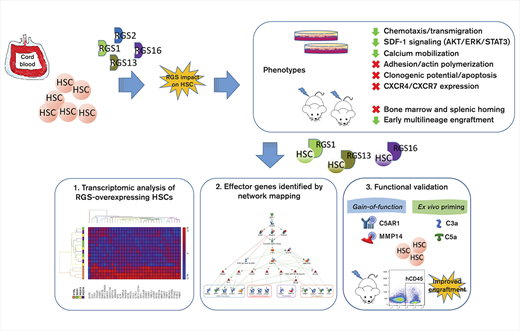
Key Points
Specific R4 RGS members are expressed in human HSPCs and regulated by the SDF-1/CXCR4 axis.
RGS1/13/16 suppress HSPC engraftment, SDF-1 signaling, and key effectors of stem cell trafficking/maintenance.
Abstract
Homing and engraftment of hematopoietic stem/progenitor cells (HSPCs) into the bone marrow (BM) microenvironment are tightly regulated by the chemokine stromal cell–derived factor-1 (SDF-1) and its G-protein–coupled receptor C-X-C motif chemokine receptor 4 (CXCR4), which on engagement with G-protein subunits, trigger downstream migratory signals. Regulators of G-protein signaling (RGS) are GTPase-accelerating protein of the Gα subunit and R4 subfamily members have been implicated in SDF-1–directed trafficking of mature hematopoietic cells, yet their expression and influence on HSPCs remain mostly unknown. Here, we demonstrated that human CD34+ cells expressed multiple R4 RGS genes, of which RGS1, RGS2, RGS13, and RGS16 were significantly upregulated by SDF-1 in a CXCR4-dependent fashion. Forced overexpression of RGS1, RGS13, or RGS16 in CD34+ cells not only inhibited SDF-1–directed migration, calcium mobilization, and phosphorylation of AKT, ERK, and STAT3 in vitro, but also markedly reduced BM engraftment in transplanted NOD/SCID mice. Genome-wide microarray analysis of RGS-overexpressing CD34+ cells detected downregulation of multiple effectors with established roles in stem cell trafficking/maintenance. Convincingly, gain-of-function of selected effectors or ex vivo priming with their ligands significantly enhanced HSPC engraftment. We also constructed an evidence-based network illustrating the overlapping mechanisms of RGS1, RGS13, and RGS16 downstream of SDF-1/CXCR4 and Gαi. This model shows that these RGS members mediate compromised kinase signaling and negative regulation of stem cell functions, complement activation, proteolysis, and cell migration. Collectively, this study uncovers an essential inhibitory role of specific R4 RGS proteins in stem cell engraftment, which could potentially be exploited to develop improved clinical HSPC transplantation protocols.
Introduction
Bone marrow (BM) transplantation has been an essential procedure for the treatment of various malignant and nonmalignant diseases, with the clinical outcome significantly dependent on the quality and quantity of hematopoietic stem and progenitor cells (HSPCs) in the graft. It has been determined that the number of CD34+ cells should be ≥1.5 × 105/kg body weight of the recipient when umbilical cord blood (CB) is used for transplantation, whereas >10 times the quantity is required when other sources of HSPCs are used.1,2 It has also been evident that higher doses of HSPCs could contribute to favorable engraftment kinetics, thus minimizing the potentially lethal complications after transplantation.3 However, the limited number of CD34+ cells in CB units and peripheral blood from poor mobilizers remains a critical issue to be addressed. A rational approach is to improve the homing and engraftment efficiency of infused HSPCs by manipulating their interaction with the BM niche.4 The chemotactic regulator stromal cell–derived factor-1 (SDF-1) and its C-X-C motif chemokine receptor 4 (CXCR4) have been well established as the prime mediators of HSPC homing, retention, and maintenance.5,6 Multiple pharmacologic approaches to enhance the SDF-1/CXCR4 axis, including priming of CB with complement component 3a (C3a),7 prostaglandin E2,8 or CD26/dipeptidyl peptidase-4 inhibitor,9 have been evaluated in clinical trials for their efficacy on improving the engraftment kinetics. Although some of these protocols might hold potentials for incorporation into standard clinical practice, it is of importance to comprehend novel mechanisms and explore new strategies to achieve the desired transplantation outcomes.
The mammalian regulators of G-protein signaling (RGS) protein family comprises 20 classical members, with a characteristic RGS domain of ∼120 amino acid residues. Based on sequence homology, these proteins are categorized into four main subfamilies (R4, R7, R12, and RZ).10 Some of them display a clear binding preference for a specific G-protein subunit, as well as subcellular and tissue locality, whereas others are more promiscuous in their selectivity and exhibit redundancy in downstream signaling and functions.11 Most studies searching for the roles of RGS proteins have been focusing on their GTPase-accelerating protein (GAP) activity, which acts downstream of G-protein–coupled receptors (GPCRs). A GPCR, on stimulation by its ligand, engages a heterotrimeric G-protein and promotes GDP to GTP exchange of the Gα subunit, resulting in dissociation from Gβγ and activation of downstream effector signals. RGS proteins bind with high affinity to the Gα subunit and accelerate the intrinsic GTPase activity by as much as 100-fold.12 By reducing the duration of GTP-Gα interaction with effectors, RGS proteins set a threshold for G-protein activation and facilitate the decay of signaling responses.13
The R4 subfamily members (RGS 1-5, 8, 13, 16, 18, and 21) are small proteins (except RGS3) with minimal N- and C-terminal extensions flanking the RGS domain. Since the discovery in the mid-1990s, incremental evidence has revealed their pivotal roles in both physiologic activities and pathologic conditions.14,15 In the hematopoietic system, specific R4 RGS proteins have been demonstrated to regulate B-cell, T-cell, and neutrophil migration,16-18 as well as platelet activation and megakaryocyte differentiation.19,20 Indeed, major chemoattractant receptors on HSPCs, including CXCR4,21 sphingosine-1-phosphate receptors (S1PRs),22 purinergic receptors,23 and complement peptide receptors,24 are GPCRs. These receptors engage specific Gα subunits to evoke migratory signals, raising the possibility of the involvement of R4 RGS proteins in the refined regulation of HSPC trafficking. In this report, we have documented the basal and SDF-1–induced expression profile of R4 RGS members in human CD34+ cells, highlighted their functional impact and mechanisms on stem cell migration, engraftment, and SDF-1/CXCR4 signaling, and laid potential strategies to enhance HSPC engraftment.
Methods
Samples
Unless otherwise specified, the sources of reagents are listed in supplemental Table 1. CB samples were collected from term infants during vaginal or cesarean deliveries. BM and granulocyte-colony stimulating factor (G-CSF) – mobilized peripheral blood (MPB) were collected from healthy donors during routine stem cell harvesting procedures before transplantation. All specimens were collected with informed written consent, following the Declaration of Helsinki. The study was approved by the Joint Chinese University of Hong Kong–New Territories East Cluster Clinical Research Ethics Committee.
Isolation of CD34+ cells
Mononuclear cells were isolated by density-gradient centrifugation on Ficoll-Paque Plus. CD34+ cells were enriched using the Indirect CD34 Microbead Kit according to the manufacturer's instructions. Enriched CD34+ cells were stained with fluorescein isothiocyanate (anti-CD34-FITC) and allophycocyanin (anti-CD45-APC) for 20 minutes at 4°C and characterized by flow cytometry (LSRFortessa; BD Biosciences, San Jose, CA). The average purities of CB, BM, and MPB CD34+ cells were 95.5 ± 4.1% (n = 96), 94.1 ± 5.5% (n = 16), and 88.7 ± 8.0% (n = 14), respectively.
Quantitative polymerase chain reaction
The basal mRNA expression of R4 RGS members in CB, BM, and MPB CD34+ cells was measured by quantitative reverse transcriptase-polymerase chain reaction (RT-qPCR). Total RNA was isolated by TRIzol reagent and RNeasy Micro Kit, and reverse transcribed using the High Capacity cDNA Reverse Transcription Kit. qPCR reactions were performed using exon-spanning Taqman probes and Taqman gene expression master mix, and run on the 7300 Real-Time PCR System (Applied Biosystems, Foster City, CA). The relative expression level of each RGS member was calculated by the comparative CT method and was normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) expression. The effects of SDF-1/CXCR4 on RGS expression were assessed by exposure of CD34+ cells to SDF-1 (100 ng/mL) in the presence or absence of AMD3100 (10 µg/mL).
Construction of lentiviral vectors
Human RGS1, RGS2, RGS13, and RGS16 full-length open reading frames (Open Biosystems, Huntsville, AL) were inserted into the pRSC-SFFV-E2A-GFP-Wpre (for all functional assays except calcium flux) or pRSC-SFFV-E2A-Crimson-Wpre lentiviral backbones by PCR cloning and verified by Sanger sequencing (ABI 3130 Genetic Analyzer; Applied Biosystem). The design of control and RGS-expressing vectors is depicted in supplemental Figure 1A. Expression of transgenes were driven by the strong SFFV promoter, which enables robust expression in human CD34+ cells.25 A standard calcium phosphate precipitation protocol was used for lentivirus production.26 Lentiviral vectors packaged in 293T cells (CRL-3216; ATCC, Manassas, VA) were concentrated 100× by high-speed centrifugation. The functional viral titers were determined by the transduction of HT1080 cells (CCL-121, ATCC) followed by flow cytometry analysis. The titers were of ≥4 × 107/mL. C3AR1, C5AR1, and MMP14 lentiviral vectors were produced with the same procedures.
Lentiviral transduction
CD34+ cells were cultured in Stemline II Hematopoietic Stem Cell Expansion Medium supplemented with 100 ng/mL of thrombopoietin, stem cell factor, and Fms-related tyrosine kinase 3 ligand. After prestimulation for 18 hours at 37°C, cells (2-5 × 105/well) were seeded in non-tissue culture (TC) treated 6-well plates precoated with 50 μg/mL of Retronectin and transduced with lentiviruses for 4 to 5 hours at a multiplicity of infection of 4 to 8. A second transduction was conducted 24 hours later. The transduction efficiency was determined by flow cytometry and expressed as the percentage of GFP+ or Crimson+ cells. With this optimized protocol, RGS1, RGS2, RGS13, and RGS16 were effectively overexpressed in CD34+ cells, with >75% transduction efficiency (supplemental Figure 1B), leading to an apparent increase in mRNA (supplemental Figure 1C) and protein levels (supplemental Figure 1D). No significant loss of cell viability (supplemental Figure 1E), proliferation kinetics (supplemental Figure 1F), and clonogenic capacity (supplemental Figure 1G) resulted from RGS overexpression.
Gene knockout
Clustered regularly interspaced short palindromic repeats, CRISPR/Cas9‐mediated gene knockout in HSPCs was performed as described elsewhere.27 Briefly, cultured CD34+ cells (1 × 106) were mixed with ribonucleoprotein complexes composing the Cas9 endonuclease and chemically modified single‐guide RNAs targeting human RGS1, RGS13, or RGS16 and electroporated using program U-008 with the Nucleofector 2b device (Lonza, Basel, Switzerland). The gene editing efficiencies were estimated at day 3 after nucleofection by Sanger sequencing of purified DNA, followed by inference of CRISPR edits analyses.28 The frequencies of resultant insertion/deletion ranged from 36% to 85%, implying considerable levels of functional knockout (supplemental Figure 2).
Migration assay
Transduced or electroporated CD34+ cells (1 × 105) were resuspended in 0.1 mL Iscove's modified Dulbecco's medium (IMDM) with 1% fetal bovine serum and seeded in the upper chamber of Transwells (6.5-mm diameter, 5-μm pore) with or without Matrigel or tumor necrosis factor (TNF)-α preactivated human umbilical vein endothelial cells (HUVECs). Assay medium (0.6 mL) containing 100 ng/mL SDF-1 was added to the bottom chamber. After 4-hour culture at 37°C, migrated cells were enumerated by flow cytometry.
Calcium flux
Transduced CD34+ cells (3 × 105) were resuspended in calcium assay buffer (Hank's Balanced Salt Solution with 20 mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid and 0.2% bovine serum albumin [BSA], pH 7.4) and loaded with 4 μM Fluo-3 AM in the presence of 0.04% pluronic acid for 30 minutes at 37°C.29 After a 1:5 dilution with assay buffer followed by incubation for 40 minutes, cells were washed and further equilibrated for 15 minutes. Fluo-3 fluorescence of the Crimson+ population at basal level and after challenge with SDF-1 (100 ng/mL) was monitored by flow cytometry over time.
Adhesion assay
Adhesion of CD34+ cells was performed on high-binding 96-well plates as previously described,30 with minor modifications. Briefly, plates were coated overnight with 20 μg/mL fibronectin at 4°C, followed by incubation for 2 hours at 37°C. Nonspecific binding was blocked by incubating with 2.5% BSA for 1 hour. Transduced CD34+ cells (1 × 105/well) were labeled with CellTracker Orange CMRA Dye and allowed to adhere to fibronectin for 45 minutes in the presence of SDF-1 (100 ng/mL). Nonadherent cells were removed by 3 washes with IMDM/0.2% BSA. Adherent cells were counted by fluorescence microscopy (EVOS XL Core; Thermo Scientific, Waltham, MA).
Actin polymerization
Transduced CD34+ cells (3 × 105) were stimulated with SDF-1 (100 ng/mL) in IMDM/0.1% BSA at 37°C for 1 minute. Reactions were stopped at 15-second intervals by adding 3 volumes of the fixation/permeabilization solution. Cells were washed and resuspended in Perm/Wash buffer, stained with phalloidin-Alexa Fluor 647 (6 U/mL) for 15 minutes, and analyzed by flow cytometry. F-actin levels of GFP+ cells are reported.
Cell proliferation, apoptosis, and colony-forming assays
Transduced CD34+ cells (1 × 104) were cultured in Stemline II/thrombopoietin/stem cell factor/Fms-related tyrosine kinase 3 ligand for 8 days. The total number of viable cells was enumerated every 2 days by Trypan blue exclusion. Apoptotic cell death was monitored right after transduction by Annexin V-APC and 7-aminoactinomycin D (7-AAD) staining. CD34+ cells (1 × 103) were resuspended in IMDM/2% fetal bovine serum, mixed with MethoCult H4434 Classic methylcellulose-based medium, and plated onto 35-mm dishes. Colonies derived from erythroid progenitors (colony-forming unit-erythroid [CFU-E] and burst-forming unit-erythroid [BFU-E]), granulocyte-macrophage progenitors (colony-forming unit − granulocyte, macrophage [CFU-GM] and (colony-forming unit − macrophage [CFU-M]), and multipotent progenitors (colony-forming unit −granulocyte, erythroid, macrophage, megakaryocyte [CFU-GEMM]) were scored at day 14.
Cell surface protein expression
Transduced CD34+ cells (1 × 105) were stained with APC-conjugated antibodies against C3AR1, C5AR1, CXCR4, CXCR7, and MMP14 for 20 minutes at 4°C. Cells were washed and resuspended in phosphate-buffered saline/0.5% BSA for flow cytometric analysis.
AKT, ERK, and STAT3 phosphorylation
Transduced CD34+ cells (3 × 105) were stimulated with SDF-1 (100 ng/mL) in IMDM/0.1% BSA at 37°C for 3 minutes. Reactions were quenched at the indicated time points. Fixed and permeabilized cells were stained with Alexa Fluor 647-conjugated antibodies against phosphorylated ERK1/2, AKT, or STAT3 and analyzed by flow cytometry. Reported values are percentages of phosphorylated species gated on GFP+ cells.
Western blotting
SDF-1–treated or transduced CD34+ cells were lysed in radioimmunoprecipitation assay buffer supplemented with protease inhibitors. Cell lysates (50 µg) were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Immunoblots were probed with antibodies against RGS1, RGS2, RGS13, or RGS16, with GAPDH as the loading control. The reactions were developed with peroxidase-conjugated secondary antibodies and the SignalFire Elite ECL Reagent. Images were captured with the Alliance Q9 Advanced Imaging Platform (Uvitec, Cambridge, UK), and analyzed by Image J (National Institutes of Health, Bethesda, MD).
Homing and engraftment assays
All animal experiments were conducted in accordance with procedures approved by the Institutional Animal Experimentation Ethics Committee. Immunodeficient NOD.CB17-Prkdcscid/J (NOD/SCID) mice were purchased from the Jackson Laboratory (Bar Harbor, ME) and bred in the Laboratory Animal Services Centre of The Chinese University of Hong Kong. Female mice of 8 to 11 weeks old were sublethally irradiated (375 cGy for homing, 300 cGy for engraftment; Gammacell 1000 Elite Irradiator, MDS Nordion, Ottawa, ON, Canada) and intravenously infused with 2.3 to 8.3 × 105 control or C3AR1-, C5AR1-, MMP14-, or RGS-overexpressing cells. In some experiments, CD34+ cells were primed ex vivo with recombinant C3a, C5a, or CCL1 (1 µg/mL) for 30 minutes or incubated with pertussis toxin (PTX; 1 µg/mL) or neutralizing antibody against VLA-4 (20 µg/mL) for 1 hour at 37°C before transplantation. Single-cell suspensions were prepared from recipient BM and spleen at 20 hours (for analysis of homing) or 8 to 16 weeks (for analysis of engraftment) after transplantation. After red blood cell lysis, nonspecific bindings were blocked by anti-mouse CD16/CD32 and human FcR Blocking Reagent. To evaluate stem cell engraftment, we analyzed total human hematopoietic cells and the multilineage subpopulations by flow cytometry using human-specific antibodies: anti-CD45-APC, anti-CD19-phycoerythrin (PE), anti-CD33-PE, anti-CD71-PE, and anti-CD34-PE. Dead cells were stained with 7-AAD and excluded from analysis. To assess stem cell homing, CD34+ cells were labeled with CellTracker Deep Red Dye before infusion. The presence of human cells in the transplant recipients was enumerated by flow cytometry and expressed as the number of CellTracker+ population per 1 × 106 cells acquired. All fluorescence-activated cell sorting data were processed and analyzed with FlowJo software.
Microarray
Amplification and labeling of RNA, hybridization to the GeneChip Human Gene 2.0 ST Array, staining, and scanning were performed according to the manufacturer's protocols (Affymetrix, Santa Clara, CA). The GeneChip Operating Software absolute analysis algorithm was applied to determine the expression of transcripts using the global scaling option. The data were normalized using the Robust Multi-array Average algorithm with Guanine and Cytosine (GC) background correction and log2 transformed by Partek Genomics Suite. Probe sets with absent calls in all conditions were excluded. Genes with consistent differential expression (fold change ≥ 1.5 between control and RGS-overexpressing CD34+ cells; P < .05) were subjected to functional enrichment and network construction using MetaCore.31 qPCR validation of differentially expressed genes was performed with CD34+ cells derived from the same samples used for microarray experiments (n = 4) and with those from independent samples (n = 4). The microarray data have been deposited in Gene Expression Omnibus (accession no. GSE98453).
Statistical analyses
Data were analyzed by paired or unpaired Student t test as indicated in the figure legends, with normality tested by the Kolmogorov-Smirnov method. All data are expressed as means ± standard error of the mean. P < .05 was considered statistically significant. Analyses were performed using SPSS.
Results
Expression profile of R4 RGS in human CD34+ cells and its regulation by SDF-1/CXCR4
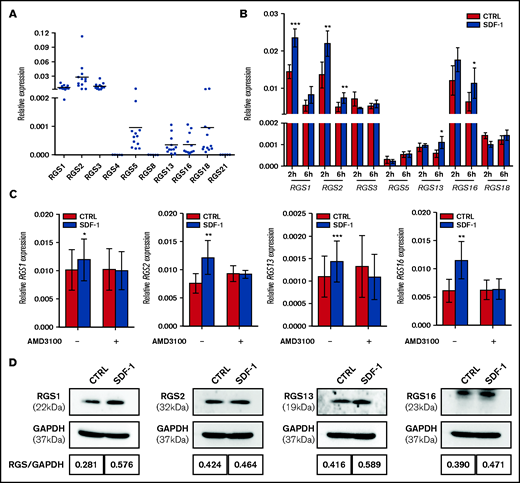
We first measured the basal mRNA expression of R4 RGS in 3 different sources of HSPCs. CD34+ cells derived from CB expressed high levels of RGS1, RGS2, and RGS3, whereas levels of RGS5, RGS13, RGS16, and RGS18 were comparatively lower. RGS4, RGS8, and RGS21 were undetectable (Figure 1A). The expression pattern of R4 RGS in BM or MPB CD34+ cells was, in general, similar to that in CB CD34+ cells. However, we observed a significantly higher expression of RGS1 (6-fold, P < .001), RGS13 (11.5-fold, P < .001), and RGS16 (63.7-fold, P = .001), as well as lower expression of RGS3 (0.5-fold, P < .001) and RGS18 (0.2-fold, P = .023) in BM CD34+ than in MPB CD34+ cells (supplemental Figure 3). A short exposure of CB CD34+ cells to the chemokine SDF-1 elevated the mRNA expression of RGS1, RGS2, RGS13, and RGS16 by 1.5- to 1.9-fold (P < .05; Figure 1B), which was averted by pretreatment with the CXCR4 antagonist AMD3100 (Figure 1C). Concordantly, SDF-1 increased the expression of these RGS members at the protein level (Figure 1D).
Expression of R4 RGS in human HSPCs and its regulation by the SDF-1/CXCR4 axis. (A) Differential mRNA levels of R4 RGS in CB-derived CD34+ cells (n = 12). (B) SDF-1 affects RGS mRNA expression. CD34+ cells were cultured in medium alone or in the presence of SDF-1 (100 ng/mL) for the indicated time points. Expression of R4 RGS was measured by RT-qPCR (n = 5). (C) AMD3100 blocks RGS induction. CD34+ cells were pretreated with AMD3100 (10 µg/mL) for 1 hour before stimulation with SDF-1 and quantified for R4 RGS expression (n = 6). Reported values are RGS expression relative to GAPDH. (D) SDF-1 affects RGS protein expression. CD34+ cells were cultured in the absence or presence of SDF-1 (100 ng/mL) for 24 hours. Expression of R4 RGS was measured by Western blotting. RGS/GAPDH intensity ratios are indicated. Statistics: 2-tailed, paired Student t test. *P < .05; **P < .01; ***P < .001.
Expression of R4 RGS in human HSPCs and its regulation by the SDF-1/CXCR4 axis. (A) Differential mRNA levels of R4 RGS in CB-derived CD34+ cells (n = 12). (B) SDF-1 affects RGS mRNA expression. CD34+ cells were cultured in medium alone or in the presence of SDF-1 (100 ng/mL) for the indicated time points. Expression of R4 RGS was measured by RT-qPCR (n = 5). (C) AMD3100 blocks RGS induction. CD34+ cells were pretreated with AMD3100 (10 µg/mL) for 1 hour before stimulation with SDF-1 and quantified for R4 RGS expression (n = 6). Reported values are RGS expression relative to GAPDH. (D) SDF-1 affects RGS protein expression. CD34+ cells were cultured in the absence or presence of SDF-1 (100 ng/mL) for 24 hours. Expression of R4 RGS was measured by Western blotting. RGS/GAPDH intensity ratios are indicated. Statistics: 2-tailed, paired Student t test. *P < .05; **P < .01; ***P < .001.
