Key Points
We have used LR-PCR and NGS to completely sequence RHD genes in a variety of blood donors.
The results show correlation between intronic SNPs and common Rh haplotypes, thus establishing reference alleles.
Abstract
The Rh blood group system (ISBT004) is the second most important blood group after ABO and is the most polymorphic one, with 55 antigens encoded by 2 genes, RHD and RHCE. This research uses next-generation sequencing (NGS) to sequence the complete RHD gene by amplifying the whole gene using overlapping long-range polymerase chain reaction (LR-PCR) amplicons. The aim was to study different RHD alleles present in the population to establish reference RHD allele sequences by using the analysis of intronic single-nucleotide polymorphisms (SNPs) and their correlation to a specific Rh haplotype. Genomic DNA samples (n = 69) from blood donors of different serologically predicted genotypes including R1R1 (DCe/DCe), R2R2 (DcE/DcE), R1R2 (DCe/DcE), R2RZ (DcE/DCE), R1r (DCe/dce), R2r (DcE/dce), and R0r (Dce/dce) were sequenced and data were then mapped to the human genome reference sequence hg38. We focused on the analysis of hemizygous samples, as these by definition will only have a single copy of RHD. For the 69 samples sequenced, different exonic SNPs were detected that correlate with known variants. Multiple intronic SNPs were found in all samples: 21 intronic SNPs were present in all samples indicating their specificity to the RHD*DAU0 (RHD*10.00) haplotype which the hg38 reference sequence encodes. Twenty-three intronic SNPs were found to be R2 haplotype specific, and 15 were linked to R1, R0, and RZ haplotypes. In conclusion, intronic SNPs may represent a novel diagnostic approach to investigate known and novel variants of the RHD and RHCE genes, while being a useful approach to establish reference RHD allele sequences.
Introduction
The Rh blood group system (ISBT004) is the second most important blood group after ABO1,2 and one of the most polymorphic blood group systems. The RHD and the RHCE genes are located on chromosome 1 (1p33.1_1p36) and encode the RhD protein and the RhCcEe protein, respectively.3,4 The D antigen is the most clinically significant antigen in the Rh system due to its high immunogenicity and to being the main cause of hemolytic disease of the fetus and newborn (HDFN).5 The RHD and the RHCE genes show 93.8% homology in their introns and coding exons.6 The similarities between these 2 genes give an indication to their evolutionary rise from the same ancestral gene through duplication.6-8 Recombination, deletion, and point mutations in these 2 genes generate the 8 most common Rh haplotypes, which include: R1 (DCe), R2 (DcE), R0 (Dce), RZ (DCE), r (dce), ry (dCE), r′ (dCe), and r″ (dcE).9
Serological testing is fast, cost-friendly, and efficient; however, it is limited by many factors: for instance, the availability of antisera,10,11 reactivity of the antibodies, and antigen status (like weak or partial expression). Current assignment of a partial or a weak D phenotype would require an extensive collection of monoclonal anti-D. Monoclonals to low-frequency Rh antigens to identify specific partial D phenotypes are unavailable. Serological testing also leads to prediction of the Rh genotype based on the most common haplotype present in the population, which, for some cases, is incorrect.12
Unlike serological testing, genotyping provides the freedom to analyze a wider range of blood antigens including low-frequency antigens, for instance: Goa, BARC, and Tar which can cause HDFN and alloimmunization.1 Complete blood group genotyping (BGG) could be widely used in transfusion practice where serology fails to clarify issues or resolve discrepancies. Extensive efforts have been made to alternatively use molecular genotyping ranging from low to high throughput.13 Different DNA microarray-based tests were introduced that enable genotyping of variant blood groups by targeting specific single-nucleotide polymorphisms (SNPs).14-19 Although these assays are very accurate, they have limitations. They are designed to target predefined nucleotides or DNA regions through polymerase chain reaction (PCR), whereas novel variants remain unknown.13,20 Complete DNA sequencing could be the most effective technique to thoroughly study blood group variations and overcome limitations in other assays.20,21
Since it was introduced in 2005, next-generation sequencing (NGS) has greatly impacted the genetic research field by elevating both throughput and data generated, and at the same time lowering significantly the cost of sequencing per nucleotide.13,14,22-24 NGS is used in HLA testing,25 which creates a strong impetus to introduce NGS for BGG.26 Genotyping could be used to genotype blood transfusion–dependent patients who are at risk of alloimmunization.27-30 It could be used to genotype donors and create a database that would make finding and recall of compatible donors for transfusion easier.14,31-33 For these databases, reference sequences for all blood group genes are critical to allow effective BGG.26
Different studies have aimed to use NGS in BGG using a variety of approaches. Dezan et al,27 Chou et al,30 and Schoeman et al34 used exome sequencing to identify Rh variants but the high similarity between the RHD and RHCE genes makes it challenging to analyze data, especially in exons 8 and 10 where there are no differences between the 2 genes. Hyland et al35 used long-range PCR (LR-PCR) to amplify the RHD gene from exon 2 to exon 7 but omitted exons 1, 8, 9, and 10. We aimed to use LR-PCR to amplify the complete gene to get a full RHD sequence including promoter, introns, and all exons. The aim was to achieve full RHD sequencing to provide utility for RHD variant detection, with a follow-up in the future of full RHCE sequencing.
Rh-associated glycoprotein gene (RHAG; ISBT030) mutations have been linked to disturbed RhD expression.36-38 Therefore, we also aimed to sequence the RHAG gene for samples that showed weak D reactivity by serology and where no mutations in the RHD gene were detected.
All samples collected in our study were tested for RHD zygosity using droplet digital PCR (dPCR) to allow us to use a large number of hemizygous RHD samples to unequivocally establish reference alleles for the RHD gene. By studying intronic SNPs and their relationship to specific Rh haplotypes, it is clear that there is a significant difference between the R2 haplotype and other haplotypes.
Materials and methods
Sample collection and processing
Donor blood samples (n = 123) were supplied in EDTA tubes by the National Health Service Blood and Transplant (NHSBT; Bristol, United Kingdom). Inclusion criteria for blood samples was either their Rh haplotype (R1, R2, R0, RZ; n = 95) or by their D reactivity (weak D; n = 28). Samples were serologically phenotyped for ABO, Rh, and other blood groups by the NHSBT and were properly consented, anonymized, and supplied with full ethical approval. Blood tubes were centrifuged at 2500g for 10 minutes at room temperature. Plasma on the top layer was carefully disposed and buffy coat was collected into a 1.5-mL tube; the remaining content was discarded.
Genomic DNA extraction and zygosity testing
Genomic DNA (gDNA) was extracted from buffy coat using the QIAamp DNA Blood Mini kit (Qiagen Ltd) following the manufacturer’s guidelines. gDNA concentration was determined on the Qubit 2.0 Fluorometer (Life Technologies) using the Qubit double-stranded DNA High Sensitivity assay kit (Life Technologies). gDNA was finally stored at −20°C. Samples were tested for zygosity with the aim of knowing the number of RHD alleles present for subsequent sequence analysis. The RHD zygosity was determined for all samples using dPCR to determine whether a sample was hemizygous (Dd) or homozygous (DD).12,39 Samples were tested for RHD exon 5 (RHD5) and RHD exon 7 (RHD7) against the reference gene AGO1 on chromosome 1.12,39 The droplet reader in combination with QuantaSoft software v1.7.4 analyzed the droplet signals and differentiated between negative and positive ones, creating an absolute concentration of DNA. The number of RHD copies per microliter present in a sample was compared with the reference gene AGO1 copies per microliter.
Primer design
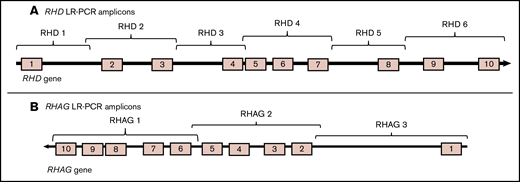
Six sets of primers (Table 1) were designed using the Primer3 software40 and CLC Main Workbench 9 software (Qiagen Ltd) to amplify the RHD gene in 6 LR-PCR amplicons (Figure 1), with ∼1 kb overlap between each of them. To eliminate amplification from the RHCE gene, primers were designed around intronic differences between the RHD and the RHCE genes positioned at the 3′ end to create RHD-specific primers. Even though exons 8 and 10 for the RHD and the RHCE genes are identical, there are intronic differences between the 2 genes that have been used to create RHD-specific primers. To ensure primer specificity, primers were assessed using Primer-BLAST on the National Center for Biotechnology Information (NCBI) website.41 In a similar manner, 3 sets of primers (Table 2) were designed to amplify the RHAG gene in 3 amplicons. The primers were ordered in a high-performance liquid chromatography purified form from Eurofins Genomics.
Sequence, exons covered, product sizes, and annealing temperature for primers used for RHD LR-PCR
| Primer name . | Sequence 5′-3′ . | Exons . | Size, bp . | Annealing temperature, °C . |
|---|---|---|---|---|
| RHD-1 forward | ATCCACTTTCCACCTCCCTGC | 1 | 10 326 | 62 |
| RHD-1 reverse | TCTTTGCACTTCTTCTGACAACA | |||
| RHD-2 forward | CTGGGAGAGTGAAGCTGGGTGTGA | 2, 3 | 13 709 | 62 |
| RHD-2 reverse | TTCATACACATCTCTACCCCCCCTC | |||
| RHD-3 forward | GTTTGAGCCCAGGAGTTAGGGACCGAG | 4 | 10 789 | 66 |
| RHD-3 reverse | CCCACTGTGACCACCCAGCATTCTA | |||
| RHD-4 forward | CATACCTTTGAATTAAGCACTTCAC | 5, 6, 7 | 9 895 | 66 |
| RHD-4 reverse | CAGAATGGCCTTTACCAGCCAT | |||
| RHD-5 forward | GTTCAAGCTGTCAAGGAGACATCACTATACA | 8 | 11 628 | 65 |
| RHD-5 reverse | CCAGTTTTAAGAATTTGTCGGCCGGTCG | |||
| RHD-6 forward | ATACATTCCATCCAGAACTGTTCACC | 9, 10 | 11 284 | 64 |
| RHD-6 reverse | AGGCCAAGAGATCCTGGTGAAACTATCC |
| Primer name . | Sequence 5′-3′ . | Exons . | Size, bp . | Annealing temperature, °C . |
|---|---|---|---|---|
| RHD-1 forward | ATCCACTTTCCACCTCCCTGC | 1 | 10 326 | 62 |
| RHD-1 reverse | TCTTTGCACTTCTTCTGACAACA | |||
| RHD-2 forward | CTGGGAGAGTGAAGCTGGGTGTGA | 2, 3 | 13 709 | 62 |
| RHD-2 reverse | TTCATACACATCTCTACCCCCCCTC | |||
| RHD-3 forward | GTTTGAGCCCAGGAGTTAGGGACCGAG | 4 | 10 789 | 66 |
| RHD-3 reverse | CCCACTGTGACCACCCAGCATTCTA | |||
| RHD-4 forward | CATACCTTTGAATTAAGCACTTCAC | 5, 6, 7 | 9 895 | 66 |
| RHD-4 reverse | CAGAATGGCCTTTACCAGCCAT | |||
| RHD-5 forward | GTTCAAGCTGTCAAGGAGACATCACTATACA | 8 | 11 628 | 65 |
| RHD-5 reverse | CCAGTTTTAAGAATTTGTCGGCCGGTCG | |||
| RHD-6 forward | ATACATTCCATCCAGAACTGTTCACC | 9, 10 | 11 284 | 64 |
| RHD-6 reverse | AGGCCAAGAGATCCTGGTGAAACTATCC |
TheRHD andRHAG genes amplified in overlapping LR-PCR amplicons. (A) Six overlapping RHD LR-PCR amplicons. (B) Three overlapping RHAG LR-PCR amplicons.
TheRHD andRHAG genes amplified in overlapping LR-PCR amplicons. (A) Six overlapping RHD LR-PCR amplicons. (B) Three overlapping RHAG LR-PCR amplicons.
Sequence, exons covered, product sizes, and annealing temperature for primers used for RHAG LR-PCR
| Primer name . | Sequence 5′-3′ . | Exons . | Size, bp . | Annealing temperature, °C . |
|---|---|---|---|---|
| RHAG-1 forward | TGGTAGGGCTGATTTCCTTGT | 6, 7, 8, 9, 10 | 10 003 | 62 |
| RHAG-1 reverse | TGGATGTTTTGGCCCAGCTT | |||
| RHAG-2 forward | GCTGATCTGAGGGTTACTCCTTT | 2, 3, 4, 5 | 10 519 | 62 |
| RHAG-2 reverse | AGGAGGATGGGAACGCTAAG | |||
| RHAG-3 forward | AATTATTCTGCAGATTTCACCCC | 1 | 15 083 | 62 |
| RHAG-3 reverse | GGAGACAAGAATTCCTCCACCTAT |
| Primer name . | Sequence 5′-3′ . | Exons . | Size, bp . | Annealing temperature, °C . |
|---|---|---|---|---|
| RHAG-1 forward | TGGTAGGGCTGATTTCCTTGT | 6, 7, 8, 9, 10 | 10 003 | 62 |
| RHAG-1 reverse | TGGATGTTTTGGCCCAGCTT | |||
| RHAG-2 forward | GCTGATCTGAGGGTTACTCCTTT | 2, 3, 4, 5 | 10 519 | 62 |
| RHAG-2 reverse | AGGAGGATGGGAACGCTAAG | |||
| RHAG-3 forward | AATTATTCTGCAGATTTCACCCC | 1 | 15 083 | 62 |
| RHAG-3 reverse | GGAGACAAGAATTCCTCCACCTAT |
LR-PCR optimization
To optimize PCR conditions, different annealing temperatures and primer concentrations were tested to ensure specific amplification from the target gene. In a 50-μL reaction, 1× master mix of LongAmp Hot Start Taq 2× Master Mix (New England Biolabs) was used with 200 ng of gDNA template; 1 μM of the forward and reverse primers was used for all amplicons except for RHD amplicon 3, where 0.2 μM of the forward and reverse primers was used. The Veriti Thermal Cycler (Applied Biosystems) program was set as follows: denaturation at 95°C for 5 minutes, 30 cycles of 95°C for 30 seconds, annealing for 30 seconds, and extension at 65°C for 10 minutes. Annealing temperature varied for each primer set (Tables 1 and 2). The last extension was at 65°C for 10 minutes; finally, samples were held at 4°C. To validate PCR amplification, PCR products were run on a 0.7% wt/vol agarose gel in 1× Tris-acetate-EDTA buffer next to a Quick-Load 1-kb Extend DNA Ladder (New England Biolabs).
Library construction, NGS, and data analysis
The LR-PCR products were purified using the Agencourt AMPure XP (Beckman Coulter). Purified amplicons were then quantified using Qubit double-stranded DNA High Sensitivity assay kit (Life Technologies) to create an equimolar pool to ensure an equal depth of coverage across the gene. Pooled amplicons were fragmented using the Ion Xpress Plus Fragment Library Kit (Life Technologies) to create a 200-base-read library and ligated to adaptors using the Ion Xpress Barcode Adapters kit (Life Technologies) following the manufacturer’s protocol. Size selection and library enrichment were carried out as by Sillence et al.12 The enriched library was then sequenced using the Ion PGM Sequencing 200 kit v2 (Life Technologies) and the Ion Torrent PGM on a 316 chip.
Data (FASTQ) were analyzed using CLC Main Workbench 9 software (Qiagen Ltd). Short reads were aligned to the human reference sequence hg38 downloaded from the NCBI database.42 The RHCE gene was masked in the RHD gene analysis by converting it into trimmed track to prevent reads from scattering. Variant detection was performed on a minimum coverage of 30 and variants detected were analyzed on a single-base basis considering different parameters including number and percentage of reads and nucleotide count.43 The reference SNP number44 was then found for each SNP detected.
Results
RHD zygosity
Samples (n = 123; Table 3) with different Rh genotypes presumed from serology results were first tested using dPCR to determine RHD zygosity. The presence or absence of the RHD amplification on the dPCR platform was used to determine whether the samples were RHD− or RHD+, respectively. Samples showing RHD5 or RHD7 to AGO1 ratios close to 1 were determined to be homozygous RHD+ and samples with ratios close to 0.5 were classified as hemizygous RHD+ (Table 3). Samples included 7 R1R1 (DCe/DCe), 21 R1r (DCe/dce), 7 R2R2 (DcE/DcE), 15 R2r (DcE/dce), 66 R1R2 (DCe/DcE), 6 R0r (Dce/dce), and 1 R2RZ (DcE/DCE) as determined by serology. Zygosity results were compatible with the serologically predicted genotype except for the following samples. Sample (004_14), previously classified by serology as being phenotypically R1r (DCe/dce), expressed ratios of 1.06 and 0.99 for the RHD5 and RHD7 multiplex reactions, respectively (Table 3). This result contradicted previous serological classifications and indicated that the sample expressed 2 copies of the RHD gene. Sample (004_42), previously classified by serology as being phenotypically R2R2, expressed ratios of 0.54 and 0.47 for the RHD5 and RHD7 multiplex reactions, respectively (Table 3). This result contradicted previous serological classifications and indicated that the sample expressed 1 copy of the RHD gene (hemizygous). In a similar manner, samples 004_35, 004_36, 004_37, 004_38, 004_39, and 004_40 were previously classified by serology as being phenotypically R1R2. However, given the ratios from the RHD5 (average 0.51) and RHD7 (average 0.51) multiplex reactions, these samples only express 1 copy of the RHD gene and are therefore classified as being RHD hemizygous. One R1R1 sample (004_07) showed discrepancy between hemizygous RHD5 (ratio 0.54) and homozygous RHD7 (ratio 1.01), indicating deletion of exon 5 in 1 of the RHD alleles.
Serologically predicted genotype, ethnicity of donors, dPCR RHD zygosity results, and RHD allele as determined by NGS for samples sequenced (n = 69)
| Sample no. . | Rh serology* . | Ethnicity* . | RHD5-to-AGO1 ratio . | RHD7-to-AGO1 ratio . | dPCR RHD zygosity . | Allele . |
|---|---|---|---|---|---|---|
| 004_01 | R1R1 | Caucasian | 1.12 | 1.05 | Homozygous | RHD*01 |
| 004_02 | R1R1 | Caucasian | 1.12 | 1.03 | Homozygous | RHD*01 |
| 004_03 | R1R1 | Other | 1.01 | 1.04 | Homozygous | RHD*01 |
| 004_04 | R1R1 | Caucasian | 1.07 | 1.03 | Homozygous | RHD*01 |
| 004_05 | R1R1 | Caucasian | 1.01 | 1.06 | Homozygous | RHD*01 |
| 004_06 | R1R1 | Caucasian | 0.99 | 1.04 | Homozygous | RHD*01 |
| 004_07 | R1R1 | Caucasian | 0.54 | 1.01 | Discrepancy† | RHD*01W.01 |
| 004_08 | R1r | Caucasian | 0.54 | 0.57 | Hemizygous | RHD*01 |
| 004_09 | R1r | Caucasian | 0.54 | 0.53 | Hemizygous | RHD*01 |
| 004_10 | R1r | Chinese | 0.51 | 0.56 | Hemizygous | RHD*01 |
| 004_11 | R1r | Caucasian | 0.53 | 0.54 | Hemizygous | RHD*01 |
| 004_12 | R1r | Caucasian | 0.55 | 0.52 | Hemizygous | RHD*01 |
| 004_13 | R1r | Caucasian | 0.54 | 0.6 | Hemizygous | RHD*01 |
| 004_14 | R1r | Caucasian | 1.06 | 0.99 | Homozygous‡ | RHD*01 |
| 004_15 | R1r | Caucasian | 0.53 | 0.50 | Hemizygous | RHD*01W.01 |
| 004_16 | R1r | Caucasian | 0.54 | 0.52 | Hemizygous | RHD*01W.01 |
| 004_17 | R1r | Caucasian | 0.58 | 0.56 | Hemizygous | RHD*01W.01 |
| 004_18 | R1r | Caucasian | 0.54 | 0.52 | Hemizygous | RHD*01W.01 |
| 004_19 | R1r | Caucasian | 0.54 | 0.47 | Hemizygous | RHD*01W.01 |
| 004_20 | R1r | Caucasian | 0.53 | 0.57 | Hemizygous | RHD*01W.01 |
| 004_21 | R1r | Caucasian | 0.52 | 0.57 | Hemizygous | RHD*01W.01 |
| 004_22 | R1r | Caucasian | 0.54 | 0.53 | Hemizygous | RHD*01W.01 |
| 004_23 | R1r | Caucasian | 0.52 | 0.53 | Hemizygous | RHD*01W.01 |
| 004_24 | R1r | Caucasian | 0.53 | 0.54 | Hemizygous | RHD*01W.01 |
| 004_25 | R1r | Caucasian | 0.53 | 0.51 | Hemizygous | RHD*01W.01 |
| 004_26 | R1r | Caucasian | 0.57 | 0.52 | Hemizygous | RHD*01W.01 |
| 004_27 | R1r | Caucasian | 0.56 | 0.54 | Hemizygous | RHD*01W.01 |
| 004_28 | R1r | Caucasian | 0.53 | 0.52 | Hemizygous | RHD*01W.03 |
| 004_29 | R1R2 | Caucasian | 1.09 | 1.03 | Homozygous | RHD*01 |
| 004_30 | R1R2 | Caucasian | 0.95 | 0.94 | Homozygous | RHD*01 |
| 004_31 | R1R2 | Caucasian | 1.08 | 1.05 | Homozygous | RHD*01 |
| 004_32 | R1R2 | Caucasian | 0.97 | 1.04 | Homozygous | RHD*01 |
| 004_33 | R1R2 | Caucasian | 1.03 | 1.08 | Homozygous | RHD*01 |
| 004_34 | R1R2 | Caucasian | 0.98 | 1.08 | Homozygous | RHD*01 |
| 004_35 | R1R2 | Caucasian | 0.46 | 0.51 | Hemizygous§ | RHD*01W.02 |
| 004_36 | R1R2 | Caucasian | 0.51 | 0.51 | Hemizygous§ | RHD*01 |
| 004_37 | R1R2 | Caucasian | 0.53 | 0.49 | Hemizygous§ | RHD*01 |
| 004_38 | R1R2 | Caucasian | 0.51 | 0.53 | Hemizygous§ | RHD*01 |
| 004_39 | R1R2 | Caucasian | 0.52 | 0.51 | Hemizygous§ | RHD*01 |
| 004_40 | R1R2 | Caucasian | 0.53 | 0.52 | Hemizygous§ | RHD*01 |
| 004_41 | R2R2 | Caucasian | 1.01 | 0.99 | Homozygous | RHD*01W.02 |
| 004_42 | R2R2 | Not disclosed | 0.54 | 0.47 | Hemizygous|| | RHD*01W.02 |
| 004_43 | R2R2 | Caucasian | 1.01 | 1.02 | Homozygous | RHD*01 |
| 004_44 | R2R2 | Caucasian | 1.01 | 0.99 | Homozygous | RHD*01 |
| 004_45 | R2R2 | Caucasian | 1.03 | 1.02 | Homozygous | RHD*01 |
| 004_46 | R2R2 | Caucasian | 1.02 | 1.01 | Homozygous | RHD*01 |
| 004_47 | R2R2 | Caucasian | 1.01 | 1 | Homozygous | RHD*01 |
| 004_48 | R2r | Caucasian | 0.53 | 0.54 | Hemizygous | RHD*01 |
| 004_49 | R2r | Caucasian | 0.53 | 0.51 | Hemizygous | RHD*01 |
| 004_50 | R2r | Caucasian | 0.53 | 0.51 | Hemizygous | RHD*01 |
| 004_51 | R2r | Caucasian | 0.52 | 0.56 | Hemizygous | RHD*01 |
| 004_52 | R2r | Caucasian | 0.48 | 0.54 | Hemizygous | RHD*01 |
| 004_53 | R2r | Caucasian | 0.53 | 0.47 | Hemizygous | RHD*01 |
| 004_54 | R2r | Caucasian | 0.52 | 0.51 | Hemizygous | RHD*01W.02 |
| 004_55 | R2r | Caucasian | 0.53 | 0.52 | Hemizygous | RHD*01W.02 |
| 004_56 | R2r | Caucasian | 0.53 | 0.5 | Hemizygous | RHD*01W.02 |
| 004_57 | R2r | Caucasian | 0.51 | 0.57 | Hemizygous | RHD*01W.02 |
| 004_58 | R2r | Caucasian | 0.58 | 0.56 | Hemizygous | RHD*01W.02 |
| 004_59 | R2r | Caucasian | 0.51 | 0.53 | Hemizygous | RHD*01W.02 |
| 004_60 | R2r | Caucasian | 0.57 | 0.52 | Hemizygous | RHD*01W.02 |
| 004_61 | R2r | Caucasian | 0.55 | 0.53 | Hemizygous | RHD*01W.02 |
| 004_62 | R2r | Caucasian | 0.58 | 0.52 | Hemizygous | RHD*01W.02 |
| 004_63 | R0r | Caucasian | 0.46 | 0.46 | Hemizygous | RHD*01 |
| 004_64 | R0r | Caucasian | 0.5 | 0.53 | Hemizygous | RHD*01 |
| 004_65 | R0r | Caucasian | 0.46 | 0.49 | Hemizygous | RHD*01 |
| 004_66 | R0r | Caucasian | 0.49 | 0.52 | Hemizygous | RHD*01 |
| 004_67 | R0r | Caucasian | 0.53 | 0.51 | Hemizygous | RHD*01 |
| 004_68 | R0r | Caucasian | 0.52 | 0.51 | Hemizygous | RHD*01 |
| 004_69 | R2RZ | Caucasian | 1.02 | 1.02 | Homozygous | RHD*01 |
| 004_70- 004_123 | R1R2 | — | 1.0¶ | 1.0¶ | Homozygous | Not sequenced |
| Sample no. . | Rh serology* . | Ethnicity* . | RHD5-to-AGO1 ratio . | RHD7-to-AGO1 ratio . | dPCR RHD zygosity . | Allele . |
