Abstract
Transcription factor GATA-2 is implicated in the survival and growth of multipotential progenitors. Here we report that the promyelocytic leukemia zinc finger (PLZF) protein can interact with GATA-2 and can modify its transactivation capacity. Fanconi anemia zinc finger (FAZF), a PLZF-homologous protein that has been variously described as ROG (repressor of GATA), and TZFP (testis zinc finger protein) also interact with GATA-2. The zinc finger region of GATA-2 is required for binding to PLZF and FAZF, but distinct interfaces on the PLZF and FAZF molecules mediate the interaction, suggesting that GATA-2 activity is controlled by these 2 homologous proteins through distinct mechanisms. GATA-2 can also physically associate with the PLZF-RARα fusion protein generated by the t(11;17) chromosomal translocation associated with acute promyelocytic leukemia (APL). Functional experiments showed that this interaction has the capacity to render GATA-dependent transcription responsive to treatment with a combination of all-trans retinoic acid and the histone deacetylase inhibitor trichostatin A (TSA). This combination of drugs has been shown to stimulate the terminal differentiation of leukemic t(11;17)-associated APL blasts, raising the possibility that GATA target genes may be involved in the molecular pathogenesis of APL.
Introduction
GATA factors comprise a family of transcriptional regulatory proteins characterized by the ability to bind a common conserved DNA sequence (WGATAR) by virtue of evolutionarily conserved C4 zinc finger domains. Hematopoietically expressed GATA factors (GATA-1, -2, and -3) have been shown to play critical, but distinct, roles in hematopoiesis.1,2 Thus, GATA-1 is expressed at a high level in erythroid cells, mast cells, megakaryocytes, and eosinophils and at a low level in multipotential progenitors. GATA-1 is functionally implicated in the maturation and differentiation of erythroid cells and megakaryocytes. GATA-3 expression within hematopoiesis is confined to T lymphocytes and is implicated in T-cell development and differentiation. GATA-2 is more broadly expressed among hematopoietic cells, with particular prominence in early progenitors and in megakaryocytes and mast cells. Phenotypes of knock-out mice suggest that GATA-2 may be critically involved in the survival and growth of multipotential progenitors.3 Forced expression studies on factor-dependent cell lines and primary cells are also consistent with an involvement of GATA-2 in these processes.4-6 GATA-2 has thus emerged as a key transcription factor in the control of cell fate outcomes of hematopoietic stem and progenitor cells.
A number of critical hematopoietic regulators, such as SCL/Tal-1, Lmo2, and AML-1, have been implicated in leukemogenic pathways.7,8 GATA-2 is often expressed in leukemia cells and cell lines,9,10 but evidence linking GATA-2 with leukemic transformation has generally been lacking. Recently, we have provided evidence that promyelocytic leukemia protein (PML) has the capacity to physically associate with GATA-2 and to modify its function.11 PML was initially identified as a fusion counterpart of retinoic acid receptor alpha (RARα) in t(15;17)-associated acute promyelocytic leukemia (APL).12,13 We were able to show that the PML-RARα chimeric fusion protein interacts with GATA-2 and renders GATA-2–dependent transcription inducible by all-transretinoic acid (ATRA). These results raised the possibility that GATA target genes may be involved in the molecular pathogenesis of APL. Although the t(15;17) translocation accounts for most APL cases, variant translocations have been described including the t(11;17) translocation, which fuses the promyelocytic zinc finger protein (PLZF) to RARα.14
Here we present evidence that GATA-2 has the capacity to physically associate with PLZF. PLZF has a pox virus and zinc finger (POZ) domain and 9 homologous C2H4 zinc fingers that mediate binding to the GTACT–AGTAC motif in DNA.15 Functions of the wild-type PLZF protein are not fully understood, though it has been implicated in apoptosis, growth suppression, differentiation,16,17 and transcriptional repression.18-21 A PLZF homologous protein has been described and initially termed FAZF-1 by virtue of its interaction with Fanconi anemia–related proteins.22 This PLZF homologue has also been described as repressor of GATA23 (ROG), reflecting its repressive interaction with GATA-3 in T cells, and as testis zinc finger protein (TZFP).24 25 In this report we have explored the possibility that GATA-2 might interact with PLZF, FAZF, or PLZF-RARα–dependent pathways because all these molecules are expressed within the progenitor compartment of hematopoiesis, where GATA-2 is thought to play a critical functional role.
Materials and methods
Expression plasmids
The human GATA-2/pMT2 expression plasmid was generously provided by S. H. Orkin (Harvard Medical School, Boston, MA).26 Flag-tagged GATA-2/pCMV and PML-RARα/pMT2 have been described previously.11 The PLZF/pSG5, PLZF-RARα/pSG5, Flag-tagged PLZF/pSG5,14 and Flag-tagged FAZF/pSG5 were generous gifts from A. Zelent (Leukaemia Research Fund Centre at the Institute of Cancer Research, London, United Kingdom). To generate expression vectors for Flag-tagged PLZF fingers 7 to 9 and Flag-tagged FAZF fingers, fragments of cDNA encoding the corresponding regions were produced by polymerase chain reaction and were cloned into pFLAG-CMV2 (Eastman-Kodak, New Haven, CT).
Cells
Cells (293T) were maintained in Dulbecco modified Eagle medium supplemented with 10% fetal calf serum. Human leukemia KG1 cells were maintained in RPMI 1640 medium supplemented with 10% fetal calf serum.