Impact of R4 RGS on SDF-mediated functions and signaling in CD34+ cells
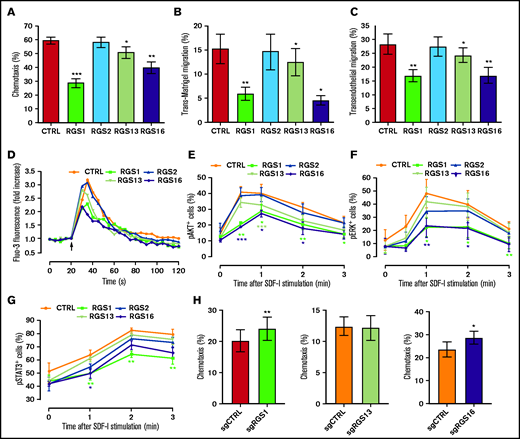
To delineate the influence of R4 RGS on HSPCs, we performed a series of SDF-1–directed, in vitro functional experiments in CB CD34+ cells after transduction with RGS-overexpressing lentiviral vectors. Of those members regulated by SDF-1, RGS1, RGS13, or RGS16 significantly reduced chemotaxis of CD34+ cells toward an SDF-1 gradient by 51.7% (P < .001), 14.3% (P = .039), and 33% (P = .006), respectively (Figure 2A) without altering the expression of SDF-1 receptors CXCR4 or CXCR7 (supplemental Figure 4A). Similarly, RGS1-, RGS13-, or RGS16-overexpressing CD34+ cells exhibited significantly reduced trans-Matrigel (Figure 2B) and transendothelial migration (Figure 2C). Adhesion to fibronectin, however, was not affected by RGS members (supplemental Figure 4B). Mobilization of intracellular calcium and polymerization of actin are early cellular responses to SDF-1 preceding HSPC migration.29,32 We observed that overexpression of RGS1 or RGS16, and to a lesser extent RGS13, suppressed SDF-1–induced calcium flux (Figure 2D) but not actin polymerization (supplemental Figure 4C) in transduced CD34+ cells. Phosphorylation of key signal transducers of the SDF-1/CXCR4 axis,33-35 including AKT (Figure 2E), ERK (Figure 2F), and STAT3 (Figure 2G) was also consistently inhibited by RGS1 or RGS16 (P < .05), whereas RGS13 only modestly affected AKT phosphorylation. None of the tested functions were influenced by RGS2. Reciprocally, knockout of RGS1 or RGS16 enhanced HSPC migration by 18.9% (P = .007) and 21.6% (P = .049; Figure 2H), further consolidating their functional importance.
R4 RGS suppress SDF-1–directed functions and signaling in HSPCs. (A-B) R4 RGS reduce HPSC motility. Chemotaxis transwell (A), trans-Matrigel (B), and transendothelial migration assays (C) were conducted using CD34+ cells transduced with control or RGS overexpression vectors in response to an SDF-1 gradient (n = 4-5). (D) R4 RGS decrease calcium flux. Transduced CD34+ cells were loaded with Fluo-3 AM and monitored for calcium mobilization before and after SDF-1 challenge. (E-G) R4 RGS inhibit SDF-1–mediated phosphorylation of signal transducers. Transduced CD34+ cells were stimulated with SDF-1 for the indicated time duration. Levels of phosphorylated AKT, ERK, and STAT3 were measured by intracellular staining with Phosflow antibodies (n = 4-5). (H) R4 RGS knockout reverted HSPC migration. CD34+ cells were electroporated with control sgRNA targeting the adeno-associated virus integration site 1 (AAVS1) or sgRNAs targeting R4 RGS to achieve loss of function, followed by chemotaxis assay (n = 4). SDF-1 at 100 ng/mL was applied for in vitro functional assays. Statistics: 2-tailed, paired Student t test. *P < .05; **P < .01; ***P < .001.
R4 RGS suppress SDF-1–directed functions and signaling in HSPCs. (A-B) R4 RGS reduce HPSC motility. Chemotaxis transwell (A), trans-Matrigel (B), and transendothelial migration assays (C) were conducted using CD34+ cells transduced with control or RGS overexpression vectors in response to an SDF-1 gradient (n = 4-5). (D) R4 RGS decrease calcium flux. Transduced CD34+ cells were loaded with Fluo-3 AM and monitored for calcium mobilization before and after SDF-1 challenge. (E-G) R4 RGS inhibit SDF-1–mediated phosphorylation of signal transducers. Transduced CD34+ cells were stimulated with SDF-1 for the indicated time duration. Levels of phosphorylated AKT, ERK, and STAT3 were measured by intracellular staining with Phosflow antibodies (n = 4-5). (H) R4 RGS knockout reverted HSPC migration. CD34+ cells were electroporated with control sgRNA targeting the adeno-associated virus integration site 1 (AAVS1) or sgRNAs targeting R4 RGS to achieve loss of function, followed by chemotaxis assay (n = 4). SDF-1 at 100 ng/mL was applied for in vitro functional assays. Statistics: 2-tailed, paired Student t test. *P < .05; **P < .01; ***P < .001.
Role of R4 RGS in homing and early engraftment of CD34+ cells
We next investigated whether the consequences of RGS overexpression observed in vitro can be recapitulated in an in vivo transplantation setting. Using the well-established NOD/SCID mouse xenotransplantation model,30,36 we found that homing of CD34+ cells to the recipient BM (supplemental Figure 5A) and spleen (supplemental Figure 5B) at 20 hours after infusion was in general not affected by RGS, except for a modest increase in BM homing of RGS2-overexpressing cells. However, overexpression of RGS1, RGS13, or RGS16, but not RGS2, markedly compromised BM engraftment of CD34+ cells at 8 weeks after transplantation by 91.3% (P = .021), 84.1% (P = .023), and 71% (P = .044), respectively, compared with animals transplanted with GFP-only control cells (Figure 3A-B). There were also trends of lower splenic engraftment in animals receiving RGS1-, RGS13-, or RGS16-overexpressing cells (Figure 3C). The lineage composition of the grafts in terms of CD19+ lymphoid cells, CD33+ myeloid cells, CD71+ erythroid cells, and CD34+ stem and progenitor cells was similar in the recipient BM (supplemental Figure 6). The suppression of engraftment by RGS1 was consistently observed following an extended monitoring time to 16 weeks after transplantation (supplemental Figure 7). To address the distinct impact of RGS on HSPC homing and engraftment, we treated CD34+ cells with PTX, a Gαi inhibitor, and tracked their short-term and long-term kinetics after transplantation. A marked inhibition of HSPC engraftment (supplemental Figure 8A) but not homing (supplemental Figure 8B) was realized, indicating that the former is a Gαi-dependent event. Furthermore, blocking integrin VLA-4 impaired homing of CD34+ cells (supplemental Figure 9), suggesting the reliance on adhesion molecules for this specific process.
