Abstract
Hereditary mutations associated with hematologic malignancies are rare. Heterozygous mutations affecting the hematopoietic transcription factor CBFA2 (also AML1/RUNX1) were recently reported to be associated with familial platelet disorder with predisposition to acute myeloid leukemia (FPD/AML, MIM 601399). A new 3-generation family with FPD/AML with a novel CBFA2 mutation is described. In this family, AML was diagnosed in a second-generation male. After allogeneic stem cell transplantation from his human leukocyte antigen–identical sister, a donor-derived, genetically identical leukemia developed in the recipient and the donor. Sequencing analysis identified a G-to-T transition within the CBFA2 gene, which involves codon 198, encoding a conserved aspartic acid within the DNA- binding Runt domain. Three of 5 siblings affected with the FPD/AML trait harbored the mutation in a heterozygous form. This experience underscores the necessity of performing mutation analysis of the CBFA2 gene before sibling allogeneic transplantation in families with FPD/AML.
Introduction
Hereditary mutations are rare in hematologic malignancies. Generally, somatic mutations are associated with leukemogenesis. The hematopoietic transcription factor CBFA2/AML1/RUNX1 is frequently affected in leukemia. Chromosomal translocations (8;21), resulting in a fusion of CBFA2 and ETO, and t(12;21), resulting in a chimerical TEL-CBFA2 gene, are detected in 15% and 25% of acute myeloid leukemia (AML) and childhood acute lymphoid leukemia (ALL), respectively. Recently, mutations in the DNA binding Runt domain of CBFA2 have been reported in hematologic (pre-) malignancies of the myeloid lineage.1-4
Familial platelet disorder (FPD) with predisposition to AML (FPD/AML, MIM 601399) is an autosomal dominant disorder characterized by thrombocytopenia, functional platelet abnormalities, and prolonged bleeding time, and it is associated with predisposition to AML.5 Recently, Gilliland and colleagues7reported haploinsufficiency of CBFA2 in 6 families with familial thrombocytopenia and propensity to the development of AML. Genetic analyses revealed heterozygous nonsense and missense mutations or intragenic deletions in one allele of the CBFA2 gene. These studies support a model for FPD/AML in which haploinsufficiency of CBFA2 is causal in leukemogenesis.
We present a newly identified family with FPD/AML, and we describe the clinical, cytogenetic and molecular features of this family harboring a novel missense mutation within the Runt domain. Our data demonstrate the necessity of performing sequence analysis of theCBFA2 gene in families with FPD/AML.
Study design
Materials were collected after informed consent of the patients. Leukocyte-derived genomic DNA was used to amplify exons 3, 4, and 5 and flanking splice sites of the CBFA2 gene by polymerase chain reaction and primers as described.2 Automated DNA sequence analysis was performed with the ABI Prism dRhodamine Terminator Cycle Sequence Ready Reaction kit (PE Biosystems, Warrington, United Kingdom), and samples were analyzed on an ABI 310 Genetic Analyzer (Applied Biosystems, Foster City, CA). Interphase fluorescence in situ hybridization (FISH) was performed using chromosome 21 probe LSI21 (Vysis, Downers Grove, IL).
Results and discussion
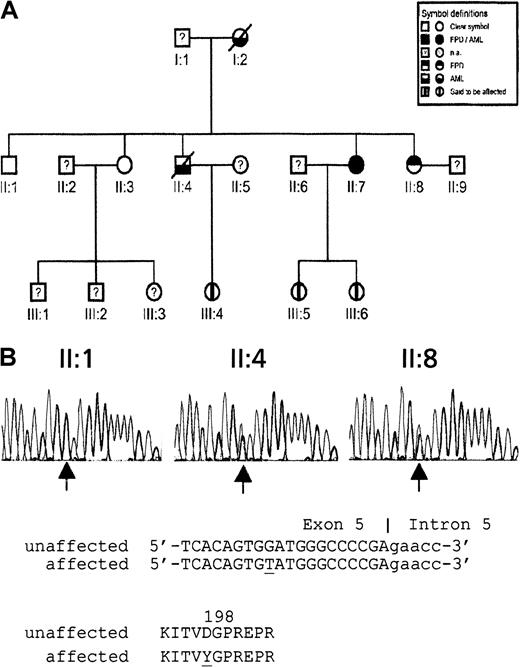
The pedigree of the family with FPD/AML is represented in Figure 1A. A 33-year-old man (patient II:4) was admitted to our hospital with AML (FAB M1) in August 1996. Cytogenetic analysis revealed a tetraploid 92,XXYY,add(7)(q31),add(7)(q31)[14]/46,XY[10] karyotype (Table 1). After remission-induction therapy, the patient underwent allogeneic bone marrow transplantation with a partial T-cell–depleted graft from his human leukocyte antigen–matched sister (patient II:7) in December 1996. During the pretransplantation work-up, a storage pool deficiency combined with slight thrombocytopenia was found in the patient II:4 and in the family donor II:7 (platelets 140 × 109/L and 110 × 109/L, respectively). Bleeding time was longer than 30 minutes, and platelet aggregation in response to adenosine diphosphate (ADP), adrenaline, and collagen was almost absent at all concentrations. Platelet ADP was decreased, whereas platelet adenosine triphosphate (ATP) and platelet serotonin showed (near) normal values (Table 2). Low-platelet ADP levels and abnormal platelet aggregations correlated with previous reports of platelet abnormalities in FDP/AML pedigrees.5 6 Subsequent bone marrow investigation of morphology and cytogenetic analysis revealed no abnormalities (Table1). In patient II:4, peripheral blood cell recovery after transplantation was slow and incomplete. One year after transplantation, hemoglobin level was 6.3 mM, white blood cell count was 2.4 × 109/L (55% neutrophils), and platelet level was 41 × 109/L. Platelet function test results were abnormal were consistent with storage pool deficiency (Table2). Five routine bone marrow investigations performed within 16 months of transplantation showed incomplete cellularity with slight dysplastic changes in all cell lineages; however, no increase in marrow blasts occurred. Twenty-one months after transplantation, 5% abnormal blasts were found. Blast infiltration increased to 20% in the following 3 months. Cytogenetic analysis revealed a 47,XX,+21[13]/47,XX,+8[7]/46,XX[4] karyotype, indicating that the leukemic cells were of donor origin (Table 1).
Pedigree of 3-generation family with FPD/AML and sequence analysis of D198Y missense mutation.
(A) Pedigree (na, not analyzed). (B) Detection by sequence analysis of a mutation in exon 5 of CBFA2 using second-generation family members II:1 (nonaffected), II:4 (affected), and II:7 (affected). Arrows indicate the position of the mutation. Genomic and encoded amino acid sequences are depicted. Affected nucleotide and amino acid are underlined.
Pedigree of 3-generation family with FPD/AML and sequence analysis of D198Y missense mutation.
(A) Pedigree (na, not analyzed). (B) Detection by sequence analysis of a mutation in exon 5 of CBFA2 using second-generation family members II:1 (nonaffected), II:4 (affected), and II:7 (affected). Arrows indicate the position of the mutation. Genomic and encoded amino acid sequences are depicted. Affected nucleotide and amino acid are underlined.
Cytogenetic and clinical features in pedigree
| Family member . | Date . | Status . | GTG karyotype . | FISH interphase (chromosome 21) . |
|---|---|---|---|---|
| II:1 | Dec 1998 | 46,XY | No + 21 | |
| II:3 | Dec 1998 | 46,XX | BT | |
| II:4 | Aug 1996 | AML M1 | 92,XXYY, add(7)(q31), add(7)(q31)[14]/46,XY[10] | |
| Apr 1997 | After BMT | 46,XX | ||
| Nov 1997 | After BMT | |||
| Sep 1998 | Relapse/secondary leukemia? | 47,XX,+21[13]/47,XX,+8[7]/46,XX[4] | + 21 (37%) | |
| II:7 | Oct 1996 | Donor | 46,XX | |
| Oct 1998 | MDS | 46,XX | BT | |
| Feb 1999 | AML M1 | 46,XX | BT | |
| Apr 1999 | Before MUD | BT | ||
| Dec 1999 | After BMT | 46,XX | ||
| Feb 2001 | After BMT | 46,XX | BT | |
| II:8 | Nov 1998 | 46,XX | BT | |
| Mar 1999 | MDS? | 46,XX | No + 21 |
| Family member . | Date . | Status . | GTG karyotype . | FISH interphase (chromosome 21) . |
|---|---|---|---|---|
| II:1 | Dec 1998 | 46,XY | No + 21 | |
| II:3 | Dec 1998 | 46,XX | BT | |
| II:4 | Aug 1996 | AML M1 | 92,XXYY, add(7)(q31), add(7)(q31)[14]/46,XY[10] | |
| Apr 1997 | After BMT | 46,XX | ||
| Nov 1997 | After BMT | |||
| Sep 1998 | Relapse/secondary leukemia? | 47,XX,+21[13]/47,XX,+8[7]/46,XX[4] | + 21 (37%) | |
| II:7 | Oct 1996 | Donor | 46,XX | |
| Oct 1998 | MDS | 46,XX | BT | |
| Feb 1999 | AML M1 | 46,XX | BT | |
| Apr 1999 | Before MUD | BT | ||
| Dec 1999 | After BMT | 46,XX | ||
| Feb 2001 | After BMT | 46,XX | BT | |
| II:8 | Nov 1998 | 46,XX | BT | |
| Mar 1999 | MDS? | 46,XX | No + 21 |
Threshold of sensitivity for FISH is at 4% to 5%.
BT indicates below threshold; BMT, bone marrow transplantation.
Results of platelet function tests in pedigree
| Family member . | Serotonin (μmol/1011platelets) . | ADP . | ATP . | Bleeding time (min/150 × 109 platelets) . | Platelet aggregation . |
|---|---|---|---|---|---|
| (μmol/1011 platelets) . | |||||
| II:1 | 439 | 3.1 | 5.2 | 5 | Good |
| II:3 | 366 | 2.5 | 5.0 | 5 | Good |
| II:4 | |||||
| Before SCT | 197 | 0.5 | 3.8 | > 30 | Very poor |
| After SCT | 189 | 1.0 | 5.2 | na | Very poor |
| II:7 | 440 | 1.4 | 5.2 | > 30 | Very poor |
| II:8 | 305 | 0.9 | 4.4 | > 30 | Very poor |
| Normal range | 200-600 | 1.7-3.0 | 3.1-7.0 | 7 | |
| Family member . | Serotonin (μmol/1011platelets) . | ADP . | ATP . | Bleeding time (min/150 × 109 platelets) . | Platelet aggregation . |
|---|---|---|---|---|---|
| (μmol/1011 platelets) . | |||||
| II:1 | 439 | 3.1 | 5.2 | 5 | Good |
| II:3 | 366 | 2.5 | 5.0 | 5 | Good |
| II:4 | |||||
| Before SCT | 197 | 0.5 | 3.8 | > 30 | Very poor |
| After SCT | 189 | 1.0 | 5.2 | na | Very poor |
| II:7 | 440 | 1.4 | 5.2 | > 30 | Very poor |
| II:8 | 305 | 0.9 | 4.4 | > 30 | Very poor |
| Normal range | 200-600 | 1.7-3.0 | 3.1-7.0 | 7 | |
SCT indicates stem cell transplantation; na, not analyzed.
Concurrently, myelodysplastic syndrome (MDS) type RAEB was diagnosed in the donor. Cytogenetic analysis showed a normal karyotype. RAEB rapidly progressed to an AML M1 phenotype. Patients II:4 and II:7 underwent transplantation using matched, unrelated donors (MUD) in March and May 1999, respectively. Patient II:4 died of Epstein-Barr virus–related lymphoma in May 2000. Patient II:7 is still in complete remission and has normal peripheral blood counts more than 2 years after transplantation. Cytogenetic analyses in December 1999 and February 2001 did not reveal abnormalities.
Cytogenetic analysis of family members II:1, II:3, and II:8 revealed normal karyotypes at the end of 1998 (Table 1). Peripheral blood cell counts and platelet functions were normal in family members II:1 and II:3. However, in family member II:8, storage pool deficiency and thrombocytopenia (platelets 90 × 109/L) were found (Table 2). Bone marrow morphology was consistent with MDS type RA with 3% to 4% myeloblasts. Third-generation family members III:4, III:5, and III:6 have anecdotal evidence of hemorrhagic diathesis. Family member I:2 had a hemorrhagic diathesis and died of leukemia.
To further elucidate the genetic origin of the FPD/AML phenotype, we sequenced exons 3, 4, and 5 of the CBFA2 gene encoding the Runt DNA-binding domain in family members II:1, II:3, II:4, II:7, and II:8. This domain is reported to be mutated in acquired and hereditary leukemia.1-3,7 As shown in Figure 1B, a missense mutation was detected within the codon encoding D198. This G-to-T transition cosegregated with the FPD/AML trait. Only the affected members II:4, II:7, and II:8 carried the mutation. Sequence analysis of 108 control subjects indicated that this was not a common polymorphism. This newly identified D198Y mutation is only 3 residues from the R201Q mutation reported by Gilliland and colleagues.7 Residue D198 is highly conserved (data not shown). It is located within a region of the Runt domain that is critical for DNA binding,8 suggesting a predicted disruption of DNA binding, analogous to the R201Q mutation. Therefore, this apparent loss-of-function D198Y mutation is unlikely to function as a dominant-negative protein, supporting a model of CBFA2 haploinsufficiency in FPD/AML.
Recently, mutations have been reported to occur within the Runt domain of CBFA2 in patients with AML with acquired trisomy 21.2 This is of particular interest because in patient II:4, in 37% of the leukemic cells of donor (patient II:7) origin, an overt trisomy 21 was detected. However, interphase FISH analysis of bone marrow cells of patients II:7 and II:8 did not reveal trisomy 21 above threshold level (Table 1). Trisomy 21 was not observed at diagnosis in patient II:4 either. Therefore, it is unclear whether an association exists between the development of MDS/AML, an acquired trisomy 21, and the CBFA2 mutation in this family with FPD/AML.
In conclusion, we report the identification of a novel CBFA2 single-nucleotide mutation within a familial leukemia. Because an association between FPD/AML and haploinsufficiency of CBFA2has been described, our report indicates that, in addition to careful phenotyping for FPD, mutation analysis of CBFA2 and its promoter and haplotype analysis are required for the screening of transplants from sibling donors in families with FPD/AML.
We thank Drs Andries Bloem and Karel Nieuwenhuis for helpful discussions. We also thank the members of the laboratories for Cellular Diagnostics and Cytogenetics for their excellent technical assistance.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Arjan Buijs, Division of Medical Genetics, University Medical Center Utrecht, Rm KC04.084.2, Lundlaan 6, 3584 EA Utrecht, The Netherlands; e-mail: a.buijs@dmg.azu.nl.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal