanol, and 1% penicillin-streptomycin, supplemented with 50 ng per milliliter M-CSF and 25 ng per milliliter IL-3), respectively. The differentiation in the bioreactor ran at 37°C with consistent gassing at 3 L per hour (21% O2; 5% CO2) and stirring at 50 rpm. The differentiation media was replaced every 7 days, whereas 1/4 volume of fresh differentiation media was added after 4 days. Macrophage production started around day 10 through 12 of differentiation and macrophages were manually collected weekly via sedimentation and subsequent filtration.
Phagocytosis assay
To assess the phagocytic activity of iMφs, 2.0 × 105 cells cultured in 24-well tissue culture plate were incubated with 10 µL pHrodo Red S aureus BioParticles Conjugate (Molecular Probes/Thermo Fisher Scientific) at 37°C, 5% CO2. Cells without BioParticles were used as control. At different time intervals of 2, 4, 6, and 24 hours, cells were harvested, washed, and analyzed by flow cytometry on a CytoFLEX S cytometer (Beckman Coulter). Data analysis was performed with FlowJo (TreeStar).
Bactericidal assay
The bactericidal ability of iMφs against S aureus strain Newman (GenBank accession no. AP009351),16 and a well-characterized MRSA (American Type Culture Collection 43300) was evaluated following standard methods. Therefore, 5.0 × 105 terminally differentiated iMφs were rinsed with phosphate-buffered saline (PBS) and cultured overnight in 1 mL RPMI 1640 medium supplemented with 10% fetal calf serum and 2 mM l-glutamine but without antibiotics, in a 12-well tissue culture plate. Subsequently, S aureus inoculum diluted in RPMI medium without antibiotics was centrifuged onto the cells (500g) and incubated at 37°C within a cell culture incubator. At different time intervals after the infection (0.5, 1, 3, 6, 9, and 24 hours), the supernatant was removed and, after rinsing the cells with PBS + 2 mM EDTA, a serial dilution of lysed cells and supernatants were plated on LB-agar. The plates were incubated overnight at 37°C and after 16 to 20 hours, the colonies were counted.
In vivo experiments
All animal experiments were approved by the local animal welfare committee and performed according to its guidelines. Male and female mice between the age of 70 and 90 days were randomized between the 3 experimental groups; infected (S aureus), infected and treated (S aureus+iMφ), and uninfected/PBS-treated (control). For intratracheal bacterial instillation, mice were anesthetized with intraperitoneal injection of ketamine/midazolam, and 1 × 107 colony-forming units (CFU) of the S aureus strain Newman were administered intratracheally through a venous catheter (Apocath Safety IV, 20G×11/4 in., Braun), as described before.17 Four hours postinfection, all mice were anesthetized using isoflurane inhalation, and 4 × 106 iMφs were intratracheally instilled through a catheter in S aureus+iMφ mice. Mice from the S aureus and control study groups were instilled with PBS. After 24 hours, the mice were sacrificed, and lungs were dissected for the analysis of bacterial loads, cell recruitment, and histology.
Statistical analysis
Statistical analysis was performed using the Prism 7 software (GraphPad) applying 1-way and 2-way analysis of variance (ANOVA). For all experiments, the mean ± standard deviation (SD) is plotted. *P < .05; **P < .01; ***P < .001; ****P < .0001.
Further methods can be found in the supplemental Information.