|---|---|---|---|---|---|---|
| 004_01 | R1R1 | Caucasian | 1.12 | 1.05 | Homozygous | RHD*01 |
| 004_02 | R1R1 | Caucasian | 1.12 | 1.03 | Homozygous | RHD*01 |
| 004_03 | R1R1 | Other | 1.01 | 1.04 | Homozygous | RHD*01 |
| 004_04 | R1R1 | Caucasian | 1.07 | 1.03 | Homozygous | RHD*01 |
| 004_05 | R1R1 | Caucasian | 1.01 | 1.06 | Homozygous | RHD*01 |
| 004_06 | R1R1 | Caucasian | 0.99 | 1.04 | Homozygous | RHD*01 |
| 004_07 | R1R1 | Caucasian | 0.54 | 1.01 | Discrepancy† | RHD*01W.01 |
| 004_08 | R1r | Caucasian | 0.54 | 0.57 | Hemizygous | RHD*01 |
| 004_09 | R1r | Caucasian | 0.54 | 0.53 | Hemizygous | RHD*01 |
| 004_10 | R1r | Chinese | 0.51 | 0.56 | Hemizygous | RHD*01 |
| 004_11 | R1r | Caucasian | 0.53 | 0.54 | Hemizygous | RHD*01 |
| 004_12 | R1r | Caucasian | 0.55 | 0.52 | Hemizygous | RHD*01 |
| 004_13 | R1r | Caucasian | 0.54 | 0.6 | Hemizygous | RHD*01 |
| 004_14 | R1r | Caucasian | 1.06 | 0.99 | Homozygous‡ | RHD*01 |
| 004_15 | R1r | Caucasian | 0.53 | 0.50 | Hemizygous | RHD*01W.01 |
| 004_16 | R1r | Caucasian | 0.54 | 0.52 | Hemizygous | RHD*01W.01 |
| 004_17 | R1r | Caucasian | 0.58 | 0.56 | Hemizygous | RHD*01W.01 |
| 004_18 | R1r | Caucasian | 0.54 | 0.52 | Hemizygous | RHD*01W.01 |
| 004_19 | R1r | Caucasian | 0.54 | 0.47 | Hemizygous | RHD*01W.01 |
| 004_20 | R1r | Caucasian | 0.53 | 0.57 | Hemizygous | RHD*01W.01 |
| 004_21 | R1r | Caucasian | 0.52 | 0.57 | Hemizygous | RHD*01W.01 |
| 004_22 | R1r | Caucasian | 0.54 | 0.53 | Hemizygous | RHD*01W.01 |
| 004_23 | R1r | Caucasian | 0.52 | 0.53 | Hemizygous | RHD*01W.01 |
| 004_24 | R1r | Caucasian | 0.53 | 0.54 | Hemizygous | RHD*01W.01 |
| 004_25 | R1r | Caucasian | 0.53 | 0.51 | Hemizygous | RHD*01W.01 |
| 004_26 | R1r | Caucasian | 0.57 | 0.52 | Hemizygous | RHD*01W.01 |
| 004_27 | R1r | Caucasian | 0.56 | 0.54 | Hemizygous | RHD*01W.01 |
| 004_28 | R1r | Caucasian | 0.53 | 0.52 | Hemizygous | RHD*01W.03 |
| 004_29 | R1R2 | Caucasian | 1.09 | 1.03 | Homozygous | RHD*01 |
| 004_30 | R1R2 | Caucasian | 0.95 | 0.94 | Homozygous | RHD*01 |
| 004_31 | R1R2 | Caucasian | 1.08 | 1.05 | Homozygous | RHD*01 |
| 004_32 | R1R2 | Caucasian | 0.97 | 1.04 | Homozygous | RHD*01 |
| 004_33 | R1R2 | Caucasian | 1.03 | 1.08 | Homozygous | RHD*01 |
| 004_34 | R1R2 | Caucasian | 0.98 | 1.08 | Homozygous | RHD*01 |
| 004_35 | R1R2 | Caucasian | 0.46 | 0.51 | Hemizygous§ | RHD*01W.02 |
| 004_36 | R1R2 | Caucasian | 0.51 | 0.51 | Hemizygous§ | RHD*01 |
| 004_37 | R1R2 | Caucasian | 0.53 | 0.49 | Hemizygous§ | RHD*01 |
| 004_38 | R1R2 | Caucasian | 0.51 | 0.53 | Hemizygous§ | RHD*01 |
| 004_39 | R1R2 | Caucasian | 0.52 | 0.51 | Hemizygous§ | RHD*01 |
| 004_40 | R1R2 | Caucasian | 0.53 | 0.52 | Hemizygous§ | RHD*01 |
| 004_41 | R2R2 | Caucasian | 1.01 | 0.99 | Homozygous | RHD*01W.02 |
| 004_42 | R2R2 | Not disclosed | 0.54 | 0.47 | Hemizygous|| | RHD*01W.02 |
| 004_43 | R2R2 | Caucasian | 1.01 | 1.02 | Homozygous | RHD*01 |
| 004_44 | R2R2 | Caucasian | 1.01 | 0.99 | Homozygous | RHD*01 |
| 004_45 | R2R2 | Caucasian | 1.03 | 1.02 | Homozygous | RHD*01 |
| 004_46 | R2R2 | Caucasian | 1.02 | 1.01 | Homozygous | RHD*01 |
| 004_47 | R2R2 | Caucasian | 1.01 | 1 | Homozygous | RHD*01 |
| 004_48 | R2r | Caucasian | 0.53 | 0.54 | Hemizygous | RHD*01 |
| 004_49 | R2r | Caucasian | 0.53 | 0.51 | Hemizygous | RHD*01 |
| 004_50 | R2r | Caucasian | 0.53 | 0.51 | Hemizygous | RHD*01 |
| 004_51 | R2r | Caucasian | 0.52 | 0.56 | Hemizygous | RHD*01 |
| 004_52 | R2r | Caucasian | 0.48 | 0.54 | Hemizygous | RHD*01 |
| 004_53 | R2r | Caucasian | 0.53 | 0.47 | Hemizygous | RHD*01 |
| 004_54 | R2r | Caucasian | 0.52 | 0.51 | Hemizygous | RHD*01W.02 |
| 004_55 | R2r | Caucasian | 0.53 | 0.52 | Hemizygous | RHD*01W.02 |
| 004_56 | R2r | Caucasian | 0.53 | 0.5 | Hemizygous | RHD*01W.02 |
| 004_57 | R2r | Caucasian | 0.51 | 0.57 | Hemizygous | RHD*01W.02 |
| 004_58 | R2r | Caucasian | 0.58 | 0.56 | Hemizygous | RHD*01W.02 |
| 004_59 | R2r | Caucasian | 0.51 | 0.53 | Hemizygous | RHD*01W.02 |
| 004_60 | R2r | Caucasian | 0.57 | 0.52 | Hemizygous | RHD*01W.02 |
| 004_61 | R2r | Caucasian | 0.55 | 0.53 | Hemizygous | RHD*01W.02 |
| 004_62 | R2r | Caucasian | 0.58 | 0.52 | Hemizygous | RHD*01W.02 |
| 004_63 | R0r | Caucasian | 0.46 | 0.46 | Hemizygous | RHD*01 |
| 004_64 | R0r | Caucasian | 0.5 | 0.53 | Hemizygous | RHD*01 |
| 004_65 | R0r | Caucasian | 0.46 | 0.49 | Hemizygous | RHD*01 |
| 004_66 | R0r | Caucasian | 0.49 | 0.52 | Hemizygous | RHD*01 |
| 004_67 | R0r | Caucasian | 0.53 | 0.51 | Hemizygous | RHD*01 |
| 004_68 | R0r | Caucasian | 0.52 | 0.51 | Hemizygous | RHD*01 |
| 004_69 | R2RZ | Caucasian | 1.02 | 1.02 | Homozygous | RHD*01 |
| 004_70- 004_123 | R1R2 | — | 1.0¶ | 1.0¶ | Homozygous | Not sequenced |
The number of RHD copies per microliter present in a sample was compared with the reference gene AGO1 copies per microliter. If a sample presented a ratio of 1, it was considered homozygous; it was considered hemizygous when present with a ratio of 0.5. Bold in the table body represents incompatible results between predicted genotype by serology and dPCR.