R4 RGS inhibit early HSPC engraftment. Control or RGS-overexpressing CD34+ cells were intravenously infused into sublethally irradiated NOD/SCID mice. (A) Representative flow cytometry plots showing the identification of engrafted human hematopoietic cells in the murine BM. Human CD45+ cells in the recipient BM (B) and spleens (C) were enumerated by flow cytometry at 8 weeks after transplantation. Each data point represents the average engraftment level of 2 animals in a single experiment, with 8 to 9 independent experiments performed (ie, 16-18 animals/group). Statistics: 2-tailed, paired Student t test. P values are indicated.
R4 RGS inhibit early HSPC engraftment. Control or RGS-overexpressing CD34+ cells were intravenously infused into sublethally irradiated NOD/SCID mice. (A) Representative flow cytometry plots showing the identification of engrafted human hematopoietic cells in the murine BM. Human CD45+ cells in the recipient BM (B) and spleens (C) were enumerated by flow cytometry at 8 weeks after transplantation. Each data point represents the average engraftment level of 2 animals in a single experiment, with 8 to 9 independent experiments performed (ie, 16-18 animals/group). Statistics: 2-tailed, paired Student t test. P values are indicated.
Potential molecular mechanisms of R4 RGS-mediated decrease in engraftment of CD34+ cells
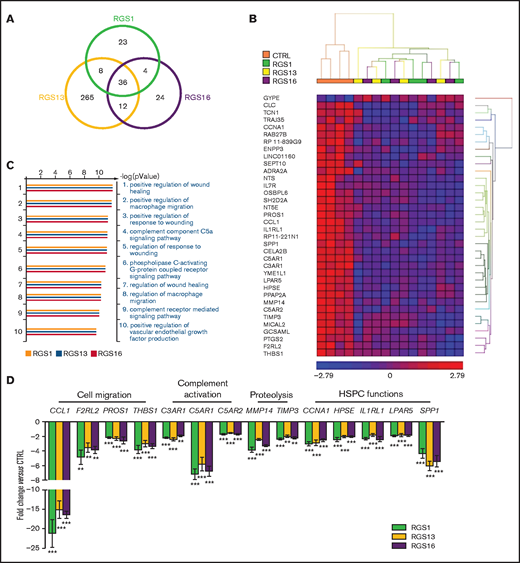
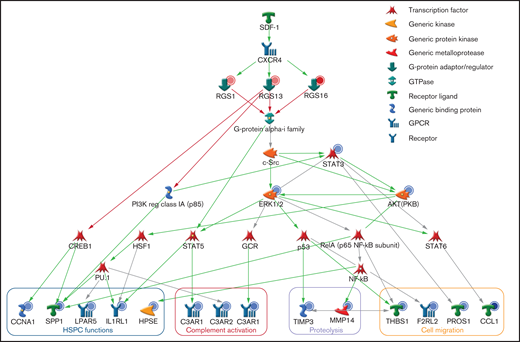
We performed a genome-wide expression microarray to explore the potential mechanisms accounting for the functional impact of R4 RGS in HSPCs. Compared with GFP-transduced control cells, overexpression of RGS1, RGS13, or RGS16 significantly altered the expression levels of 71, 321, and 76 genes in CD34+ cells (Figure 4A), of which, 23 genes (12 up- and 11 downregulated), 265 genes (115 up- and 150 downregulated), and 24 genes (10 up- and 14 downregulated) were exclusively altered by RGS1, RGS13, or RGS16, respectively, with their enriched functions shown in supplemental Table 2. Notably, there were 36 genes (1 up- and 35 downregulated) commonly altered by these RGS members (Figure 4B; supplemental Table 3), which possibly contain hits that can explain the phenotypic changes of CD34+ cells on RGS overexpression. Accordingly, gene ontology analyses revealed that these genes were enriched in functions related to cell migration, G-protein, and complement signaling (Figure 4C). Differential expression of 14 genes, chosen based on their relevance to stem cell trafficking/maintenance (supplemental Table 4), were validated by RT-qPCR. Levels of these target genes, subdivided into 4 major functional categories, were decreased by 1.5- to 21.3-fold in RGS-overexpressing cells (P < .01; Figure 4D). By incorporating our gene expression and functional data into a well-annotated database, we constructed an evidence-based network of RGS1, RGS13, and RGS16 regulatory signals along the SDF-1/CXCR4 axis (Figure 5). Based on this analysis, we propose a putative model as follows: Facilitated by initial binding to the Gαi complex, these RGS members could link to inactivation of signal transducers AKT, ERK, and STAT3, as well as various transcription factors, and mediate downregulation of effectors involved in stem cell functions (CCNA1, HPSE, IL1RL1, LPAR5, SPP1), complement activation (C3AR1, C5AR1, C5AR2), proteolysis (MMP14, TIMP3), and cell migration (CCL1, F2RL2, PROS1, THBS1). The concerted action of these cellular events could ultimately lead to impaired HSPC engraftment.
Gene expression profiles of RGS-overexpressing HSPCs. Expression microarray was performed on control or RGS1-, RGS13-, and RGS16-overexpressing CD34+ cells (n = 4). (A) Venn diagram showing the number of significantly deregulated genes in RGS-overexpressing cells. (B) Heat map showing the 36 differentially expressed genes commonly altered by the 3 RGS members. (C) Top 10 enriched gene ontology (biological processes) of RGS-regulated genes. (D) Differential expression of 14 target genes with relevance to stem cell trafficking/maintenance was validated by RT-qPCR (n = 8). Statistics: 2-tailed, paired Student t test. **P < .01; ***P < .001.
Gene expression profiles of RGS-overexpressing HSPCs. Expression microarray was performed on control or RGS1-, RGS13-, and RGS16-overexpressing CD34+ cells (n = 4). (A) Venn diagram showing the number of significantly deregulated genes in RGS-overexpressing cells. (B) Heat map showing the 36 differentially expressed genes commonly altered by the 3 RGS members. (C) Top 10 enriched gene ontology (biological processes) of RGS-regulated genes. (D) Differential expression of 14 target genes with relevance to stem cell trafficking/maintenance was validated by RT-qPCR (n = 8). Statistics: 2-tailed, paired Student t test. **P < .01; ***P < .001.