Results
iMφs demonstrate efficient phagocytosis and antibacterial activity in vitro
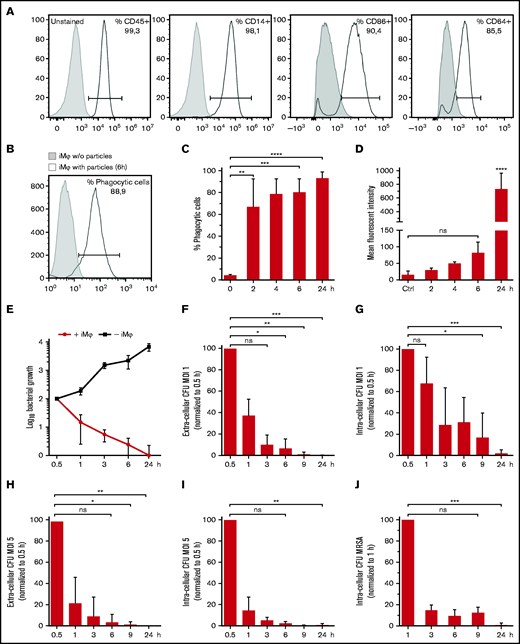
Using a scalable production platform,14 we generated iMφs representing a highly pure population coexpressing hematopoietic and monocyte/macrophage markers (CD45 and CD14, respectively). Moreover, CD64, the high‐affinity IgG receptor FcγR with function in both innate and adaptive immune responses and CD86, a costimulatory receptor for T-cell activation and survival, were expressed by iMφs (Figure 1A). Evaluating iMφs antibacterial activity in vitro revealed highly efficient phagocytic uptake of S aureus BioParticles conjugated to a pH-sensitive fluorescent dye (pHrodo). After 6 hours of incubation, 80 ± 13% (mean ± SD, n = 3) of iMφs had phagocytosed S aureus BioParticles (Figure 1B). Ongoing phagocytic activity was proven by an increasing frequency and mean fluorescence intensity over 24 hours (P < .0001, Figure 1C and 1D, respectively). Incubating iMφs with the viable S aureus strain Newman resulted in significantly restricted bacterial growth, whereas logarithmic bacterial growth in the absence of iMφs was observed (Figure 1E). In addition, iMφs infected with different multiplicities of infection (MOIs) of S aureus strain Newman (MOI 1 and 5) effectively cleared the extra- and intracellular bacteria within 24 hours postinfection (P < .001 and P < .01, respectively; Figure 1F-I). Likewise, iMφs demonstrated efficient intracellular clearance of a MRSA strain within 24 hours postinfection (P < .001; Figure 1J).
Characterization of iMφs and in vitro antibacterial activity of iMφs against S aureus. (A) Representative flow cytometric analysis of iMφs (gray filled: unstained; black line: respective antibody, a representative experiment of n = 3). (B-D) Phagocytosis of pHrodo red S aureus BioParticles. (B) Representative flow cytometry analysis (gray filled: iMφs without BioParticles; black line: iMφs with BioParticles after 6 hours at 37°C). (C) Frequency and (D) mean fluorescent intensity of BioParticle-positive iMφs 6 hours after incubation with pHrodo BioParticles at 37°C. (E) Growth of viable Newman S aureus bacteria in the presence and absence of iMφs. (F-J) Phagocytic and bactericidal ability of iMφs against S aureus Newman and MRSA. (F) Extracellular and (G) intra-cellular bacterial CFU after infecting cells with Newman at a MOI 1. (H) Extracellular and (I) intracellular bacterial CFU after infecting cells with Newman at a MOI 5. (J) Intracellular bacterial CFU after infecting cells with MRSA at a MOI 2. (C-J) Mean ± SD, n = 3. *P < .05; **P < .01; ***P < .001; ****P < .0001; ns, not significant, as determined by ordinary 1-way ANOVA with Dunnett’s multiple comparisons test (C,D) or Kruskal-Wallis ANOVA with Dunn’s correction analysis (F-J).
Characterization of iMφs and in vitro antibacterial activity of iMφs against S aureus. (A) Representative flow cytometric analysis of iMφs (gray filled: unstained; black line: respective antibody, a representative experiment of n = 3). (B-D) Phagocytosis of pHrodo red S aureus BioParticles. (B) Representative flow cytometry analysis (gray filled: iMφs without BioParticles; black line: iMφs with BioParticles after 6 hours at 37°C). (C) Frequency and (D) mean fluorescent intensity of BioParticle-positive iMφs 6 hours after incubation with pHrodo BioParticles at 37°C. (E) Growth of viable Newman S aureus bacteria in the presence and absence of iMφs. (F-J) Phagocytic and bactericidal ability of iMφs against S aureus Newman and MRSA. (F) Extracellular and (G) intra-cellular bacterial CFU after infecting cells with Newman at a MOI 1. (H) Extracellular and (I) intracellular bacterial CFU after infecting cells with Newman at a MOI 5. (J) Intracellular bacterial CFU after infecting cells with MRSA at a MOI 2. (C-J) Mean ± SD, n = 3. *P < .05; **P < .01; ***P < .001; ****P < .0001; ns, not significant, as determined by ordinary 1-way ANOVA with Dunnett’s multiple comparisons test (C,D) or Kruskal-Wallis ANOVA with Dunn’s correction analysis (F-J).
Adoptive transfer of iMφs ameliorates pulmonary S aureus infection in immunodeficient mice
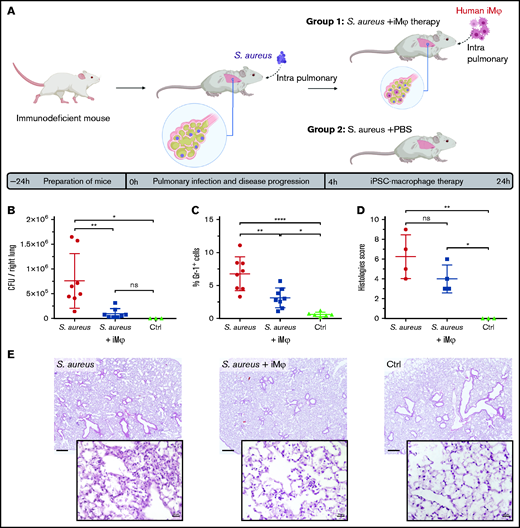
After showing profound antimicrobial activity in vitro, the therapeutic effect of iMφs on pulmonary infection was evaluated in vivo in Rag2tm1.1Flv Csf2/Il3tm1.1(CSF2,IL3)Flv Il2rgtm1.1Flv/J (hIL-3/GM-CSF KI) mice. Human IL-3/GM-CSF KI mice represent an immunodeficient mouse strain lacking B, T, and NK cells. Moreover, because of the absence of murine IL-3 and GM-CSF, myeloid cell development and function, in particular alveolar macrophages’ development, is hampered.18 Acute pulmonary infection was established in hIL-3/GM-CSF KI mice via intratracheal instillation of 1 × 107 CFU of S aureus strain Newman per mouse. Four hours postinfection, either 4 × 106 iMφs (S aureus+iMφ group) or PBS (S aureus and control groups) were intratracheally applied to animals. Mice were analyzed 24 hours postinfection (Figure 2A). Infected animals receiving iMφs (S aureus+iMφ) demonstrated a significant reduction in pulmonary bacterial load (P < .01). CFUs recovered from the lungs of iMφ-treated mice were reduced to approximately 10% of the S aureus load recovered from the lungs of untreated mice (9.8 ± 10.0 × 104 and 7.6 ± 5.5 × 105, respectively, mean ± SD, n = 8), and comparable to CFU levels in noninfected control mice (Figure 2B). Likewise, a significant decrease of pulmonary infiltration with murine Gr-1+ monocytes/granulocytes was observed in transplanted mice (S aureus+iMφ group) compared with the S aureus study group (P < .01, 3.1 ± 1.4% and 6.7 ± 2.5%, respectively, mean ± SD, n = 8; Figure 2C). Moreover, histopathological studies of the lungs showed less edema, less infiltration of neutrophils, and less tissue damage associated with a lower histology score in S aureus+iMφ mice compared with the S aureus group (Figure 2D,E).
Therapeutic effect of adoptively transferred iMφs in pulmonary infected immunodeficient mice. (A) Scheme of pulmonary infection and therapeutic transplantation of iMφs in hIL-3/GM-CSF KI immunodeficient mice. Animals were infected by intrapulmonary administration of 1 × 107 CFU of S aureus Newman. Four hours postinfection, the treatment group received 4 × 106 human iMφs intrapulmonary (S aureus+iMφ), whereas control mice received PBS (S aureus+PBS). (B) CFU per right lung and (C) frequency of murine monocyte/granulocyte (mGr-1+ cells) infiltration 24 hours postinfection (data from 2 independent experiments, individual values and mean ± SD, 3-8 animals/group). (D) Histopathology scores of the left lung tissue 24 hours postinfection (individual values and mean ± SD) and (E) representative microscopy images of hematoxylin-eosin staining of the lung tissue (original magnification ×2.5 for the top and ×40 for the bottom rows, scale bar = 500 and 20 µm, respectively). *P < .05; **P < .01; ****P < .0001; ns, not significant, as determined by 1-way ANOVA with Tukey’s multiple comparisons test.
Therapeutic effect of adoptively transferred iMφs in pulmonary infected immunodeficient mice. (A) Scheme of pulmonary infection and therapeutic transplantation of iMφs in hIL-3/GM-CSF KI immunodeficient mice. Animals were infected by intrapulmonary administration of 1 × 107 CFU of S aureus Newman. Four hours postinfection, the treatment group received 4 × 106 human iMφs intrapulmonary (S aureus+iMφ), whereas control mice received PBS (S aureus+PBS). (B) CFU per right lung and (C) frequency of murine monocyte/granulocyte (mGr-1+ cells) infiltration 24 hours postinfection (data from 2 independent experiments, individual values and mean ± SD, 3-8 animals/group). (D) Histopathology scores of the left lung tissue 24 hours postinfection (individual values and mean ± SD) and (E) representative microscopy images of hematoxylin-eosin staining of the lung tissue (original magnification ×2.5 for the top and ×40 for the bottom rows, scale bar = 500 and 20 µm, respectively). *P < .05; **P < .01; ****P < .0001; ns, not significant, as determined by 1-way ANOVA with Tukey’s multiple comparisons test.
iMφs demonstrate a strong and dynamic regulation of pro-inflammatory gene sets after pathogen contact
Of note, the scalable and continuous bioreactor production pipeline applied here for the generation of human iMφs can, in principle, be further developed for clinical-scale production of therapeutic cells. Thus, we aimed to further characterize the antimicrobial response of iMφs in comparison with PBMφs by analyzing cytokine secretion as well as transcriptomic changes by microarray analysis. Secretion of the pro-inflammatory cytokine IL-6, which also represents an important driver of immunity against S aureus infections,19 was measured after stimulating macrophages with lipopolysaccharide (LPS). Comparing iMφs and PBMφ pro-inflammatory response to LPS stimulation showed that iMφs were activated stronger and secreted higher levels of IL-6 (P < .05; Figure 3A).
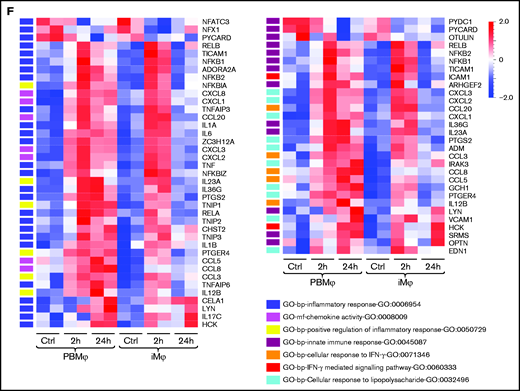
Whole transcriptome analysis of iMφs and PBMφs pre- and postinfection with S aureus. (A) Secretion of the proinflammatory cytokine IL-6 by iMφs and PBMφs before and 1 hour after stimulation with LPS. (B) Principal component analysis (PCA) of uninfected control (green) and infected (red) iMφs and PBMφs 2 hours postinfection. (C) Hierarchal heatmap clustering of differentially expressed genes (DEG, P < .05 and >2-fold change) between uninfected and 2 hour postinfection cells, and corresponding Venn diagram of shared upregulated genes 2 hours postinfection. (D) Word Cloud of the most prominent upregulated genes in iMφs associated with pro-inflammatory gene ontologies (regulation of NF-kappaB import into nucleus-GO:0042345; response to IL-1-GO:0070555; cellular response to lipopolysaccharide-GO:0032496; I-kappaB kinase/NF-kappaB signaling-GO:0007249; I-kappaB kinase/NF-kappaB signaling-inflammatory response-GO:0006954). Genes represented in more GOs are designated with a larger font. (E) Hierarchical heatmap clustering of 195 DEGs in cells 2 and 24 hours postinfection compared with uninfected cells (P < .05, >2-fold change). (F) Heatmaps representing the expression pattern of genes linked to selected GOs, comparing 2- and 24-hour infected cells to uninfected macrophages (P < .01, >2-fold change).