—, individual ethnicities not given.
As supplied by the NHSBT, Bristol, United Kingdom.
Sample shows discrepancy between hemizygous RHD5 and homozygous RHD7 meaning that 1 of the RHD alleles has a deletion in exon 5.
Eight samples show incompatible dPCR results with serologically predicted genotypes indicating incorrectly predicted genotypes by serology; these samples include:
R1r sample shows the homozygous RHD gene.
6 R1R2 samples show the hemizygous RHD gene.
R2R2 sample shows the hemizygous RHD gene.
Average ratio.
NGS data
To establish reference RHD allele sequences, we aimed to sequence hemizygous RHD samples; nevertheless, RHD homozygous samples were also included in the sequence analysis to detect weak D that could be undetectable by serological testing due to the presence of a wild-type copy of the RHD allele. We purposely included the 6 R1R2 samples (004_35, 004_36, 004_37, 004_38, 004_39, 004_40) that tested as hemizygous for the RHD gene and included another set of 6 homozygous R1R2 samples (004_29, 004_30, 004_31, 004_32, 004_33, 004_34) for a comparison, which were randomly chosen from the remaining 60 homozygous R1R2 samples.
Samples (n = 69; Table 3) with different Rh serologically predicted genotypes were sequenced on the Ion PGM, including 7 R1R1 (DCe/DCe), 21 R1r (DCe/dce), 7 R2R2 (DcE/DcE), 15 R2r (DcE/dce), 12 R1R2 (DCe/DcE), 6 R0r (Dce/dce), and 1 R2RZ (DcE/DCE). Data were aligned to the hg38 reference sequence using CLC Workbench 9 software (Qiagen Ltd). It is noteworthy that the RHD reference sequence (NC_000001.11)42 is RHD*DAU0 (RHD*10.00), presenting a SNP in exon 8 (1136C>T), causing amino acid change Thr379Met; therefore, all 69 samples presented a SNP in exon 8 (1136T>C) Met379Thr.
Three exonic SNPs and 519 intronic SNPs were detected across the 69 samples. Of the 28 samples that were serologically phenotyped as weak D, 26 of them were confirmed to be weak D by NGS and the RHD allele was determined. One R1r sample (004_28) showed a SNP in exon 1 (8C>G) Ser3Cys that encodes weak D type 3 (RHD*01W.3). Thirteen R1r samples (004_15, 004_16, 004_17, 004_18, 004_19, 004_20, 004_21, 004_22, 004_23, 004_24, 004_25, 004_26, 004_27) and 1 R1R1 sample (004_07) showed a SNP in exon 6, (809T>G) Val270Gly that encodes weak D type 1 (RHD*01W.1). Nine R2r samples (004_54, 004_55, 004_56, 004_57, 004_58, 004_59, 004_60, 004_61, 004_62), 2 R2R2 samples (004_41, 004_42), and 1 R1R2 sample (004_35) showed the exon 9 (1154G>C) SNP that causes amino acid change Gly385Ala, which encodes weak D type 2 (RHD*01W.02).
One R1r sample (004_14) and the R2RZ (004_69) sample were serologically predicted to be weak D but no SNPs in the RHD gene causing amino acid changes in the RhD protein were detected by sequencing. For these 2 samples (004_14 and 004_69), the RHAG gene was sequenced to test whether there were any mutations in the RHAG gene that could be leading to weak D expression. One RHAG exon 6 mutation 808G>A was detected in sample (004_14), causing the Val270Ile change that encodes for the RHAG*04 allele. Sample (004_69) showed a wild-type RHAG*01 allele predicting no amino acid changes.
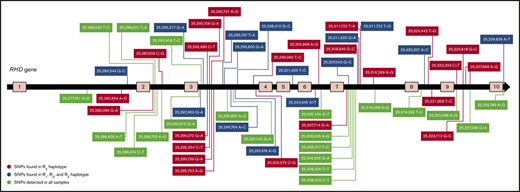
Intronic SNPs
Due to RHD*DAU0 (RHD*10.00) being the reference sequence hg38, 21 homozygous SNPs were detected in all 69 samples (Table 4) that are specific to the reference allele, that is, RHD*DAU0 (RHD*10.00). Multiple intronic SNPs are suspected to be haplotype specific, for example, 23 SNPs (Table 5) were homozygous SNPs in all samples with the R2 haplotype. They were detected in R2R2, R2r, and in 3 of the 6 R1R2 samples (004_35, 004_36, 004_37), which were determined by dPCR to be hemizygous for RHD gene. These SNPs were also present in 6 R1R2 samples (D homozygous; 004_29, 004_30, 004_31, 004_32, 004_33, 004_34), and in the R2RZ sample (004_69) as heterozygous SNPs.
Position of intronic variations and their reference SNP number detected in all samples sequenced
| Position . | hg38 (RHD*DAU0) . | All samples . | Location . | Reference SNP no.* . |
|---|---|---|---|---|
| 25 277 761 | A | G | Intron 1 | rs28661958 |
| 25 286 520 | T | C | Intron 2 | rs183024534 |
| 25 286 601 | T | A | Intron 2 | NA† |
| 25 286 605 | A | T | Intron 2 | NA† |
| 25 286 674 | C | T | Intron 2 | NA† |
| 25 286 732 | A | G | Intron 2 | NA† |
| 25 290 908 | T | C | Intron 3 | rs28521909 |
| 25 290 915 | G | A | Intron 3 | rs28562109 |
| 25 295 850 | A | G | Intron 3 | rs28451966 |
| 25 297 140 | G | A | Intron 3 | rs28786680 |
| 25 305 164 | G | T | Intron 6 | rs28703207 |
| 25 308 306 | T | C | Intron 7 | rs28374144 |
| 25 308 317 | T | C | Intron 7 | rs28719684 |
| 25 308 325 | G | A | Intron 7 | rs71493569 |
| 25 308 326 | C | T | Intron 7 | rs71493569 |
| 25 308 403 | C | T | Intron 7 | rs1801096 |
| 25 316 058 | A | G | Intron 7 | rs28453868 |
| 25 319 292 | T | C | Intron 8 | rs28397158 |
| 25 322 588 | A | G | Intron 9 | rs28435180 |
| 25 327 036 | G | A | Intron 9 | rs61777612 |
| 25 329 789 | A | G | Intron 10 | rs28654325 |
| Position . | hg38 (RHD*DAU0) . | All samples . | Location . | Reference SNP no.* . |
|---|---|---|---|---|
| 25 277 761 | A | G | Intron 1 | rs28661958 |
| 25 286 520 | T | C | Intron 2 | rs183024534 |
| 25 286 601 | T | A | Intron 2 | NA† |
| 25 286 605 | A | T | Intron 2 | NA† |
| 25 286 674 | C | T | Intron 2 | NA† |
| 25 286 732 | A | G | Intron 2 | NA† |
| 25 290 908 | T | C | Intron 3 | rs28521909 |
| 25 290 915 | G | A | Intron 3 | rs28562109 |
| 25 295 850 | A | G | Intron 3 | rs28451966 |
| 25 297 140 | G | A | Intron 3 | rs28786680 |
| 25 305 164 | G | T | Intron 6 | rs28703207 |
| 25 308 306 | T | C | Intron 7 | rs28374144 |
| 25 308 317 | T | C | Intron 7 | rs28719684 |
| 25 308 325 | G | A | Intron 7 | rs71493569 |
| 25 308 326 | C | T | Intron 7 | rs71493569 |
| 25 308 403 | C | T | Intron 7 | rs1801096 |
| 25 316 058 | A | G | Intron 7 | rs28453868 |
| 25 319 292 | T | C | Intron 8 | rs28397158 |
| 25 322 588 | A | G | Intron 9 | rs28435180 |
| 25 327 036 | G | A | Intron 9 | rs61777612 |
| 25 329 789 | A | G | Intron 10 | rs28654325 |
Intronic SNPs present in all samples with R2 haplotype
| Position . | SNP . | Location . | Reference SNP no.* . |
|---|---|---|---|
| 25 282 654 | A>G | Intron 1 | rs3866916 |
| 25 285 089 | G>A | Intron 2 | rs675072 |
| 25 287 909 | C>G | Intron 2 | rs28718098 |
| 25 295 072 | G>A | Intron 3 | rs372986392 |
| 25 295 354 | C>T | Intron 3 | rs2904840 |
| 25 295 489 | C>T | Intron 3 | rs190056379 |
| 25 295 708 | G>A | Intron 3 | rs182346769 |
| 25 295 731 | A>G | Intron 3 | rs201512625 |
| 25 295 739 | G>A | Intron 3 | rs200682399 |
| 25 295 753 | A>G | Intron 3 | rs143670081 |
| 25 298 980 | T>C | Intron 3 | rs2904843 |
| 25 300 575 | C>G | Intron 3 | rs2986167 |
| 25 305 898 | A>G | Intron 6 | rs12126031 |
| 25 307 714 | G>A | Intron 7 | rs2257611 |
| 25 308 845 | G>C | Intron 7 | rs2478025 |
| 25 311 722 | T>A | Intron 7 | rs796579065 |
| 25 316 269 | A>G | Intron 7 | rs2427767 |
| 25 320 442 | T>G | Intron 8 | rs3927482 |
| 25 321 858 | T>C | Intron 8 | rs28669938 |
| 25 323 393 | C>T | Intron 9 | rs77160738 |
| 25 323 618 | G>C | Intron 9 | rs201304363 |
| 25 323 713 | G>C | Intron 9 | rs202154122 |
| 25 327 668 | A>G | Intron 9 | NA† |
| Position . | SNP . | Location . | Reference SNP no.* . |
|---|---|---|---|
| 25 282 654 | A>G | Intron 1 | rs3866916 |
| 25 285 089 | G>A | Intron 2 | rs675072 |
| 25 287 909 | C>G | Intron 2 | rs28718098 |
| 25 295 072 | G>A | Intron 3 | rs372986392 |
| 25 295 354 | C>T | Intron 3 | rs2904840 |
| 25 295 489 | C>T | Intron 3 | rs190056379 |
| 25 295 708 | G>A | Intron 3 | rs182346769 |
| 25 295 731 | A>G | Intron 3 | rs201512625 |
| 25 295 739 | G>A | Intron 3 | rs200682399 |
| 25 295 753 | A>G | Intron 3 | rs143670081 |
| 25 298 980 | T>C | Intron 3 | rs2904843 |
| 25 300 575 | C>G | Intron 3 | rs2986167 |
| 25 305 898 | A>G | Intron 6 | rs12126031 |
| 25 307 714 | G>A | Intron 7 | rs2257611 |
| 25 308 845 | G>C | Intron 7 | rs2478025 |
| 25 311 722 | T>A | Intron 7 | rs796579065 |
| 25 316 269 | A>G | Intron 7 | rs2427767 |
| 25 320 442 | T>G | Intron 8 | rs3927482 |
| 25 321 858 | T>C | Intron 8 | rs28669938 |
| 25 323 393 | C>T | Intron 9 | rs77160738 |
| 25 323 618 | G>C | Intron 9 | rs201304363 |
| 25 323 713 | G>C | Intron 9 | rs202154122 |
| 25 327 668 | A>G | Intron 9 | NA† |
Intronic SNPs (hg38) and their reference SNP number were present in R2r, R2R2, R1R2, and R2RZ samples. Intronic SNPs were present as homozygous in all 9 weak D type 2 R2r samples, all 6 R2r samples and all 7 R2R2 samples, and in 3 of the 6 R1R2 samples that tested as hemizygous for the RHD gene by dPCR. These SNPs were also present as heterozygous SNPs in all 6 homozygous R1R2 samples and in the R2RZ sample.
From the database of SNPs.44
Not applicable. Not found in the database of SNPs.44
Fifteen SNPs (Table 6) were detected as homozygous in all R1R1, R1r and in 3 of 6 R1R2 samples (004_38, 004_39, 004_40), which were shown by dPCR to be hemizygous for the RHD gene. They were also detected in all 6 R0r samples (004_63, 004_64, 004_65, 004_66, 004_67, 004_68). These SNPs were also found as heterozygous SNPs in 6 R1R2 samples (D homozygous) (004_29, 004_30, 004_31, 004_32, 004_33, 004_34), and in the R2RZ sample (004_69). Table 7 shows the different intronic SNPs detected and their correspondence in R2 and R1, R0, RZRHD alleles in comparison with the reference sequence. From the 519 intronic SNPs detected, most were not conserved across each haplotype (data not shown). Most of these SNPs have been reported and show corresponding reference numbers in the database of SNPs.44
Intronic SNPs present in all samples with R1, R0, and RZ haplotypes
| Position . | SNP . | Location . | Reference SNP no.* . |
|---|---|---|---|
| 25 284 544 | G>C | Intron 1 | rs2301153 |
| 25 292 953 | G>A | Intron 3 | rs28645510 |
| 25 295 317 | G>A | Intron 3 | rs2986157 |
| 25 295 797 | T>A | Intron 3 | rs2986163 |
| 25 295 800 | G>A | Intron 3 | rs2986164 |
| 25 296 764 | A>C | Intron 3 | rs599792 |
| 25 297 476 | A>G | Intron 3 | rs1830962 |
| 25 298 410 | G>C | Intron 3 | rs1293267 |
| 25 301 905 | T>G | Intron 5 | rs28510210 |
| 25 304 945 | A>T | Intron 6 | rs28685153 |
| 25 307 040 | G>C | Intron 7 | rs3118453 |
| 25 311 520 | G>A | Intron 7 | rs2478028 |
| 25 311 722 | T>G | Intron 7 | rs796579065 |
| 25 320 257 | A>C | Intron 8 | rs28628791 |
| 25 329 839 | A>T | Intron 10 | rs28668998 |
| Position . | SNP . | Location . | Reference SNP no.* . |
|---|---|---|---|
| 25 284 544 | G>C | Intron 1 | rs2301153 |
| 25 292 953 | G>A | Intron 3 | rs28645510 |
| 25 295 317 | G>A | Intron 3 | rs2986157 |
| 25 295 797 | T>A | Intron 3 | rs2986163 |
| 25 295 800 | G>A | Intron 3 | rs2986164 |
| 25 296 764 | A>C | Intron 3 | rs599792 |
| 25 297 476 | A>G | Intron 3 | rs1830962 |
| 25 298 410 | G>C | Intron 3 | rs1293267 |
| 25 301 905 | T>G | Intron 5 | rs28510210 |
| 25 304 945 | A>T | Intron 6 | rs28685153 |
| 25 307 040 | G>C | Intron 7 | rs3118453 |
| 25 311 520 | G>A | Intron 7 | rs2478028 |
| 25 311 722 | T>G | Intron 7 | rs796579065 |
| 25 320 257 | A>C | Intron 8 | rs28628791 |
| 25 329 839 | A>T | Intron 10 | rs28668998 |
SNPs were present as homozygous in all 6 R1R1 samples, 1 R1R1 weak D type 1 sample, all 13 R1r weak D type 1 samples, 1 R1r weak D type 3 sample, all 6 R1r samples, 6 R0r samples. These SNPs were also present as hemizygous in 3 of the 6 R1R2 samples that tested as hemizygous for the RHD gene by dPCR. SNPs were also detected as heterozygous SNPs in the 6 homozygous R1R2 samples and 1 R2RZ sample.
From the database of SNPs.44
Position of intronic variations determined by NGS and their corresponding nucleotide in R2 and R1, R0, RZRHD alleles in comparison with the reference sequence (hg38)
| Intronic position . | Reference SNP no.* . | Intronic location . | hg38 . | R1, R0, RZ . | R2 . |
|---|---|---|---|---|---|
| 25 277 761 | rs28661958 | Intron 1 | A | G | G |
| 25 282 654 | rs3866916 | Intron 1 | A | A | G |
| 25 284 544 | rs2301153 | Intron 1 | G | C | G |
| 25 285 089 | rs675072 | Intron 2 | G | G | A |
| 25 286 520 | rs183024534 | Intron 2 | T | C | C |
| 25 286 601 | NA† | Intron 2 | T | A | A |
| 25 286 605 | NA† | Intron 2 | A | T | T |
| 25 286 674 | NA† | Intron 2 | C | T | T |
| 25 286 732 | NA† | Intron 2 | A | G | G |
| 25 287 909 | rs28718098 | Intron 2 | C | C | G |
| 25 290 908 | rs28521909 | Intron 3 | T | C | C |
| 25 290 915 | rs28562109 | Intron 3 | G | A | A |
| 25 292 953 | rs28645510 | Intron 3 | G | A | G |
| 25 295 072 | rs372986392 | Intron 3 | G | G | A |
| 25 295 317 | rs2986157 | Intron 3 | G | A | G |
| 25 295 354 | rs2904840 | Intron 3 | C | C | T |
| 25 295 489 | rs190056379 | Intron 3 | C | C | T |
| 25 295 708 | rs182346769 | Intron 3 | G | G | A |
| 25 295 731 | rs201512625 | Intron 3 | A | A | G |
| 25 295 739 | rs200682399 | Intron 3 | G | G | A |
| 25 295 753 | rs143670081 | Intron 3 | A | A | G |
| 25 295 797 | rs2986163 | Intron 3 | T | A | T |
| 25 295 800 | rs2986164 | Intron 3 | G | A | G |
| 25 295 850 | rs28451966 | Intron 3 | A | G | G |
| 25 296 764 | rs599792 | Intron 3 | A | C | A |
| 25 297 140 | rs28786680 | Intron 3 | G | A | A |
| 25 297 476 | rs1830962 | Intron 3 | A | G | A |
| 25 298 410 | rs1293267 | Intron 3 | G | C | G |
| 25 298 980 | rs2904843 | Intron 3 | T | T | C |
| 25 300 575 | rs2986167 | Intron 3 | C | C | G |
| 25 301 905 | rs28510210 | Intron 5 | T | G | T |
| 25 304 945 | rs28685153 | Intron 6 | A | T | A |
| 25 305 164 | rs28703207 | Intron 6 | G | T | T |
| 25 305 898 | rs12126031 | Intron 6 | A | A | G |
| 25 307 040 | rs3118453 | Intron 7 | G | C | G |
| 25 307 714 | rs2257611 | Intron 7 | G | G | A |
| 25 308 306 | rs28374144 | Intron 7 | T | C | C |
| 25 308 317 | rs28719684 | Intron 7 | T | C | C |
| 25 308 325 | rs71493569 | Intron 7 | G | A | A |
| 25 308 326 | rs71493569 | Intron 7 | C | T | T |
| 25 308 403 | rs1801096 | Intron 7 | C | T | T |
| 25 308 845 | rs2478025 | Intron 7 | G | G | C |
| 25 311 520 | rs2478028 | Intron 7 | G | A | G |
| 25 311 722‡ | rs796579065 | Intron 7 | T | G | A |
| 25 316 058 | rs28453868 | Intron 7 | A | G | G |
| 25 316 269 | rs2427767 | Intron 7 | A | A | G |
| 25 319 292 | rs28397158 | Intron 8 | T | C | C |
| 25 320 257 | rs28628791 | Intron 8 | A | C | A |
| 25 320 442 | rs3927482 | Intron 8 | T | T | G |
| 25 321 858 | rs28669938 | Intron 8 | T | T | C |
| 25 322 588 | rs28435180 | Intron 9 | A | G | G |
| 25 323 393 | rs77160738 | Intron 9 | C | C | T |
| 25 323 618 | rs201304363 | Intron 9 | G | G | C |
| 25 323 713 | rs202154122 | Intron 9 | G | G | C |
| 25 327 036 | rs61777612 | Intron 9 | G | A | A |
| 25 327 668 | NA† | Intron 9 | A | A | G |
| 25 329 789 | rs28654325 | Intron 10 | A | G | G |
| 25 329 839 | rs28668998 | Intron 10 | A | T | A |
| Intronic position . | Reference SNP no.* . | Intronic location . | hg38 . | R1, R0, RZ . | R2 . |
|---|---|---|---|---|---|
| 25 277 761 | rs28661958 | Intron 1 | A | G | G |
| 25 282 654 | rs3866916 | Intron 1 | A | A | G |
| 25 284 544 | rs2301153 | Intron 1 | G | C | G |
| 25 285 089 | rs675072 | Intron 2 | G | G | A |
| 25 286 520 | rs183024534 | Intron 2 | T | C | C |
| 25 286 601 | NA† | Intron 2 | T | A | A |
| 25 286 605 | NA† | Intron 2 | A | T | T |
| 25 286 674 | NA† | Intron 2 | C | T | T |
| 25 286 732 | NA† | Intron 2 | A | G | G |
| 25 287 909 | rs28718098 | Intron 2 | C | C | G |
| 25 290 908 | rs28521909 | Intron 3 | T | C | C |
| 25 290 915 | rs28562109 | Intron 3 | G | A | A |
| 25 292 953 | rs28645510 | Intron 3 | G | A | G |
| 25 295 072 | rs372986392 | Intron 3 | G | G | A |
| 25 295 317 | rs2986157 | Intron 3 | G | A | G |
| 25 295 354 | rs2904840 | Intron 3 | C | C | T |
| 25 295 489 | rs190056379 | Intron 3 | C | C | T |
| 25 295 708 | rs182346769 | Intron 3 | G | G | A |
| 25 295 731 | rs201512625 | Intron 3 | A | A | G |
| 25 295 739 | rs200682399 | Intron 3 | G | G | A |
| 25 295 753 | rs143670081 | Intron 3 | A | A | G |
| 25 295 797 | rs2986163 | Intron 3 | T | A | T |
| 25 295 800 | rs2986164 | Intron 3 | G | A | G |
| 25 295 850 | rs28451966 | Intron 3 | A | G | G |
| 25 296 764 | rs599792 | Intron 3 | A | C | A |
| 25 297 140 | rs28786680 | Intron 3 | G | A | A |
| 25 297 476 | rs1830962 | Intron 3 | A | G | A |
| 25 298 410 | rs1293267 | Intron 3 | G | C | G |
| 25 298 980 | rs2904843 | Intron 3 | T | T | C |
| 25 300 575 | rs2986167 | Intron 3 | C | C | G |
| 25 301 905 | rs28510210 | Intron 5 | T | G | T |
| 25 304 945 | rs28685153 | Intron 6 | A | T | A |
| 25 305 164 | rs28703207 | Intron 6 | G | T | T |
| 25 305 898 | rs12126031 | Intron 6 | A | A | G |
| 25 307 040 | rs3118453 | Intron 7 | G | C | G |
| 25 307 714 | rs2257611 | Intron 7 | G | G | A |
| 25 308 306 | rs28374144 | Intron 7 | T | C | C |
| 25 308 317 | rs28719684 | Intron 7 | T | C | C |
| 25 308 325 | rs71493569 | Intron 7 | G | A | A |
| 25 308 326 | rs71493569 | Intron 7 | C | T | T |
| 25 308 403 | rs1801096 | Intron 7 | C | T | T |
| 25 308 845 | rs2478025 | Intron 7 | G | G | C |
| 25 311 520 | rs2478028 | Intron 7 | G | A | G |
| 25 311 722‡ | rs796579065 | Intron 7 | T | G | A |
| 25 316 058 | rs28453868 | Intron 7 | A | G | G |
| 25 316 269 | rs2427767 | Intron 7 | A | A | G |
| 25 319 292 | rs28397158 | Intron 8 | T | C | C |
| 25 320 257 | rs28628791 | Intron 8 | A | C | A |
| 25 320 442 | rs3927482 | Intron 8 | T | T | G |
| 25 321 858 | rs28669938 | Intron 8 | T | T | C |
| 25 322 588 | rs28435180 | Intron 9 | A | G | G |
| 25 323 393 | rs77160738 | Intron 9 | C | C | T |
| 25 323 618 | rs201304363 | Intron 9 | G | G | C |
| 25 323 713 | rs202154122 | Intron 9 | G | G | C |
| 25 327 036 | rs61777612 | Intron 9 | G | A | A |
| 25 327 668 | NA† | Intron 9 | A | A | G |
| 25 329 789 | rs28654325 | Intron 10 | A | G | G |
| 25 329 839 | rs28668998 | Intron 10 | A | T | A |
Discussion
RHD reference sequences
We have established a methodology to fully sequence the RHD gene including promotor, introns, and all exons that can be used to study the different RHD alleles in the population to establish reference RHD allele sequences. We sequenced hemizygous (1 copy) RHD genes in samples that were confirmed to be hemizygous RHD samples by dPCR, and compared those sequences with homozygous (2 copy) RHD genes in samples confirmed as homozygous RHD by dPCR.
Two RHD reference sequences were submitted to GenBank and registered with accession numbers MG944308 and MG944309 for the R1, R0, RZ haplotypes and the R2 haplotype, respectively. We are additionally working on establishing the method for fully sequencing the homologous RHCE gene. In many cases when serology fails to determine an RhD variant and other platforms cannot detect the RHD allele, follow-up work would only require RHD sequencing to determine the exact nucleotide changes and the RHCE gene sequencing would not be needed.
The RHD gene was fully sequenced on the Ion PGM through LR-PCR amplification. Although LR-PCR is an efficient technique in amplifying the gene for sequencing, the LR-PCR approach is limited. Hybrid RHD-RHCE alleles or partial D alleles may not amplify if a primer position is compromised by deletion or mutations. The RHD-specific primers in the current study were subsequently tested with different weak and partial D samples including: RHD*DVI.01, RHD*DNB, RHD*DIV.04, RHD*DVII.01, DFR1, DFR2, and RHD*DIIIa (data not shown). Amplification for all 6 PCR amplicons was achieved in all samples except for samples with the RHD*DVI.01 allele, in which amplicon 4 did not amplify successfully (data not shown). This issue could be resolved in the future using a hybrid primer approach, for example, an RHD-specific forward primer and an RHCE-specific reverse primer.
Data analysis revealed 3 exonic SNPs that encode 3 RHD alleles, which include RHD*01W.1, RHD*01W.02, and RHD*01W.3. Weak D type 1 RHD*01W.1 was detected in 14 samples with R1 haplotype, whereas weak D type 2 RHD*01W.02 was found in 12 R2 haplotype samples. These results support the hypothesis that different weak D alleles are linked to a specific haplotype, in which weak D type 1 is linked to the R1 (DCe) haplotype, and weak D type 2 is linked to the R2 (DcE) haplotype.45 One R1r (DCe/dce) sample (004_28) was genotyped by NGS as weak D type 3 (RHD*01.03), which is linked to R1 haplotype.45
Rh haplotype-specific SNPs
Analyzing intronic SNPs (Table 7) revealed 21 homozygous SNPs (Table 4) present in all samples sequenced. These represent SNP variants of the RHD*DAU0 (RHD*10.00) allele, which the hg38 reference sequence encodes. Some intronic SNPs were found to be present in a specific haplotype (R2), 23 SNPs were homozygous in all R2r, R2R2 and in 3 of the 6 R1R2 samples that tested as hemizygous by dPCR (Table 5). They were also detected as heterozygous SNPs in all R1R2 samples tested as homozygous by dPCR and in the R2RZ sample. Homozygous intronic mutations were detected in all R1R1, R1r, and in the other 3 of the 6 R1R2 samples tested as hemizygous in dPCR (Table 6). These SNPs were also present in 6 R0r samples, and detected as heterozygous SNPs in 6 R1R2 samples tested as homozygous by dPCR and in the R2RZ sample. The similarities of the intronic SNPs pattern between different haplotypes (R1, R0, and RZ) suggest that these haplotypes might have risen from the same ancestral gene. There were no intronic SNPs specific to each of the R1, R0, or RZ alleles.
RHAG NGS
Two samples (004_14 and 004_69) were serologically phenotyped as weak D; however, no amino acid changes were predicted from sequencing of the RHD gene. Different mutations in the RHAG gene (ISBT030) have been reported that disturb the expression of the Rh proteins.36-38 Therefore, we sequenced the RHAG gene for these samples (004_14 and 004_69) that showed weak D reactivity without finding any alterations in the RHD gene. Sample (004_14) showed a SNP 808G>A in exon 6 of the RHAG gene leading to Val270Ile that encodes the RHAG*04 allele. In this sample, this mutation could be the main cause for the weak D reactivity, hence no changes were detected from the sequencing of the RHD gene in this sample to explain the weak D reactivity.
dPCR discrepant results characterized by RHD NGS
dPCR was used to test for 2 targets in the RHD gene against the reference gene AGO1 on chromosome 1. dPCR has demonstrated high sensitivity when used as a detection method for RHD genotyping.12,39 All samples included in this cohort demonstrated compatible zygosity results with the serologically predicted genotype except for 9 samples. Eight samples showed incompatible results with the predicted genotype by serological testing; they include: 1 R1r sample (004_14), which showed the presence of a homozygous RHD gene; 6 R1R2 samples (004_35, 004_36, 004_37, 004_38, 004_39, 004_40), which showed the presence of a hemizygous RHD gene; and 1 R2R2 sample (004_42), which showed as hemizygous for the RHD gene (Table 3). One R1R1 sample (004_07) showed a discrepancy between the RHD5 and RHD7 results. Sample (004_07) presented a ratio of 0.54 for RHD5 against the reference gene AGO1, indicating a hemizygous result; a ratio of 1.0 for RHD7 against the reference gene AGO1 indicated a homozygous result. This discrepancy between hemizygous RHD5 and homozygous RHD7 means that 1 of the RHD alleles has a deletion in exon 5. This gene deletion could not be detected through the NGS due to the presence of a wild-type copy of the other RHD allele masking the probable failed amplification of the variant allele.
The 6 R1R2 (DCe/DcE) samples, which showed only 1 copy of the RHD gene (hemizygous), had their genotypes predicted by serology findings based on the probability of the gene in the population, but in these cases the genotypes are in fact less frequent or occurring with a lower probability. These samples are expected to be either R1r′′ (DCe/dcE), RZr (DCE/dce), R2r′ (DcE/dCe), or R0ry (Dce/dCE) from zygosity information, which all could be inappropriately assigned by serology as R1R2 (DCe/DcE) due to gene frequencies in the population. Three of the R1R2 (004_35, 004_36, 004_37) samples have the intronic SNPs suspected to be linked to the R2 haplotype and are missing all the other intronic SNPs that are linked to the R1, R0, RZ haplotypes. Sample 004_35 was genotyped as weak D type 2, and due to the link between the R2 haplotype and weak D type 2, this sample could only be R2r′ (DcE/dCe). The other 2 samples could also be genotyped as R2r′ (DcE/dCe) as inferred by their intronic SNP pattern. The correct genotype of the other 3 hemizygous R1R2 samples (004_38, 004_39, 004_40) missing the R2-specific SNPs could be either R1r″ (DCe/dcE), RZr (DCE/dce), or R0ry (Dce/dCE). Considering the frequency of these alleles46 in the population, in which R1r″ is 1%, RZr is 0.19%, and R0ry is <0.01%, it is very likely for these samples to be R1r″ (DCe/dcE). Based on our zygosity results, the frequency of R1r″ seems to be higher than anticipated46 in the population. Definitive genotypes for these samples could be confirmed by sequencing the RHCE gene, in addition to the RHD gene, and hence only having the RHD gene sequencing to date is a limitation of this study. In ongoing work to sequence the RHCE gene, multiple primer sets have been designed to amplify the gene in LR-PCR amplicons but the regions surrounding introns 2 and 8 of the RHCE gene are problematic. We have sequenced 35 samples for the RHCE gene (data not shown) that had poor depth of coverage for amplicons covering introns 2 and 8, which has made data analysis and variant calling from these regions challenging. Successful and robust sequencing of the RHCE gene would add to the data set and aid identification of particular alleles in samples. It will also be of interest to sequence the RHCE gene in samples lacking the RHD gene, for example, rr (dce/dce) samples.
The R1r (004_14) sample that was homozygous RHD by dPCR showed R1/R0/RZ-related SNPs and was missing all R2-related SNPs, suggesting that the correct genotype could be R1R0 (DCe/Dce). The hemizygous R2R2 sample (004_42) was genotyped as weak D type 2 and showed R2-specific SNPs, therefore, its correct genotype could only be R2r″ (DcE/dcE).
We sequenced the RHD gene in 69 samples using NGS to study RHD mutations, assessed variations present in the population and identified reference RHD allele sequences (Table 7). Intronic SNPs were used to determine their relation to specific haplotypes. We found that 21 intronic SNPs were present in all samples indicating their specificity to the RHD*DAU0 (RHD*10.00) haplotype, which the hg38 reference sequence encodes. Twenty-three intronic SNPs were found to be R2 specific, and 15 were related to R1, R0, and RZ haplotypes. In future work, we aim to identify the pattern of intronic SNPs in the RHCE gene. Intronic SNPs may represent a novel diagnostic approach to investigate known and novel variants of the RHD and RHCE genes.
Acknowledgments
The authors thank Michele Kiernan of the University of Plymouth Systems Biology Centre (Plymouth, United Kingdom) for carrying out the sequencing run and for support and assistance in this work. The authors thank Amr Halawani of Jazan University, Jazan, Saudi Arabia (formerly based at the University of Plymouth, Plymouth, United Kingdom), for training in NGS library preparation and bioinformatics.
This work was supported by King Abdulaziz University (Jeddah, Saudi Arabia).
Authorship
Contribution: W.A.T. performed experiments, analyzed data, and wrote the manuscript; and T.E.M. and N.D.A. supervised the study and revised the manuscript.
Conflict-of-interest disclosure: A patent relating to the Rh specificity of the intronic polymorphisms identified in this study has been filed (P120661GB) (T.E.M. and N.D.A.). The laboratory also received funding from Biofortuna for aspects of blood-group genotyping and next-generation sequencing work. N.D.A. was an expert witness for Premaitha in their UK high-court case, Premaitha vs Illumina, July 2017, relating to noninvasive prenatal diagnosis. W.A.T. declares no competing financial interests.
Correspondence: Tracey E. Madgett, School of Biomedical Sciences, Faculty of Medicine and Dentistry, University of Plymouth, Plymouth PL4 8AA, United Kingdom; e-mail: tracey.madgett@plymouth.ac.uk.