Antibodies
Mouse monoclonal anti-PLZF antibody was a generous gift from A. Zelent. Anti-Flag antibody M2, biotinylated M2, streptavidin–agarose beads, and protein G–agarose beads were purchased from Sigma (St Louis, MO). Anti–GATA-2 polyclonal antibody was purchased from Santa Cruz Biotechnology (Santa Cruz, CA).
Protein interaction assays in cells
Using a standard calcium phosphate coprecipitation method, 293T cells (1 × 106) grown in 10-cm diameter dishes were transfected with the indicated expression plasmids. The total amount of plasmids was equalized by the addition of corresponding empty vectors. Forty-eight hours later, nuclear extracts were prepared as described elsewhere27 and were immunoprecipitated with biotinylated anti-Flag antibody M2 in combination with streptavidin–agarose beads or anti-PLZF antibody in combination with protein G–agarose beads in the binding buffer (20 mM HEPES KCl, pH 7.9, 140 mM NaCl, 1 mM EDTA, 1 mM dithiothreitol, 5 mg bovine serum albumin/mL, 5% protease inhibitor cocktail [Sigma], and 0.1% NP-40). After 5 washes with the binding buffer, immune complexes were analyzed by Western blotting using the indicated antibodies.
DNA binding assays
Nuclear extracts from the 293T cells transfected with the indicated expression plasmids or KG1 cells were incubated with 40 pmol biotinylated double-strand oligonucleotide probes containing the recognition sites for GATA-2 (TATTTTTATCTGATAGGAAGT) in combination with streptavidin–agarose beads in the binding buffer described above, essentially as described in the literature.28 29 After 5 washes with the binding buffer, proteins captured by the probe were analyzed by Western blotting using the indicated antibodies. Biotinylated double-strand oligonucleotide probe mutated in a core recognition sequence from GATA to TTTA was used as a control.
Protein interaction assays in solution
Fragments of cDNA encoding GATA-2 and PLZF were produced using convenient restriction enzymes and polymerase chain reaction methods and then were cloned into the glutathione S-transferase (GST) fusion vector pGEX5x-1 (Pharmacia, Uppsala, Sweden). Bacterially expressed GST fusion proteins were purified according to the manufacturer's instructions. Nuclear extracts of 293T cells transfected with an expression plasmid for Flag–GATA-2 or Flag–PLZF were incubated with the indicated GST fusion proteins bound to the resin in the binding buffer (50 mM Tris HCl, pH 7.5, 140 mM NaCl, 1 mM EDTA, 1 mM dithiothreitol, 5 mg bovine serum albumin/mL, 5% protease inhibitor cocktail, and 0.1% NP-40) as described previously.11 The resin was washed 5 times with the binding buffer, and the bound protein was analyzed by Western blotting with anti-Flag antibody M2.
Transactivation assays
A luciferase reporter plasmid in which 2 copies of back-to-back double GATA sites in the mouse CD34 promoter were placed upstream of the β-globin minimal promoter–driving luciferase gene (designated CD34 × 2/Luc) has been described.11 A luciferase reporter plasmid in which a murine GATA-1 promoter (positions −798 to −574) containing a double GATA site30 was arrayed upstream of the β-globin minimal promoter (designated GATA-1/Luc) was a gift from M. Yamamoto (Tsukuba University, Tsukuba, Japan). Mutant reporters in which core recognition sequences were changed from GATA to TTTA (designated mutant CD34 × 2/Luc and mutant GATA-1/Luc) were described previously.11 Luciferase reporter assays were conducted as described11 with pRL–CMV–renilla luciferase plasmids (Promega) monitoring transfection efficiencies. ATRA (Sigma) or trichostatin A (TSA; Sigma) was added to the culture medium 24 hours after transfection where indicated, and luciferase activities were measured after another 24 hours. Relative luciferase activities reflect duplicate values from a representation of no fewer than 2 independent experiments.
Results
Interaction of GATA-2 and PLZF
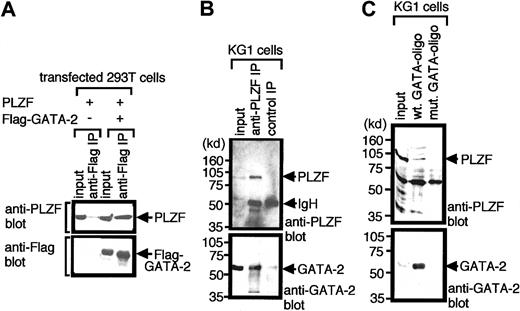
To examine whether GATA-2 could interact with PLZF in living cells, we conducted co-immunoprecipitation experiments using the 293T transient transfection system. 293T cells were cotransfected with expression plasmids for PLZF and Flag-tagged GATA-2 or an empty vector. Two days after transfection, nuclear extracts were prepared and subjected to immunoprecipitation using anti-Flag antibody. Immunoprecipitated materials were fractionated by polyacrylamide gel electrophoresis and subsequently analyzed by Western blotting using an anti-PLZF or anti-Flag antibody (Figure1A). Results show that PLZF coprecipitated with GATA-2 in this system, suggesting that these 2 proteins can complex in vivo.
Interaction of GATA-2 with PLZF in mammalian cells.
(A) Nuclear extracts of 293T cells transfected with expression plasmids encoding PLZF or Flag-GATA-2 were immunoprecipitated (IP) with anti-Flag antibody and were analyzed by Western blotting with anti-PLZF (top panel) or anti-Flag (lower panel) antibodies. Nonimmunoprecipitated material (10% input) was analyzed as a control for appropriate expression of proteins programmed by transfected plasmids. (B) Nuclear extracts of hematopoietic KG1 cells were immunoprecipitated with anti-PLZF antibody and analyzed by Western blotting with anti-PLZF (top panel) or anti-GATA-2 (lower panel) antibody. Input (10%) was used as a control for appropriate expression of the proteins. Numbers on the left indicate positions of molecular weight markers in kilodaltons. (C) Nuclear extracts of KG1 cells were incubated with biotinylated oligonucleotides harboring GATA motifs (wt GATA-oligo) or biotinylated mutant oligonucleotides in which GATA motifs were changed to TTTA (mut GATA-oligo). Oligonucleotides were then recovered by streptavidin–agarose beads, and the precipitated proteins were analyzed by Western blotting with anti-PLZF (top panel) or anti-GATA-2 (lower panel) antibody. Input (10% input) was used as a control. Note that GATA-2 and PLZF coprecipitated in transfected cells (A) and native hematopoietic cells (B). Endogenous GATA-2 had a capacity to recruit PLZF to GATA motifs in DNA (C).
Interaction of GATA-2 with PLZF in mammalian cells.
(A) Nuclear extracts of 293T cells transfected with expression plasmids encoding PLZF or Flag-GATA-2 were immunoprecipitated (IP) with anti-Flag antibody and were analyzed by Western blotting with anti-PLZF (top panel) or anti-Flag (lower panel) antibodies. Nonimmunoprecipitated material (10% input) was analyzed as a control for appropriate expression of proteins programmed by transfected plasmids. (B) Nuclear extracts of hematopoietic KG1 cells were immunoprecipitated with anti-PLZF antibody and analyzed by Western blotting with anti-PLZF (top panel) or anti-GATA-2 (lower panel) antibody. Input (10%) was used as a control for appropriate expression of the proteins. Numbers on the left indicate positions of molecular weight markers in kilodaltons. (C) Nuclear extracts of KG1 cells were incubated with biotinylated oligonucleotides harboring GATA motifs (wt GATA-oligo) or biotinylated mutant oligonucleotides in which GATA motifs were changed to TTTA (mut GATA-oligo). Oligonucleotides were then recovered by streptavidin–agarose beads, and the precipitated proteins were analyzed by Western blotting with anti-PLZF (top panel) or anti-GATA-2 (lower panel) antibody. Input (10% input) was used as a control. Note that GATA-2 and PLZF coprecipitated in transfected cells (A) and native hematopoietic cells (B). Endogenous GATA-2 had a capacity to recruit PLZF to GATA motifs in DNA (C).
We next asked whether GATA-2 could complex with PLZF in hematopoietic cells. For these experiments we made use of the human leukemia cell line KG1, which is thought to represent the lymphomyeloid stem cell compartment and which expresses PLZF31 and GATA-2.10 KG1 nuclear extracts were immunoprecipitated using an anti-PLZF antibody, and the immunoprecipitated materials were analyzed by Western blotting with anti-PLZF or anti–GATA-2 antibodies. Results shown in Figure 1B reveal that within native hematopoietic cells, the endogenous GATA-2 and PLZF proteins can complex together.
We next tested whether GATA-2 could recruit PLZF to a GATA recognition sequence in DNA in hematopoietic cells (Figure 1C). KG1 cell nuclear extract was incubated with a biotinylated double-strand DNA oligonucleotide containing a GATA recognition motif, and then the oligonucleotide was captured using streptavidin–agarose beads. The proteins, which coprecipitated with the oligonucleotide, were analyzed by Western blotting using anti–GATA-2 and anti-PLZF antibodies. Results showed that the oligonucleotides that contained GATA motifs captured GATA-2 in the nuclear extracts and that PLZF was also contained in the DNA–protein complex. In control experiments, biotinylated mutant oligonucleotides in which GATA motifs were mutated to TTTA captured neither GATA-2 nor PLZF, indicating that the capture of PLZF by these oligonucleotides was stringently GATA site–dependent.
PLZF inhibits transactivation capacity of GATA-2
We next asked whether the association of PLZF with GATA-2 had any functional consequences for GATA-2 activity. Because PLZF has been shown to complex with corepressors of transcription (such as mSin3A and HDAC1),18-21,32,33 we examined the effects of PLZF on the ability of GATA-2 to potentiate the transcriptional activity of a GATA-dependent luciferase reporter. Two copies of the back-to-back double GATA motifs in the mouse CD34 promoter have been successfully used as a reporter of GATA-2 activity when arrayed upstream of β-globin minimal promoter driving luciferase.11 Although PLZF somewhat inhibited activity from this reporter in 293T cells, even in the absence of GATA-2 expression, the inhibitory effects of PLZF on this reporter were considerably more evident on GATA-2–dependent activity from the reporter than on its GATA-2–independent basal activity (Figure2). These results, coupled with the observation that endogenous GATA-2 can recruit endogenous PLZF to a GATA motif in DNA, suggest that PLZF may function as a negative regulator for GATA-2, inhibiting its transactivation capacity.
PLZF inhibits GATA-2–dependent reporter activity in transient transfection assays.
293T cells were transfected with a luciferase reporter plasmid containing 2 copies of double GATA sites in the mouse CD34 promoter (0.5 μg), together with expression plasmids for GATA-2 (GATA-2/pMT2, 200 ng; solid bars) and the indicated amounts of PLZF/pSG5 plasmids. Luciferase activities were standardized against renilla luciferase activity from a cotransfected control reporter (pRL-CMV–renilla luciferase) and were expressed as fold increases of the activity of reporter alone.
PLZF inhibits GATA-2–dependent reporter activity in transient transfection assays.
293T cells were transfected with a luciferase reporter plasmid containing 2 copies of double GATA sites in the mouse CD34 promoter (0.5 μg), together with expression plasmids for GATA-2 (GATA-2/pMT2, 200 ng; solid bars) and the indicated amounts of PLZF/pSG5 plasmids. Luciferase activities were standardized against renilla luciferase activity from a cotransfected control reporter (pRL-CMV–renilla luciferase) and were expressed as fold increases of the activity of reporter alone.
Mapping the sites of interaction between GATA-2 and PLZF
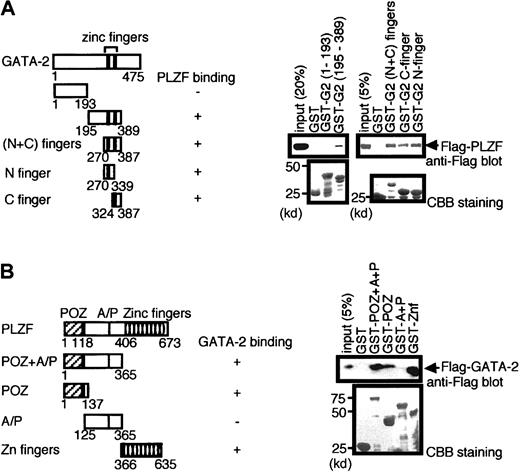
We next determined the region in GATA-2 that was required for its interaction with PLZF using GST pull-down experiments. Various portions of GATA-2 were fused to GST, produced in bacteria, and tested for binding to Flag-tagged PLZF expressed in 293T cells (Figure3A). Zinc fingers of GATA-2 were found to bind PLZF, with either N- or C-finger alone sufficient to retain PLZF. The amino-terminal portion (aa 1 to 193) did not display any affinity for PLZF. Preparations of the most carboxyl portion of GATA-2 (aa 390-475) fused to GST were consistently highly degraded and thus unsuitable for analysis in this assay (data not shown).11
Mapping of regions of GATA-2 and PLZF involved in the interaction by GST pull-down experiments.
(A) GST fusion proteins containing the indicated portions of GATA-2 were tested for the ability to bind Flag-tagged PLZF contained in 293T cell nuclear extract programmed with Flag-PLZF expression plasmids. The first and last amino acids of GATA-2 region present in the various GST fusions are indicated, and the abilities of the proteins to bind PLZF are summarized. Western blot analysis of the pull-down materials using anti-Flag antibody is shown on the right (top panel), and Coomassie brilliant blue (CBB) staining is presented in the lower panel to allow assessment of the quality and quantity of the various GST–GATA-2 proteins used. Numbers on the left indicate positions of molecular weight markers in kilodaltons. (B) Reciprocal pull-down analysis in which various GST–PLZF fusion proteins were analyzed for the ability to bind Flag-tagged GATA-2.
Mapping of regions of GATA-2 and PLZF involved in the interaction by GST pull-down experiments.
(A) GST fusion proteins containing the indicated portions of GATA-2 were tested for the ability to bind Flag-tagged PLZF contained in 293T cell nuclear extract programmed with Flag-PLZF expression plasmids. The first and last amino acids of GATA-2 region present in the various GST fusions are indicated, and the abilities of the proteins to bind PLZF are summarized. Western blot analysis of the pull-down materials using anti-Flag antibody is shown on the right (top panel), and Coomassie brilliant blue (CBB) staining is presented in the lower panel to allow assessment of the quality and quantity of the various GST–GATA-2 proteins used. Numbers on the left indicate positions of molecular weight markers in kilodaltons. (B) Reciprocal pull-down analysis in which various GST–PLZF fusion proteins were analyzed for the ability to bind Flag-tagged GATA-2.
Similar pull-down experiments were conducted to determine the region within PLZF involved in binding to GATA-2 (Figure 3B). These experiments showed that POZ domain or zinc finger domain alone was sufficient to retain GATA-2. We conclude that the interaction of GATA-2 and PLZF involves the zinc finger regions of GATA-2 and POZ domain and zinc finger domains of PLZF.