Whole transcriptome analysis of iMφs and PBMφs pre- and postinfection with S aureus. (A) Secretion of the proinflammatory cytokine IL-6 by iMφs and PBMφs before and 1 hour after stimulation with LPS. (B) Principal component analysis (PCA) of uninfected control (green) and infected (red) iMφs and PBMφs 2 hours postinfection. (C) Hierarchal heatmap clustering of differentially expressed genes (DEG, P < .05 and >2-fold change) between uninfected and 2 hour postinfection cells, and corresponding Venn diagram of shared upregulated genes 2 hours postinfection. (D) Word Cloud of the most prominent upregulated genes in iMφs associated with pro-inflammatory gene ontologies (regulation of NF-kappaB import into nucleus-GO:0042345; response to IL-1-GO:0070555; cellular response to lipopolysaccharide-GO:0032496; I-kappaB kinase/NF-kappaB signaling-GO:0007249; I-kappaB kinase/NF-kappaB signaling-inflammatory response-GO:0006954). Genes represented in more GOs are designated with a larger font. (E) Hierarchical heatmap clustering of 195 DEGs in cells 2 and 24 hours postinfection compared with uninfected cells (P < .05, >2-fold change). (F) Heatmaps representing the expression pattern of genes linked to selected GOs, comparing 2- and 24-hour infected cells to uninfected macrophages (P < .01, >2-fold change).
General transcriptome changes in macrophages upon S. aureus infection was assessed by principal component analysis (PCA; >5-fold change and P < .01). Macrophages from both cell sources separated into 2 clusters (1) before and (2) 2 hours postinfection with S aureus, suggesting a similar transcriptome for macrophages from different sources and similar inflammatory responses to pathogen contact (Figure 3B). However, substantial differences in the inflammatory transcriptomic profile were revealed upon detailed analysis. Compared with uninfected cells (control), the number of upregulated genes in iMφs 2 hours postinfection (>2-fold change and P < .05) was more than 10-fold higher than in PBMφ (288 vs 27, respectively; Figure 3C), with 276 genes being uniquely upregulated in iMφs. Upregulated genes in iMφs were associated with different inflammatory-related gene ontologies (GOs) (adjusted P value, 10-9-10-6), which included several upregulated pro-inflammatory chemokines and cytokines (eg, CCL3, CCL5, CCL16, CCL20, CXCL1, CXCL2, CXCL3, CXCL8, IL12B, IL36, IL23A, TNF, IL1A/B, and IL-6), as well as the inflammatory response transcription factor NF-κB (Figure 3D).
Interestingly, iMφs and PBMφs presented different gene expression patterns when analyzed 24 hours postinfection. Here, iMφs 24 hours postinfection showed a similar gene expression profile to uninfected iMφs and clustered together in the hierarchical heatmap analysis (P < .05 and >2-fold change). In bright contrast, PBMφ 24 hours postinfection maintained an inflammatory gene expression profile similar to cells 2 hours postinfection (Figure 3E), which becomes even more obvious when restricting the analysis to genes associated with inflammatory and innate immune responses (eg, GO-bp-inflammatory response-GO:0006954, GO-bp-innate immune response-GO:0045087, GO-bp-cellular response to interferon-γ -GO:0071346, and GO-bp-cellular response to lipopolysaccharide-GO:0032496; Figure 3F). Differentially expressed genes (DEGs) between iMφs and PBMφs 24 hours postinfection comprised inflammatory chemokines (CXCL1, CXCL2, CXCL3, CXCL8, and CCL3), cytokines (eg, IL1A/B, IL-6, and TNF), and NF-κB, which remained highly upregulated in PBMφs, thereby highlighting the persistence of a pro-inflammatory status.
The different pattern of inflammatory responses in iMφs and PBMφs 24 hours postinfection led us to a more detailed analysis to further prove the ability of iMφs to restore the stationary state, whereas PBMφs, still being pro-inflammatory activated. The expression of prominent pro-inflammatory cytokines and chemokines such as IL-6 and IL-1β, CXCL1, and CXCL8 reflected the different patterns between iMφs and PBMφs. The relative expression of those genes in iMφs was almost reduced to the level of control samples 24 hours postinfection, whereas PBMφs 2 and 24 hours postinfection represented similar expression values (supplemental Figure 1). The same inflammatory response pattern between iMφs and PBMφs was observed when the expression level of IL-6, IL-1β, and CXCL8 was evaluated with quantitative reverse transcriptase polymerase chain reaction using the same RNA samples applied in microarray analysis (Figure 4A).