Proposed regulatory pathways of R4 RGS in HSPCs. The molecular network of RGS1, RGS13, and RGS16 downstream of SDF-1/CXCR4 and Gαi was generated by MetaCore with the integration of our gene expression and functional data. The red and blue circles represent up- and downregulation of gene expression/protein activity. The green, red, and gray arrows indicate positive, negative, and unspecified interactions between objects, respectively. The effector genes were categorized by functions.
Proposed regulatory pathways of R4 RGS in HSPCs. The molecular network of RGS1, RGS13, and RGS16 downstream of SDF-1/CXCR4 and Gαi was generated by MetaCore with the integration of our gene expression and functional data. The red and blue circles represent up- and downregulation of gene expression/protein activity. The green, red, and gray arrows indicate positive, negative, and unspecified interactions between objects, respectively. The effector genes were categorized by functions.
Genetic and pharmacologic means to alter specific effectors for enhancing HSPC engraftment
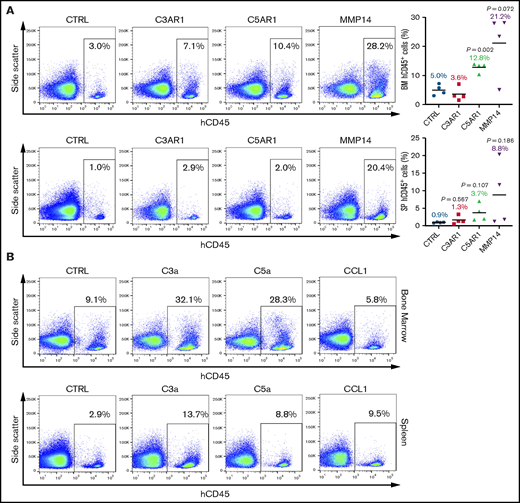
RGS-mediated downregulation of representative targets with predominant roles in HSPC trafficking, including C3AR1,24,37 C5AR1,38,39 and MMP14,40,41 were further validated at the protein level (supplemental Figure 10). To investigate whether reversing the expression of these effectors could improve hematopoietic reconstitution, we transduced CD34+ cells with lentiviral vectors (supplemental Figure 11A) to drive specific overexpression (supplemental Figure 11B) and transplanted the resulting cells into NOD/SCID mice to assess their engraftment potential. Convincingly, overexpression of C5AR1 and MMP14, but not C3AR1, markedly enhanced BM engraftment by 2.6- and 4.2-fold, and splenic engraftment by 4.1- and 9.8-fold, respectively (Figure 6A). Furthermore, a brief priming of CD34+ cells with the complement receptor ligands, namely C3a and C5a, also yielded a significant improvement of HSPC engraftment in the recipient BM by 3.5- and 3.1-fold and in spleens by 4.7- and 3.0-fold, respectively, whereas CCL1 priming only enhanced splenic engraftment (Figure 6B).
Overexpression of RGS-regulated effectors or priming with their ligands improve HSPC engraftment. CD34+ cells were (A) transduced with C3AR1, C5AR1, or MMP14 lentiviral vectors or (B) stimulated with C3a, C5a, or CCL1 (1 μg/mL; 30 minutes), and intravenously infused into sublethally irradiated NOD/SCID mice. Human CD45+ cells were enumerated in the recipient BM and spleens at 8 weeks after transplantation. Shown are representative flow cytometry plots illustrating the detection of engrafted hematopoietic cells. (A) Four mice per group. (B) Two mice per group. Statistics: 2-tailed, paired Student t test. P values are indicated.
Overexpression of RGS-regulated effectors or priming with their ligands improve HSPC engraftment. CD34+ cells were (A) transduced with C3AR1, C5AR1, or MMP14 lentiviral vectors or (B) stimulated with C3a, C5a, or CCL1 (1 μg/mL; 30 minutes), and intravenously infused into sublethally irradiated NOD/SCID mice. Human CD45+ cells were enumerated in the recipient BM and spleens at 8 weeks after transplantation. Shown are representative flow cytometry plots illustrating the detection of engrafted hematopoietic cells. (A) Four mice per group. (B) Two mice per group. Statistics: 2-tailed, paired Student t test. P values are indicated.
Discussion
This study has provided a glance on the expression profile of R4 RGS and their regulation by the SDF-1/CXCR4 axis in human CD34+ cells. In addition, functional and mechanistic evaluation have revealed their previously unrecognized roles in HSPC migration, SDF-1 signaling, and hematopoietic reconstitution that could be linked to the modulation of established effectors of stem cell engraftment, some of which with alterable genetic or pharmacologic approaches. Our data not only lay new knowledge on the biological functions of RGS family proteins but also shed light on the future design of new strategies to enhance clinical HSPC transplantation.
We observed a consistent expression pattern of R4 RGS in CD34+ cells, with detectable mRNA level of 7 members (RGS1, RGS2, RGS3, RGS5, RGS13, RGS16, and RGS18) regardless of the stem cell sources. However, substantially higher expression of RGS1, RGS13, and RGS16, as well as lower expression of RGS3 and RGS18, was evident in BM-resident than in G-CSF–mobilized HSPCs, suggesting that they are prone to regulation by niche or mobilization factors. Indeed, exposure of CD34+ cells to the BM-enriched chemokine SDF-1, albeit modest, induced a CXCR4-dependent elevation in RGS1, RGS2, RGS13, and RGS16. The different kinetics of RGS induction raises the possibility that these RGS members are interrelated and may differentially control SDF-1/CXCR4 signaling. It has been reported that other environmental cues in the stem cell niche, including hypoxia and transforming growth factor-β, could enhance the expression of RGS1 and RGS16 in CB CD34+ cells.42 Hypoxic conditions could also upregulate RGS1 in normal and malignant B cells.43 These data, together with ours, collectively suggest that the expression of specific RGS members is dynamically regulated in the BM niche during entry and exit of HSPCs and is likely driven by multiple factors known to maintain stem cell quiescence.44-46
In search of the functional significance of R4 RGS in HSPC, we primarily adopted a gain-of-function approach by overexpressing selected members in CD34+ cells, coupled with a comprehensive series of in vitro and in vivo experiments related to stem cell trafficking. We demonstrated that RGS1, RGS13, and RGS16 inhibit SDF-1-directed transmigration, calcium mobilization, and phosphorylation of AKT, ERK, and STAT3 without affecting CXCR4 or CXCR7 expression. These findings are in line with the only report addressing the role of hypoxia-induced RGS1 in CB CD34+ cells42 and consistent with emerging evidence pinpointing the definitive functions of R4 RGS members in migration of mature blood cells through negative regulation of the SDF-1/CXCR4 axis.16,18,19,47 Through xenotransplantation experiments, we provided compelling data revealing the profound influence of RGS members on the reconstitution of HSPCs, as demonstrated by the significantly compromised BM engraftment in NOD/SCID mice receiving RGS1-, RGS13-, or RGS16-overexpressing CD34+ cells without direct alterations in HSPC survival, proliferation, and clonogenic capacity. The lack of bias toward the production of specific lineages suggests that RGS may act by maintaining stem cell renewal at the expense of hematopoietic differentiation, which could be further tested through serial transplantation and limiting dilution assays. Interestingly, RGS did not affect adhesion and homing kinetics of CD34+ cells, possibly because of the occurrence of alternative mechanisms such as integrins VLA-4/VLA-5/LFA-1 and protein kinase C that were independent of Gα proteins,48,49 which we have provided confirmatory evidence through PTX and anti–VLA-4 blocking experiments. Another possibility is that homed HSPCs might not reach the BM parachyme and fail to lodge in their specialized niches for hematopoietic recovery. It should also be noted that RGS2, known to interact with Gαq but not Gαi,50,51 on the contrary, exhibited modest enhancement of HSPC homing, suggesting the existence of selectivity of RGS1, RGS13, and RGS16 to a particular Gα subunit, likely Gαi,12 for governing tracfficking of HSPCs.
By transcriptome profiling of CD34+ cells following overexpression of RGS1, RGS13, or RGS16, we provided new evidence that, in addition to their GAP activity, these RGS members could suppress the expression of myriad effectors putatively influencing HSPCs. The only gene that appeared to be upregulated by RGS was GYPE that encodes a poorly characterized red cell membrane glycophorin. Its functional significance remains to be determined as no preferential erythroid output was witnessed subsequent to RGS transduction. Notably, the complement cascade (C3AR1, C5AR1, C5AR2), as well as proteolytic enzymes and regulators (MMP14, TIMP3) are endowed with established functions in stem cell trafficking. Pharmacologic inhibition or genetic ablation of C3aR impaired engraftment of human and murine HSPCs24 and, conversely, enhanced G-CSF–induced mobilization.37 C5aR stimulation, on the other hand, could enhance egress of HSPCs by inducing granulocytic secretion of proteolytic enzymes.38,39 It has been demonstrated that MMP14-deficient mice displayed severe pancytopenia,40 and HSPCs from these animals exhibited poor engraftment in syngeneic transplantation.41 Although TIMP3 possesses inhibitory activity toward matrix metalloproteinases, it could enhance HSPC proliferation via stimulation of cell cycle entry.52 RGS1, RGS13, and RGS16 also inhibited CCNA1, HSPE, IL1R1, LPAR5, and SPP1, which have known roles in HSPC reconstitution,53 retention,54 migration,55 survival,56 and adherence to the endosteal niche.57 RGS expression, driven by niche factors, could therefore affect HSPC engraftment by suppressing a network of downstream effectors with diverse but essential functions. Indeed, we have provided corroborating data showing the probable mechanisms underlying RGS-controlled activities, as revealed by the marked enhancement of HSPC engraftment through overexpression of the identified effectors C5AR1 or MMP14 or priming with receptor agonists C3a or C5a, which warrant further exploitation.
In summary, this study revealed, for the first time, that the dynamic expression profiles of R4 RGS in human HSPCs dictated their pivotal roles in stem cell migration and reconstitution through modulation of critical effectors in the SDF-1 signaling pathway and delivered new methods to enhance HSPC engraftment. Considering primary trafficking receptors on HSPC are GPCRs, we envision that targeting novel GPCR regulators such as RGS could simultaneously alter multiple pathways, leading to accelerated hematopoietic recovery. Furthermore, the recent discovery of small molecule inhibitors of specific RGS members10,58 lays the foundation for configuring practical strategies to improve stem cell transplantation in the clinics.
Acknowledgments
This study was supported by research grants from the Health and Medical Research Fund, Food and Health Bureau, Hong Kong (project 01120706), the Children's Thalassaemia Foundation, Hong Kong (project 2017/01), the Innovation and Technology Fund, Innovation and Technology Commission, Hong Kong (project ITS/208/16FX), the Direct Grant for Research from The Chinese University of Hong Kong, Hong Kong (project 4054366), the H.K. Paediatric Bone Marrow Transplant Fund from The Chinese University of Hong Kong, Hong Kong (project 7105113), and the Sanming Project of Medicine, Shenzhen, China (project SZSM202011004).
The funding bodies were not involved in the study design, the collection, analysis and interpretation of data, or the decision to submit the manuscript for publication.
Authorship
Contribution: K.Y.Y.C., C.Z., Y.T.S.W., W.H.N., S.P.F., and H.W. performed experiments and analyzed data; X.-B.Z., P.M.K.T., W.K., B.F., E.N.Y.P., and C.C. provided advice on study design and contributed to essential laboratory reagents; C.C.W., K.Y.L., C.K.L., T.Y.L., M.H.L.N., K.F.T., and H.S.L. provided clinical samples; P.C.N., P.M.P.Y., K.L., A.W.K.L., C.K.L., and K.T.L. conceived the study, interpreted data and wrote the manuscript; and all authors reviewed and approved the final paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Chi Kong Li, and Kam Tong Leung, Doctor’s Office, 9/F, Tower B, Research Office, 8/F, Tower A, Hong Kong Children’s Hospital, Kowloon Bay, Kowloon, Hong Kong; e-mails: ckli@cuhk.edu.hk and ktleung@cuhk.edu.hk.
References
Author notes
K.Y.Y.C., C.Z., and Y.T.S.W. contributed equally to this study.
The microarray data have been deposited in Gene Expression Omnibus (Accession no. GSE98453).
The full-text version of this article contains a data supplement.