Interaction of GATA-2 and FAZF
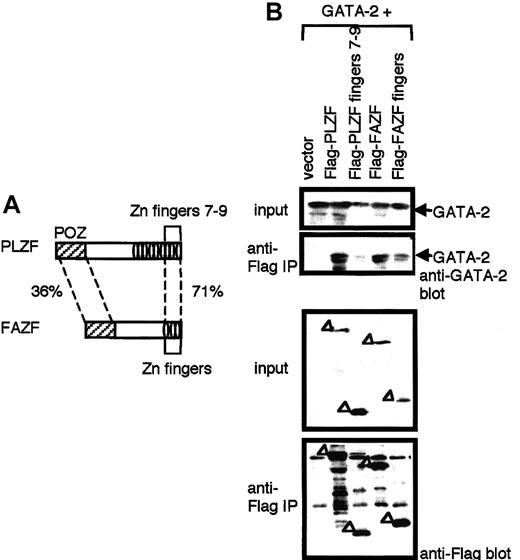
FAZF displays a strong homology to the PLZF protein in the DNA-binding zinc finger region22 and has a POZ domain and 3 zinc fingers, and it is identical to the recently identified molecule, ROG.23 The amino acid sequence of FAZF is 36% and 71% homologous to POZ and to the last 3 zinc fingers (fingers 7-9), respectively, of PLZF (Figure 4A). The zinc finger domain of FAZF has been shown to have an ability to bind GATA-3.23 We therefore tested the binding activity of FAZF and its zinc finger domain to GATA-2 by co-immunoprecipitation experiments using the 293T transient transfection system (Figure 4B). Results showed that FAZF, like PLZF, has an ability to bind GATA-2 and that the interaction was mediated through the zinc fingers of FAZF. Although the last 3 zinc fingers (fingers 7-9) of PLZF are highly homologous to the zinc fingers of FAZF, the terminal zinc fingers 7-9 of PLZF did not bind GATA-2 (Figure 4B). Thus it appears that though PLZF and FAZF can bind GATA-2 by its zinc fingers, they do so through different regions on the PLZF–FAZF moieties. These results suggest that GATA-2 activity is regulated by these 2 homologous proteins through distinct interaction mechanisms.
Interaction of GATA-2 with FAZF.
(A) Schematic drawing of PLZF and FAZF. Percentages represent the homology between the indicated domains. (B) 293T cells transfected with an expression plasmid for GATA-2 in combination with an expression plasmid for Flag-tagged versions of PLZF, PLZF zinc fingers 7 to 9, FAZF, or FAZF zinc fingers. Cell lysates were prepared and immunoprecipitated (IP) with anti-Flag antibody. Immunoprecipitated materials were analyzed by Western blot analysis with anti–GATA-2 (top) and anti-Flag (bottom) antibodies. Input (10%) was used as a control for appropriate expression of the proteins indicated.
Interaction of GATA-2 with FAZF.
(A) Schematic drawing of PLZF and FAZF. Percentages represent the homology between the indicated domains. (B) 293T cells transfected with an expression plasmid for GATA-2 in combination with an expression plasmid for Flag-tagged versions of PLZF, PLZF zinc fingers 7 to 9, FAZF, or FAZF zinc fingers. Cell lysates were prepared and immunoprecipitated (IP) with anti-Flag antibody. Immunoprecipitated materials were analyzed by Western blot analysis with anti–GATA-2 (top) and anti-Flag (bottom) antibodies. Input (10%) was used as a control for appropriate expression of the proteins indicated.
Interaction of GATA-2 and PLZF-RARα
The POZ domain of PLZF is retained in the PLZF-RARα chimera,14 raising the possibility that PLZF-RARα may have the potential to associate with GATA-2; GATA-2 is the predominant GATA factor expressed in early myeloid progenitor cells9,10 34 that represent the cellular target for transformation in APL.
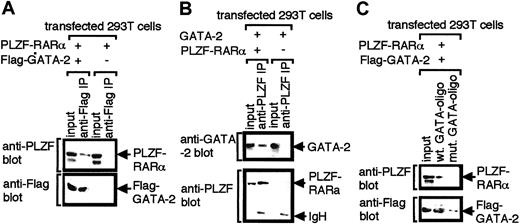
We examined this possibility by co-immunoprecipitation experiments using 293T cells transfected with expression plasmids for Flag–GATA-2 and PLZF-RARα. Nuclear extracts were prepared and immunoprecipitated with anti-Flag antibody. Immunoprecipitated materials were subsequently analyzed by Western blotting with anti-Flag and anti-PLZF antibodies. The results showed that PLZF-RARα coprecipitated with GATA-2 (Figure5A). Reciprocal immunoprecipitation experiments were also conducted. Nuclear extracts programmed with expression plasmids for PLZF-RARα and GATA-2 were immunoprecipitated with anti-PLZF antibody (Figure 5B). GATA-2 was found to coprecipitate with PLZF-RARα, confirming the specificity of the association of the 2 proteins.
Interaction of GATA-2 with PLZF-RARα.
Nuclear extracts of 293T cells transfected with the indicated expression plasmids were immunoprecipitated with anti-Flag (A) or anti-PLZF (B) antibody and were analyzed by Western blotting with anti-Flag and anti-PLZF antibodies (A) or anti- PLZF and anti-GATA-2 antibodies (B). (C) Nuclear extracts of 293T cells transfected with expression plasmids for PLZF-RARα and Flag-tagged GATA-2 were incubated with biotinylated oligonucleotides harboring GATA motifs (wt GATA-oligo) or biotinylated mutant oligonucleotides in which GATA motifs were substituted with TTTA (mut GATA-oligo). Oligonucleotides were recovered by incubating with streptavidin–agarose beads, and precipitated proteins were analyzed by Western blotting with anti-PLZF or anti-Flag antibody.
Interaction of GATA-2 with PLZF-RARα.
Nuclear extracts of 293T cells transfected with the indicated expression plasmids were immunoprecipitated with anti-Flag (A) or anti-PLZF (B) antibody and were analyzed by Western blotting with anti-Flag and anti-PLZF antibodies (A) or anti- PLZF and anti-GATA-2 antibodies (B). (C) Nuclear extracts of 293T cells transfected with expression plasmids for PLZF-RARα and Flag-tagged GATA-2 were incubated with biotinylated oligonucleotides harboring GATA motifs (wt GATA-oligo) or biotinylated mutant oligonucleotides in which GATA motifs were substituted with TTTA (mut GATA-oligo). Oligonucleotides were recovered by incubating with streptavidin–agarose beads, and precipitated proteins were analyzed by Western blotting with anti-PLZF or anti-Flag antibody.
Importantly, PLZF-RARα was recruited to GATA motifs in DNA through interaction with GATA-2 (Figure 5C). Nuclear extracts of 293T cells programmed with expression plasmids for Flag–GATA-2 and PLZF-RARα were incubated with biotinylated oligonucleotide harboring GATA motifs. These oligonucleotides were then captured by streptavidin–agarose beads, and the precipitated proteins were analyzed by Western blotting with anti-Flag and anti-PLZF antibodies. Results showed that GATA-2 was captured by the oligonucleotides and that PLZF-RARα was also contained in the GATA-2–DNA complex. A mutant oligonucleotide in which GATA motifs were mutated to TTTA captured neither GATA-2 nor PLZF-RARα. These results indicate that PLZF-RARα–GATA-2 complex can bind GATA motifs in DNA.
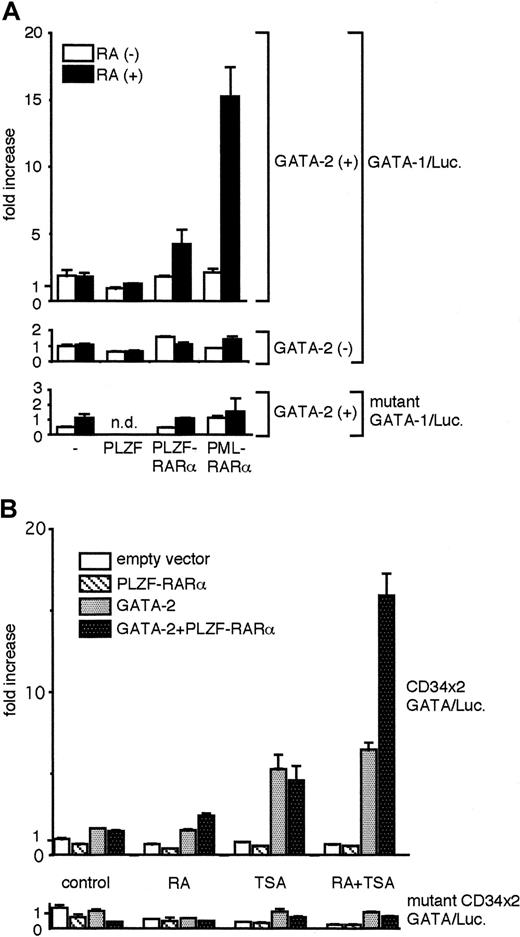
We next conducted reporter-based transactivation analysis in 293T cells to examine whether the expression of PLZF-RARα could functionally modulate GATA-2–dependent transactivation. Because we recently showed that GATA-2 activity can be strongly stimulated by ATRA in the presence of PML-RARα,11 we compared the effects of ATRA on GATA-2 activity in combination with PLZF, PLZF-RARα, and PML-RARα. As indicated in Figure 6A, 293T cells were transfected with the GATA-dependent luciferase reporter (GATA-1/Luc) together with expression plasmids for either PLZF, PLZF-RARα, or PML-RARα. The experiments were conducted in the presence or absence of GATA-2 expression, and, in each case, cells were treated with ATRA or diluent alone. In the absence of additional cotransfected expression plasmids, ATRA had no effect on GATA-2 activity. PLZF inhibited GATA-2 activity, and ATRA did not change GATA-2 activity in the presence of PLZF. In the presence of PLZF-RARα, ATRA stimulated GATA-2 activity weakly (approximately 2.2-fold), compared with the stimulation observed in the presence of PML-RARα (approximately 10-fold). These results suggest functional differences between the PLZF portion of PLZF-RARα and the PML portion of PML-RARα. PLZF-RARα and PML-RARα have been shown to bind the retinoic acid responsive element (RARE) in DNA. ATRA (1 μM) alone is sufficient to activate the transcription by PML-RARα from RARE, whereas TSA, a histone deacetylase inhibitor, is needed in addition to ATRA to activate transcription by PLZF-RARα.19-21,32 Therefore, we next tested the effects of these reagents in our assays. GATA-dependent luciferase reporter constructs were transfected into 293T cells, either alone or together with a PLZF-RARα expression plasmid as indicated in Figure 6B. Control experiments demonstrated that RAR alone had no transcriptional effect on the GATA reporter construct (data not shown). The experiments were conducted in the presence or absence of a GATA-2 expression plasmid. In each case, cells were treated with diluent alone or with ATRA, TSA, or ATRA plus TSA. In the absence of PLZF-RARα, GATA-2 activity was not affected by ATRA, but it was stimulated approximately 3.5-fold by TSA. ATRA plus TSA produced an effect on GATA-2 activity similar to that of TSA alone. In the presence of PLZF-RARα, ATRA stimulated GATA-2 activity slightly, but significantly, consistent with the results shown in Figure 6A. TSA stimulated GATA-2 activity to a level comparable to that seen when TSA was added in the absence of PLZF-RARα. Strikingly, in the presence of PLZF-RARα, ATRA plus TSA stimulated GATA-2 activity by approximately 10-fold, significantly higher than the simple sum of the effects of ATRA or TSA alone and comparable to the level achieved by the combination of GATA-2 and PML-RARα in the presence of ATRA (data not shown and11). These results suggest the enhanced activity of GATA-2 produced by ATRA plus TSA is strictly dependent on the presence of PLZF-RARα. Any combination of ATRA and TSA in the presence or absence of PLZF-RARα had no stimulatory effects on reporter activity in the absence of GATA-2, and effects of ATRA or TSA on GATA-2 activity were abrogated when GATA motifs in the reporter were disrupted (Figure 6A-B). These results confirm the strict dependence of the effects observed on the presence of GATA-2 protein and GATA motifs in DNA
PLZF-RARα has a capacity to modulate GATA-2 activity.