Gene expression profile of iMφs and PBMφs 24 hours postinfection. (A) Quantitative reverse transcriptase polymerase chain reaction analysis of pro-inflammatory cytokines and chemokine; IL1-β, IL-6, and CXCL8 expression in noninfected macrophages (Ctrl), and S aureus infected macrophages 2 and 24 hours postinfection (2h and 24h), normalized to GAPDH (n = 2 biological replicates, mean ± SD). (B) Principal component analysis (PCA) of uninfected control (green) and infected (blue) iMφs and PBMφs 24 hours postinfection. (C) Hierarchal heatmap clustering of differentially expressed genes (DEGs, P < .05 and >2-fold change) between uninfected and 24-hour postinfection cells, and Word Cloud of upregulated genes 24 hours postinfection (all 15 genes in iMφs and top 27 genes in PBMφs). Genes represented in more GOs are designated with a larger font. Bar graph of top-ranked GOs (adjusted P values 10-3-10-5) related to genes upregulated in PBMφs 24 hours postinfection. (D) Number of DEGs between iMφs and PBMφ 2 and 24 hours postinfection in comparison with respective uninfected control cells.
Gene expression profile of iMφs and PBMφs 24 hours postinfection. (A) Quantitative reverse transcriptase polymerase chain reaction analysis of pro-inflammatory cytokines and chemokine; IL1-β, IL-6, and CXCL8 expression in noninfected macrophages (Ctrl), and S aureus infected macrophages 2 and 24 hours postinfection (2h and 24h), normalized to GAPDH (n = 2 biological replicates, mean ± SD). (B) Principal component analysis (PCA) of uninfected control (green) and infected (blue) iMφs and PBMφs 24 hours postinfection. (C) Hierarchal heatmap clustering of differentially expressed genes (DEGs, P < .05 and >2-fold change) between uninfected and 24-hour postinfection cells, and Word Cloud of upregulated genes 24 hours postinfection (all 15 genes in iMφs and top 27 genes in PBMφs). Genes represented in more GOs are designated with a larger font. Bar graph of top-ranked GOs (adjusted P values 10-3-10-5) related to genes upregulated in PBMφs 24 hours postinfection. (D) Number of DEGs between iMφs and PBMφ 2 and 24 hours postinfection in comparison with respective uninfected control cells.
Moreover, PCA analysis revealed clear segregation of uninfected PBMφs and PBMφs 24 hours postinfection, reflecting the high difference in gene expression. In contrast, control iMφs and 24-hour postinfection samples clustered close together (Figure 4B). Hierarchal heatmap analysis (P < .05 and >2-fold change) of these samples showed that 447 genes were differentially expressed between control and 24 hours postinfection PBMφ samples, which was more than 10-fold higher compared with iMφs (447 vs 36 genes). The majority of upregulated genes in 24-hours postinfection PBMφs compared with control samples were pro-inflammatory genes such as CXCL, IL-6, or TNF (see Word Cloud) and associated with GOs such as response to molecules of bacterial origin, response to cytokine or, regulation of NF-κB signaling. Although the 15 upregulated genes in 24 hours postinfection iMφs still comprised few pro-inflammatory genes (eg, CCL15 or EDN1 (see Word Cloud)), no clear association with inflammatory GOs was observed (Figure 4C).
Taken together, the number of DEGs between control and 2-hour postinfection samples was approximately 15-fold higher in iMφs compared with PBMφs, reflecting a higher pro-inflammatory activation of iMφs. On the other hand, 24 hours postinfection, the number of DEGs compared with control samples was approximately 13-fold lower in iMφs, reflecting the potential of iMφs to recover the stationary state after resolving the infection (Figure 4D).
Discussion
Antibiotic therapy represents the current gold standard for the treatment of bacterial infections. However, the rapid emergence of antibiotic-resistant strains together with the lack of innovation in the development of new antibiotics, highlight the need for new treatment strategies. Especially vulnerable groups, such as patients with primary or secondary immunodeficiencies are at a high risk of developing life-threatening infections. Given the important role of macrophages as the first line of cellular host defense, we developed an immunotherapy approach to complement antibiotic therapy and offer novel treatment options for these patients. Our data show that adoptive transfer of iMφs into the lungs of pulmonary infected mice can be a promising approach to enhance a debilitated immunity and to combat S aureus-mediated infections.
Although enhancing the immune system by cell-based approaches has revolutionized cancer immunotherapies in past years, the potential of macrophage-based cell therapies enhancing the immune system to combat (bacterial) infection has not yet been sufficiently explored. In fact, macrophage-based immunotherapies against bacterial infections offer the advantage that these innate immune cells, in contrast to T cells, recognize pathogens via unspecific receptors (eg, pattern recognition receptors)20 and are thus expected to induce an efficient immune response against a wide variety of pathogens. However, macrophage recognition patterns, bactericidal function, and cytokine secretion are different in response to gram-positive or gram-negative bacteria.21 We observed that besides similarities in the inflammatory response of iMφs to gram-negative and gram-positive bacteria, more inflammatory genes were upregulated after contact with S aureus compared with P aeruginosa. However, more detailed analysis would be needed to specifically address the mode of action of pathogen recognition and clearance because the observed differences in our studies could also be influenced by different time points of analysis (1 vs 2 hours postinfection). Importantly, showing antimicrobial efficacy of iMφs against the gram-positive pathogen S aureus in an immunodeficient mouse model extended on previous data demonstrating treatment of pulmonary infections caused by the gram-negative P aeruginosa14 highlights the broad applicability of this immunotherapy concept.
Enhancing the endogenous immunity by the adoptive transfer of monocyte/macrophages has also been proven for peritoneal infections caused by different (antibiotic resistant) bacteria such as Klebsiella pneumoniae or MRSA.22 Of note, the authors used preactivated primary murine bone marrow and human PB-derived macrophages, whereas we highlight the use of iPSC-derived macrophages, which can be generated in large quantities using scalable, industry-compatible differentiation systems14 as an off-the-shelf product. Importantly, iPSC technology also reduces the need for the availability of suitable donors and minimizes problems with a donor-to-donor variability by constantly providing macrophages in a defined quality. A further benefit of using iPSC-derived macrophages would also be the increased pro-inflammatory response of the cells to the bacterial stimulus. We demonstrate that our iMφs possess a higher antimicrobial activity (such as IL-6 secretion) and stronger induction of a pro-inflammatory transcriptome after pathogen contact compared with PBMφs. Notably, iMφs also quickly terminate this inflammatory program and return to a stationary state after the infection is resolved, which represents an important safety aspect for future clinical translation.
To prove our immunotherapeutic concept, we used hIL-3/GM-CSF KI mice, which represent a unique mouse model that lacks murine alveolar macrophages, but support the development and engraftment of human macrophages and permit the study of human mucosal immune responses to lung pathogens.18 Although the role of macrophages in the initiation, coordination, and termination of immune responses is manifold and involves the interplay with innate and adaptive immune cells,23 the immunodeficiency background of hIL-3/GM-CSF KI mice suggests phagocytosis and killing as the major mode of action of transplanted iMφs in our immunotherapy approach. Therefore, a macrophage-based therapy approach could be clinically relevant in patients suffering from primary or secondary immunodeficiencies and other diseases associated with dysfunctional macrophages. In fact, alveolar macrophages can also be paralyzed following, for example, sepsis, trauma)24 or from systemic immunosuppressive treatment, favoring the adoptive transfer of fully functional cells into the respiratory system to regenerate the immunity of the affected lungs also in these patient populations. Similarly, iMφs could also be applied to patients suffering from neutropenia-associated infections. Because the immunocompromised hosts are highly vulnerable to pulmonary infection following preconditioning, defined administration scenarios could be developed to (1) prevent or (2) rescue a transplanted host from severe infections. Following this concept and the broad clinical spectrum and cause of primary or secondary immunodeficiencies, either preventive or therapeutic administration of iMφs could be envisioned for the individual patient.
Of note, we showed profound therapeutic efficacy of iMφs in immunodeficient mice; however, further studies evaluating the effect of a macrophage-based therapy in a humanized mouse model (eg, reconstituted with human CD34+ hematopoietic stem/progenitor cells)18 would be of interest. This model would allow studying the interaction of the transplanted iMφs with the endogenous innate and adaptive immunity and would additionally support the applicability of the introduced immunotherapy concept beyond immunodeficient patients.
Given the primitive origin of alveolar macrophages and iMφs,25,26 the activation of gene sets associated with the alveolar macrophage phenotype has been shown in iMφs following intrapulmonary transfer,27,28 which proposes the iMφs as a suitable cell type for short- and long-term therapeutic interventions. The present study expands the field of cell-based immunotherapies targeting gram-positive bacteria; however, their use against other pathogens such as fungi and/or viral infections remains unexplored. The unique transcriptome profile of iMφs described here, in addition to other studies describing iMφs with anti-(myco) bacterial properties,29 suggest adoptive transfer of either naive or activated iMφs as an alternative treatment approach for infectious diseases, which could accompany standardized antibiotic therapy. In combination with unique attempts of universal iPSCs,30 and the potential to generate also other cells of the innate immunity,31,32 the production of a GMP-compliant and “off-the-shelf” cell products pave the way for new antimicrobial therapies.
Acknowledgments
The authors thank T. Buchegger, I. Gensch, D. Kloos, and V. Lutscher for excellent technical support and V. Woods for editing the text. Microarray data used or referred to in this publication were generated by the Research Core Unit Genomics at Hannover Medical School.
This work was supported by the Else Kröner-Fresenius-Stiftung (2016_A146) to M. Ackermann and the Federal Ministry of Education and Research in Germany (BMBF) (01EK1602A) to N.L. and R.Z. This study was funded by the REBIRTH Cluster of Excellence (EXC62, Deutsche Forschungsgemeinschaft [DFG]), the REBIRTH Research Center for Translational Regenerative Medicine (ZN3440, State of Lower Saxony, Ministry of Science and Culture [Nieders. Vorab]) to N.L. and R.Z. as well as European Society of Clinical Microbiology and Infectious Diseases to N.L. and the DFG under Germany’s Excellence Strategy – EXC 2155 ''RESIST'' (39087428) to D.V. and N.L. Further, this project has received funding from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation program (852178) to N.L.; additionally, R.Z. received funding for this project from the German Research Foundation (DFG) (ZW64/4-1 and KFO311/ZW64/7-1), the German Ministry for Education and Science (BMBF) (13N14086, 01EK1601A, 13XP5092B and 031L0249) and the European Union H2020 program to the project TECHNOBEAT (66724) and A.M. was supported by the “Deutsches Zentrum für Lungenfoschung DZL” (82DZL002B1).
Authorship
Contribution: A.R.H. performed research, analyzed and interpreted the data, and wrote the manuscript; B.F., M.H., J.L.S., and A.M. contributed in vivo experiments and edited the manuscript; F.M. contributed to the generation of macrophages from iPSC in bioreactor platform; S.G. and M. Abeln performed the histology study; R.Z. and D.V. contributed expertise and edited the manuscript; and M. Ackermann and N.L. contributed to study design, data interpretation, and writing the manuscript.
Conflict-of-interest disclosure: N.L., M. Ackermann, and R.Z. have filed a patent application (PCT/EP2018/061574) on the generation and use of iPSC-derived macrophages. The remaining authors declare no competing financial interests.
Correspondence: Nico Lachmann, Hannover Medical School, Carl-Neuberg-Str. 1, 30625 Hannover, Germany; e-mail: Lachmann.Nico@mh-hannover.de.
References
Author notes
M.A. and N.L. contributed equally to this work.
The authors declare that all data supporting the findings of this study are available within the article and its supplementary Information files or from the corresponding author on reasonable request: lachmann.nico@mh-hannover.de. Microarray data are deposited under accession number GSE179815 in Gene Expression Omnibus (GEO) database repository (https://www.ncbi.nlm.nih.gov/geo/).
The full-text version of this article contains a data supplement.