Luciferase reporter gene assays using 293T cells were conducted as described in the legend to Figure 2. (A) Expression plasmids for GATA-2 (GATA-2/pMT2, 100 ng), PLZF (PLZF/pSG5; 500 ng), PLZF-RARα (PLZF-RARα/pSG5, 500 ng), and PML-RARα (PML-RARα/pMT2, 500 ng) were used as indicated, in combination with GATA-1/Luc or mutant GATA-1/Luc reporter plasmid (0.5 μg). ATRA (RA; 1 μΜ) (solid bar) or solvent (dimethyl sulfoxide) (open bar) was added to the culture media 24 hours after transfection, and luciferase activities were measured 24 hours later. (B) Expression plasmids for GATA-2 (GATA-2/pMT2, 100 ng) and PLZF-RARα (PLZF-RARα/pSG5, 500 ng) were used in combination with CD34 × 2/Luc or mutant CD34 × 2/Luc reporter plasmids (0.5 μg), as indicated. ATRA (RA; 1 μM) trichostatin A (TSA, 100 nM), or both, was added to the culture media 24 hours after transfection, and luciferase activities were measured 24 hours later. Luciferase activities were normalized as described in the legend to Figure 2 and were presented as fold increase in activity from the wild-type GATA-1/Luc (A) or CD34 × 2/Luc reporter (B) alone in the absence of the chemical reagents.
PLZF-RARα has a capacity to modulate GATA-2 activity.
Luciferase reporter gene assays using 293T cells were conducted as described in the legend to Figure 2. (A) Expression plasmids for GATA-2 (GATA-2/pMT2, 100 ng), PLZF (PLZF/pSG5; 500 ng), PLZF-RARα (PLZF-RARα/pSG5, 500 ng), and PML-RARα (PML-RARα/pMT2, 500 ng) were used as indicated, in combination with GATA-1/Luc or mutant GATA-1/Luc reporter plasmid (0.5 μg). ATRA (RA; 1 μΜ) (solid bar) or solvent (dimethyl sulfoxide) (open bar) was added to the culture media 24 hours after transfection, and luciferase activities were measured 24 hours later. (B) Expression plasmids for GATA-2 (GATA-2/pMT2, 100 ng) and PLZF-RARα (PLZF-RARα/pSG5, 500 ng) were used in combination with CD34 × 2/Luc or mutant CD34 × 2/Luc reporter plasmids (0.5 μg), as indicated. ATRA (RA; 1 μM) trichostatin A (TSA, 100 nM), or both, was added to the culture media 24 hours after transfection, and luciferase activities were measured 24 hours later. Luciferase activities were normalized as described in the legend to Figure 2 and were presented as fold increase in activity from the wild-type GATA-1/Luc (A) or CD34 × 2/Luc reporter (B) alone in the absence of the chemical reagents.
Discussion
In this report, we have presented evidence that transcription factor GATA-2 can physically associate with PLZF in vitro and in hematopoietic cells in vivo. This interaction resulted in decreased functional activity of GATA-2, as judged by transactivation assays. The mechanisms whereby PLZF modulates GATA factor-dependent transactivation are, however, unclear from the current study. PLZF has been shown to complex with transcriptional corepressors, including N-CoR (nuclear corepressor) and HDAC1 (histone deacetylase 1).18-21,32 33Our experiments (Figure 1C) showed that PLZF could associate with GATA-2 bound to a GATA motif in DNA, indicating that GATA-2 could fall under the control of PLZF together with its attendant corepressors.
Experiments directed at mapping the region of GATA-2 involved in its interaction with PLZF identified the zinc finger region of GATA-2 as playing a critical role. Reciprocal mapping experiments performed with PLZF identified the POZ domain and the zinc fingers of PLZF as critical determinants for its interaction with GATA-2. The zinc finger region of the GATA factors, in addition to its role in DNA binding, has been implicated as a protein–protein interaction domain. GATA factors have been shown to associate with other regulatory proteins (such as Sp1,35 Lmo2,36 CBP,37 other GATAs,38 and PML 11) by virtue of their C4C4 zinc finger regions. Because the zinc finger region has been highly evolutionarily conserved throughout the GATA family,39 it is not surprising that GATA-1 and GATA-3 also have the potential to bind PLZF (data not shown), though PLZF expression within the hematopoietic system is largely confined to immature cells,14,40 in which GATA-2 is thought to be the predominant GATA factor.10,34 Recently ROG was identified as a binding partner of GATA-3.23 ROG has a POZ domain and 3 zinc fingers, and it inhibits GATA-3 activity. ROG is identical to FAZF22 and TZFP.24,25 The zinc fingers of FAZF have been identified as critical for its interaction with GATA-3. In line with this, we presented evidence that FAZF could associate with GATA-2 through the zinc fingers of FAZF (Figure 4B). Although the zinc fingers of FAZF were highly homologous to the last 3 zinc fingers (fingers 7-9) of PLZF (71% at the amino acid level), zinc fingers 7 to 9 of PLZF did not retain the capacity to bind GATA-2 (Figure 4A-B). Interestingly, Guidez et al41 have reported that key lysines in the last 3 zinc fingers of PLZF can be acetylated and that this acetylation alters the functional properties of PLZF.41 These residues are not present in the terminal zinc fingers of FAFZ, and this may provide a mechanistic explanation for the differences in binding activities observed. Nevertheless, taken together, our findings suggest that GATA-2 activity is controlled by these 2 homologous proteins (PLZF and FAZF) by distinct interaction mechanisms.
It is interesting to speculate on what the biologic consequences of the interaction of GATA-2 with PLZF and FAZF might be. GATA-2 has been implicated in the growth and survival of hematopoietic progenitor cells. Knockout experiments support GATA-2 functioning as a positive regulator of these processes.3 Forced-expression experiments have revealed positive and negative effects on progenitor cell proliferation and differentiation.4-6 These different results may reflect differences in cell context. They may also, in part, be attributable to the nature of the GATA-2 moieties involved given that some of the studies made use of GATA-2/ER (estrogen receptor) fusion molecules, which may not retain all the properties of the native GATA-2 molecule. PLZF has also been shown to be involved in growth control, by which it functions as a negative regulator of the cell cycle.16,17 PLZF knockout has apparently normal hematopoiesis, though the cycling activity of blood progenitors has not to our knowledge been closely examined.42 FAZF is also expressed within hematopoietic progenitors (A. Zelent, unpublished observations, January 1998) and has been shown to be a negative regulator of GATA-3 activity in T cells.23 Its function within hematopoietic progenitors is not as yet understood. Although it is difficult at this stage to speculate on the biologic consequences of the association of GATA-2 with PLZF or FAZF, it seems likely that PLZF and FAZF will function to repress GATA-2 activity. The observation that PLZF and FAZF use distinct regions to interact with GATA-2 raises the possibility that their interactions may be differentially regulated, providing additional points for control. Expectations as to whether the repressive effects of PLZF or FAZF on GATA-2 will manifest as pro- or antiproliferative signals depends on one's view of GATA-2 activities on these processes. In truth, thinking of GATA-2 simply as a pro- or antiproliferative activity is probably inadequate. The function of any given transcription factor is the sum of the activity of the entire network of genes whose activity it is involved in regulating in that particular cell context or cell state. Similarly, depending on the local transcriptional context, individual factors may function positively or negatively with respect to the transcription of particular genes.
The interaction of PLZF-RARα with GATA-2 raises the intriguing possibility that GATA-2 activity may be deregulated in leukemia. APL is associated with chromosomal translocations involving RARα, typically giving rise to PML-RARα12,13 or PLZF-RARα fusion proteins.14 Transcription from ATRA-responsive elements (RARE) by PML-RARα is known to be stimulated by pharmacologic concentrations of ATRA (1 μM), but not by physiological concentrations (1 nM). PLZF-RARα, on the other hand, is a weaker activator but does respond to ATRA.43 Thus, in the case of PLZF-RARα, RARE-dependent transcription is most evidently stimulated by the combination of ATRA (1 μM) and TSA, but only weakly by ATRA alone. This observation may reflect the release of co-repressors by the combination of both drugs, but not sufficiently by either one. Given that the activity of GATA-2 complexed with PLZF-RARα was evidently stimulated by ATRA plus TSA but only weakly by ATRA alone, one can speculate that similar mechanisms operate on PLZF-RAR in RARE-dependent transcription and in GATA-dependent transcription. These drug treatments induce the differentiation of APL cells associated with PML-RARα or PLZF-RARα, arguing for RARE-driven transcription as primarily important for the induction of differentiation.43 We have recently presented evidence showing that through its interaction with PML-RARα, the activity of GATA-2 can be stimulated by pharmacologic (1 μM), but not by physiologic (1 nM), concentrations of ATRA.11 In this report we have shown that GATA-2 activity associated with PLZF-RARα is most evidently stimulated by treatment with ATRA (1 μM) plus TSA. These findings raise the possibility that some GATA-2 target genes are activated or de-repressed during chemically induced differentiation of APL cells. Understanding the functional significance of interactions between GATA-2 and wild-type or chimeric PLZF proteins will require a better understanding of GATA-2 target genes in normal and leukemic cells.
We thank A. Zelent for useful suggestions and Janine Harris for secretarial assistance.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Tariq Enver, Section of Gene Function and Regulation, Institute of Cancer Research, Chester Beatty Laboratories, 237 Fulham Rd, London SW3 6JB, United Kingdom; e-mail:tariq@icr.ac.uk.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal