Abstract
T-cell receptor (TCR) gene rearrangements are mediated via V(D)J recombination, which is strictly regulated during lymphoid differentiation, most probably through the action of specific transcription factors. Investigated was whether cotransfection ofRAG1 and RAG2 genes in combination with lymphoid transcription factors can induce TCR gene rearrangements in nonlymphoid human cells. Transfection experiments showed that basic helix-loop-helix transcription factors E2A and HEB induce rearrangements in the TCRD locus (Dδ2-Dδ3 and Vδ2-Dδ3) and TCRG locus (ψ Vγ7-Jγ2.3 and Vγ8-Jγ2.3). Analysis of these rearrangements and their circular excision products revealed some peculiar characteristics. The Vδ2-Dδ3 rearrangements were formed by direct coupling without intermediate Dδ2 gene segment usage, and most Dδ2-Dδ3 recombinations occurred via direct coupling of the respective upstream and downstream recombination signal sequences (RSSs) with deletion of the Dδ2 and Dδ3 coding sequences. Subsequently, the E2A/HEB–induced TCR gene recombination patterns were compared with those in early thymocytes and acute lymphoblastic leukemias of T- and B-lineage origin, and it was found that the TCR rearrangements in the transfectants were early (immature) and not necessarily T-lineage specific. Apparently, some parts of theTCRD (Vδ2-Dδ region) and TCRG genes are accessible for recombination not only in T cells, but also in early B-cells and even in nonlymphoid cells if the appropriate transcription factors are present. The transfection system described here appeared to be useful for studying the accessibility of immunoglobulin and TCR genes for V(D)J recombination, but might also be applied to study the induction of RSS-mediated chromosome aberrations.
Introduction
Antigen recognition by lymphocytes is dependent upon successful rearrangement of immunoglobulin and T-cell receptor (TCR) genes from variable (V), diversity (D), and joining (J) gene segments through the process of V(D)J recombination.1The rearrangement processes are mediated by the recombination-activating gene (RAG)–1 and RAG2 proteins, which specifically recognize the recombination signal sequences (RSSs) that flank the coding regions of the V, D, and J gene segments.2-4 RSSs are consensus sequences consisting of a heptamer and nonamer separated by a 12–base pair (bp) or 23-bp spacer. Site-specific cleavage at the borders of RSSs and coding elements by the RAG proteins is followed by a process of rejoining of DNA ends in which the double-strand break-repair enzymes play a central role. Antigen receptor assembly is critically dependent upon expression of the lymphoid-specific RAG proteins; this is further illustrated by the fact that ectopic RAG expression results in site-specific recombination both in vitro and in vivo.2-4
The whole process of V(D)J recombination is ordered and tightly regulated during lymphoid differentiation.5 The hierarchical order is apparent at different levels. First, cells committed to the B-cell lineage undergo immunoglobulin rearrangements, whereas TCR genes rearrange in T-cell precursors. Nevertheless, so-called cross-lineage rearrangements might occasionally occur in precursor B and T cells; this phenomenon is particularly evident from malignantly transformed lymphoid precursor cells, ie, B- and T-lineage acute lymphoblastic leukemias (ALLs).6,7 Furthermore, immunoglobulin (Ig) heavy chain (IGH) rearrangements are known to precede Ig kappa (IGK) and Ig lambda light-chain recombination.8 Similarly, data from human T-ALL and sorted human thymocyte subpopulations indicate that TCRδ(TCRD) recombination occurs prior to TCRγ(TCRG) and TCRβ (TCRB) rearrangement, whereas the TCRα (TCRA) locus is rearranged at a late stage only.9-11 The ordered process is also apparent from the finding that DH-JH or Dβ-Jβ rearrangements generally precede the coupling of their respective V gene segments.12,13 Finally, regulation can occur even at the level of gene segment usage, as is apparent from murine thymic ontogeny where Vγ and Vδ gene rearrangements occur in waves with different V segments being used in fetal and adult thymocytes.14 15
It has been suggested that the tight and hierarchical regulation of the rearrangement processes can be explained by differential chromatin accessibility to the V(D)J recombinase,16 which in turn is controlled by transcription factors binding to promoters and enhancers. E proteins are an important class of transcription factors in lymphoid differentiation. They consist of a helix-loop-helix (HLH) dimerization motif and a basic DNA binding domain that binds to conserved E-box motifs, as identified in immunoglobulin, TCRB, and CD4 enhancers. Members of the E-protein family include E2-2, HEB, and the E12 and E47 splice variant products of the E2Agene.17 E12 and E47 are differentially expressed in a wide variety of tissues, but exist as homodimers only in B-lineage cells.18,19 In T cells, E-box binding complexes are heterodimers of E2A and HEB. E2A−/− mice have a block in B-cell differentiation prior to the start of the immunoglobulin recombination process.20,21 Induction of DH-JH and IGK rearrangements upon ectopic E2A expression in a pre–T-cell line or even nonlymphoid human cells illustrates its involvement in regulation of V(D)J recombination.22,23 E2A−/− mice also have a severe, though less complete, defect in T-cell differentiation at the double-negative stage. Interestingly, in E2A−/− mice, certain TCRγδ T-cell subsets are lacking, owing to an impaired ability to rearrange particular TCRG and TCRDgene segments.15 Targeted disruption of theHEB gene gives rise to a partial block in early T-cell differentiation prior to the development of double-positive cells.24
It is now generally accepted that V(D)J recombination processes are also involved in the formation of particular chromosome aberrations in human leukemias.25-27 This especially concerns aberrations in human T-ALL, in which TCRB or TCRD gene segments are translocated to oncogenes, resulting in activation of these oncogenes through TCR regulatory elements.28Although little is known about the exact molecular processes, it is tempting to speculate that transcription factor–induced accessibility of the involved loci is a critical step in the formation of these aberrations.
Here we show that 2 types of E proteins, E2A and HEB, have the ability to target the recombination machinery to TCR loci in nonlymphoid cells. Expression of E2A or HEB in the presence of RAG1/RAG2 appeared to induce immature types of TCRDrearrangements, several TCRG rearrangements, but noTCRB recombination.
Materials and methods
Cell culture
Nonlymphoid BOSC 23 cells were cultured in Dulbecco modified Eagle medium with 10% fetal bovine serum at 37°C with 5% CO2.29
DNA constructs
The E12 and E47 variants of the human E2A transcription factor gene were cloned into the pHβAPneo vector, which has been previously described.30 The human HEB transcription factor gene was cloned in the pXS vector, which is derived from pcDL-Sra296.31 The pEBB-Rag1 and pEBB-Rag2 expression vectors have been described before.32
Transfection protocol
Transfections were performed via calcium phosphate precipitation as described.23 BOSC 23 cells were plated on the day prior to transfection at a density of 4.5 × 106 cells per 10-cm dish. On the day of the transfection, 18 to 24 μg total DNA, including 6 μg each expression vector or carrier DNA, was used per transfection. The cells were harvested 3 days after transfection.
Polymerase chain reaction analysis of TCR rearrangements (coding and signal joints)
By polymerase chain reaction (PCR), 200 ng genomic DNA, isolated from the various BOSC 23–transfected cell cultures, was analyzed in a 50-μL reaction volume containing 1 × Taq Gold buffer (Applied Biosystems, Foster City, CA), 1.5 mM MgCl2, 12.5 pmol each primer, 200 μM deoxy–nucleoside 5′ triphosphate, and 1 U AmpliTaq Gold (Applied Biosystems). PCR reactions were performed on an ABI480 machine as follows: 10 minutes preactivation at 94°C, 40 cycles of 45 seconds at 94°C, 90 seconds at 60°C, 2 minutes at 72°C, followed by a 10-minute extension at 72°C. The primers used to detect TCRD rearrangements as well as their circular excision products locus are listed in Table1. TCRG gene rearrangements were studied with the use of VγI-3′, VγII-3′, VγIII-3′, VγIV-3′, Jγ1.1/2.1-3′, Jγ1.2-3′, and Jγ1.3/2.3-3′ primers.33,TCRB analysis was performed with Vβ2 and Vβ5A family primers (A.W.L., unpublished data, 2001) or Dβ1 and Dβ2 primers (T. Szczepanski, unpublished results, 2001) in combination with Jβ1(2) and Jβ2(2) primers.34 Deletional rearrangements in the TCRD locus were studied with the use of δREC-3′ and ψJα-3′ primers (Table 1).35 In all reactions, proper positive controls for the various types of TCR rearrangements were included: well-defined leukemic cell DNA and/or total thymus DNA. We used mock-transfected BOSC 23 cells and/or HeLa genomic DNA as template containing negative controls. We analyzed 20 μL each PCR on a 2% agarose gel, followed by ethidium bromide staining. If TCR rearrangements were detected in the agarose gels, the remainder of the PCR products were subjected to heteroduplex analysis to discriminate between monoclonal and polyclonal rearrangements.36 In short, heteroduplex analysis consisted of 5 minutes denaturation at 94°C and 60 minutes renaturation at 4°C prior to electrophoresis on 6% nondenaturing polyacrylamide gels (polyacrylamide to bisacrylamide, 29:1) in 0.5 × TBE buffer.36 Ethidium bromide–stained homoduplex or heteroduplex PCR products were visualized with UV light.
Primers and probes used for polymerase chain reaction amplification of TCRD rearrangements and real-time quantitative polymerase chain reaction
| Primer . | Sequence (5′-3′) . | Reference . |
|---|---|---|
| Coding joint PCR | ||
| Vδ1-5′⁗ | ACT CAA GCC CAG TCA TCA GTA TCC | 33 |
| Vδ2-5′ | ACC AAA CAG TGC CTG TGT CAA TAG G | 33 |
| Vδ3-5′ | GAC CAG ACG GTG GCG AGT GGC | 33 |
| Dδ2-5′ RO2 | CCA CAT TGG GAG TGT CAA CAT TT | This study |
| Dδ3-3′ N | CTT CCT GCT ATC CCT TCC AGG | 33 |
| Dδ3-5′ | CGC GTC GAC CAT ATA GTG GAA ACC GAG GGG | This study |
| Jδ1-3′ | ACC TCT TCC CAG GAG TCC TCC | 33 |
| Jδ3-3′ | CGC GTC GAC TCA AAT TAT CCC AGA AAT ATA GG | 44 |
| δREC-3′ | GCA ACA TCA CTC TGT GTC TAG C | 35 |
| ψJα-3′ | CCT GAA GCT TAA GGC ACA TTA GAA TCT CTC ACT G | 35 |
| Signal joint PCR | ||
| Vδ2-sj 3′ | CTG GTC AGT GGT TTT TGA GCT GCT | This study |
| Dδ2-5′ XBg | GTA GAT CTA GAA GAG GGT TTT TAT ACT GAT GTG | This study |
| Dδ2-lower | TCC CAA TGC TGA GAC ATA CAT | This study |
| Dδ3-upper | CAG GGG CCA TAT AGT TGT GAA | This study |
| Dδ3-5′ S | CGC GTC GA CCA TAT AGT GTG AAA CCG AGG GG | This study |
| Dδ3-3′ N | CTT CCT GCT ATC CCT TCC AGG | 33 |
| Jδ1 sj 5′ | GTC CCT ACC TGC AGA TGA TTA ACC | This study |
| Jδ3 sj 5′ | CCC TTG GTC TCA TCA AGA GCA GC | This study |
| Real-time PCR | ||
| F-DD2-KLON | GAA GAA GAG GGT TTT TAT ACT GAT GTG TT | This study |
| R-DD3-CONS4 | TTG CCC CTG CAG TTT TTG TAC | This study |
| T-DD3-CONS2 | ATA CGC ACA GTG CTA CAA AAC CTA CAG AGA CCT | This study |
| F-VG8-KLON | TCT ATT ACT GTG CCA CCT GGG ATA | This study |
| R-JG13/23-KLON | TTC CTG CCT TCC CTC TAT TAC CTT | This study |
| T-JG13/23-CONS3 | TGT CAC AGG TAA GTA TCG GAA GAA TAC AAC ATT TCC | This study |
| Primer . | Sequence (5′-3′) . | Reference . |
|---|---|---|
| Coding joint PCR | ||
| Vδ1-5′⁗ | ACT CAA GCC CAG TCA TCA GTA TCC | 33 |
| Vδ2-5′ | ACC AAA CAG TGC CTG TGT CAA TAG G | 33 |
| Vδ3-5′ | GAC CAG ACG GTG GCG AGT GGC | 33 |
| Dδ2-5′ RO2 | CCA CAT TGG GAG TGT CAA CAT TT | This study |
| Dδ3-3′ N | CTT CCT GCT ATC CCT TCC AGG | 33 |
| Dδ3-5′ | CGC GTC GAC CAT ATA GTG GAA ACC GAG GGG | This study |
| Jδ1-3′ | ACC TCT TCC CAG GAG TCC TCC | 33 |
| Jδ3-3′ | CGC GTC GAC TCA AAT TAT CCC AGA AAT ATA GG | 44 |
| δREC-3′ | GCA ACA TCA CTC TGT GTC TAG C | 35 |
| ψJα-3′ | CCT GAA GCT TAA GGC ACA TTA GAA TCT CTC ACT G | 35 |
| Signal joint PCR | ||
| Vδ2-sj 3′ | CTG GTC AGT GGT TTT TGA GCT GCT | This study |
| Dδ2-5′ XBg | GTA GAT CTA GAA GAG GGT TTT TAT ACT GAT GTG | This study |
| Dδ2-lower | TCC CAA TGC TGA GAC ATA CAT | This study |
| Dδ3-upper | CAG GGG CCA TAT AGT TGT GAA | This study |
| Dδ3-5′ S | CGC GTC GA CCA TAT AGT GTG AAA CCG AGG GG | This study |
| Dδ3-3′ N | CTT CCT GCT ATC CCT TCC AGG | 33 |
| Jδ1 sj 5′ | GTC CCT ACC TGC AGA TGA TTA ACC | This study |
| Jδ3 sj 5′ | CCC TTG GTC TCA TCA AGA GCA GC | This study |
| Real-time PCR | ||
| F-DD2-KLON | GAA GAA GAG GGT TTT TAT ACT GAT GTG TT | This study |
| R-DD3-CONS4 | TTG CCC CTG CAG TTT TTG TAC | This study |
| T-DD3-CONS2 | ATA CGC ACA GTG CTA CAA AAC CTA CAG AGA CCT | This study |
| F-VG8-KLON | TCT ATT ACT GTG CCA CCT GGG ATA | This study |
| R-JG13/23-KLON | TTC CTG CCT TCC CTC TAT TAC CTT | This study |
| T-JG13/23-CONS3 | TGT CAC AGG TAA GTA TCG GAA GAA TAC AAC ATT TCC | This study |
PCR indicates polymerase chain reaction; sj, signal joint.
Cloning of PCR products and sequencing
Following amplification, PCR products were purified by means of QIAquick PCR purification kits (Qiagen, Hilden, Germany) and cloned into pGEM-T Easy vector (Promega, Madison, WI) according to the manufacturer's instructions. Clones containing insert were sequenced on the ABI377 fluorescent sequencer, by means of the dye terminator cycle sequencing kit and AmpliTaq FS (Applied Biosystems).7
Real-time quantitative PCR of TCR rearrangements
Levels of particular rearranged TCRD andTCRG PCR products were quantified by real-time quantitative (RQ) PCR, by means of TaqMan technology on the ABI Prism 7700 Sequence Detection System (Applied Biosystems), as described earlier.37-39 To this end, forward (F-DD2-KLON, F-VG8-KLON) and reverse primers (R-DD3-CONS4, R-JG13/23-KLON) (Table 1) were designed by means of Primer Express (Applied Biosystems) and Oligo6.2 (Dr W. Rychlik, Molecular Biology Insights, Cascade, CO) software to select melting temperature values of 58°C to 60°C and to exclude hairpin formation, dimer formation, and false priming. Design of the primers was performed so that the primers could be used with already present dual-labeled TaqMan probes (T-DD3-CONS2, T-JG13/23-CONS3) (Table 1). An albumin primer/probe RQ-PCR set (Applied Biosystems) was used to quantitate and normalize the amount of DNA used in the various transfections.37
Results
E2A and HEB induce incomplete TCRD gene rearrangements in nonlymphoid cells
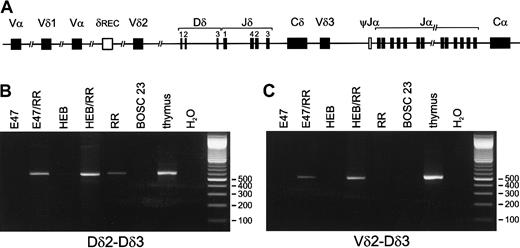
To study the effect of E-box proteins on recombination events in the various human TCR loci, we employed the model system described by Romanow et al.23 Nonlymphoid BOSC 23 cells, which harbor their TCR loci in germline configuration, were transfected with E2A, splice variants (E12 or E47), or HEB, either alone or in combination with the RAG1/RAG2 proteins. PCR analysis of genomic DNA, isolated 3 days posttransfection, was performed by means of specific primers for the most frequently occurring types of incomplete and completeTCRD gene rearrangements (Dδ2-Dδ3, Vδ2-Dδ3, Dδ2-Jδ1, Vδ1-Jδ1, Vδ2-Jδ1, Vδ3-Jδ1 recombinations) (Figure 1A). Transfection of E2A or HEB alone, or mock transfection, did not result in activation of any of these rearrangements. However, Dδ2-Dδ3 and Vδ2-Dδ3 rearrangements were clearly induced upon transfection of either E2A or HEB in combination with the 2 RAG genes (Figure 1B-C). In contrast to Vδ2-Dδ3 rearrangements, Dδ2-Dδ3 recombination products were also detectable in transfectants with the 2 RAG genes only. However, quantification by RQ-PCR with the TaqMan technology revealed that the Dδ2-Dδ3 rearrangement levels were essentially higher (5- to 10-fold) in the HEB-plus-RAG transfectants than in the RAG1/RAG2–only transfectants. Further cloning and sequencing of the PCR products from the various transfectants showed heterogeneous Vδ2-Dδ3 junctional regions in all transfection combinations, with variable numbers of deleted nucleotides at both sides and occasionally introduction of palindromic (P) nucleotides (Table2). Strikingly, virtually all sequenced Dδ2-Dδ3 rearrangements of the transfected cells were found to be identical, showing complete deletion of the Dδ2 and Dδ3 gene segments and direct coupling of the upstream RSS of the Dδ2 segment to the downstream RSS of the Dδ3 segment (Table 2, Figure2). This so-called signal joint, which is normally present in excision circles, was only occasionally found in normal thymocytes (Table 2, Figure 2).
Induction of TCRDrecombination by E2A or HEB in the presence of RAG1 and RAG2.
Both E2A and HEB in combination with RAG1 and RAG2 have the ability to induce TCRD recombination in nonlymphoid cells. (A) Schematic diagram of the human TCRD/TCRA locus. Presented are the Vδ, Dδ, and Jδ gene segments that are positioned between the Vα and Jα-Cα regions in the TCRA locus. The δREC and ψJα gene segments that flank the TCRD locus are involved in deletion of the TCRD gene prior toTCRA recombinations. (B) (C) BOSC 23 cells were transfected with E47, HEB, RAG1, and RAG2 expression vectors. Following genomic DNA isolation, 200 ng DNA of the various transfectants was used for PCR amplification by means of Dδ2-5′ RO2 plus Dδ3-3′ N (panel B) or Vδ2-5′ plus Dδ3-3′ N (panel C) primers, which are used for specific detection of Dδ2-Dδ3 and Vδ2-Dδ3 rearrangements, respectively. Thymus DNA was used as positive control, and mock-transfected BOSC 23 as nonspecific template control. PCR products were run on a 2% agarose gel and stained with ethidium bromide. Rearranged Dδ2-Dδ3 and Vδ2-Dδ3 PCR products were observed upon combined transfection of either E2A or HEB together with RAG1 plus RAG2 (RR). Dδ2-Dδ3 PCR products were also detectable in genomic DNA derived from transfectants expressing RR only (lane RR), though at much lower levels as quantitated via RQ-PCR (see “Results”).
Induction of TCRDrecombination by E2A or HEB in the presence of RAG1 and RAG2.
Both E2A and HEB in combination with RAG1 and RAG2 have the ability to induce TCRD recombination in nonlymphoid cells. (A) Schematic diagram of the human TCRD/TCRA locus. Presented are the Vδ, Dδ, and Jδ gene segments that are positioned between the Vα and Jα-Cα regions in the TCRA locus. The δREC and ψJα gene segments that flank the TCRD locus are involved in deletion of the TCRD gene prior toTCRA recombinations. (B) (C) BOSC 23 cells were transfected with E47, HEB, RAG1, and RAG2 expression vectors. Following genomic DNA isolation, 200 ng DNA of the various transfectants was used for PCR amplification by means of Dδ2-5′ RO2 plus Dδ3-3′ N (panel B) or Vδ2-5′ plus Dδ3-3′ N (panel C) primers, which are used for specific detection of Dδ2-Dδ3 and Vδ2-Dδ3 rearrangements, respectively. Thymus DNA was used as positive control, and mock-transfected BOSC 23 as nonspecific template control. PCR products were run on a 2% agarose gel and stained with ethidium bromide. Rearranged Dδ2-Dδ3 and Vδ2-Dδ3 PCR products were observed upon combined transfection of either E2A or HEB together with RAG1 plus RAG2 (RR). Dδ2-Dδ3 PCR products were also detectable in genomic DNA derived from transfectants expressing RR only (lane RR), though at much lower levels as quantitated via RQ-PCR (see “Results”).
Sequences of cloned TCRD recombination products
| Sample . | 5′ gene segment* . | P nucleotides . | 3′ gene segment* . |
|---|---|---|---|
| Vδ2-Dδ3 rearrangements | |||
| Germline† | TGT GCC TGT GAC ACC | ACT GGG GGA TAC G cac agt g | |
| E47/RR transfectant | 0 | −3 | |
| 0 | −4 | ||
| HEB/RR transfectant | −2 | −3 | |
| −1 | −2 | ||
| −1 | −3 | ||
| −2 | T | 0 | |
| 0 | −3 | ||
| Dδ2-Dδ3 rearrangements‡ | |||
| Germline2-153 | c att gtg CCT TCC TAC | ACT GGG GGA TAC G cac agt g | |
| E12/RR transfectant (4/4)# | c att gtg | cac agt g | |
| E47/RR transfectant (10/11)# | c att gtg | cac agt g | |
| E47/RR transfectant (1/11)# | c att gtg CCT TCC TAC | CT GGG GGA TAC G cac agt g | |
| HEB/RR transfectant (7/7)# | c att gtg | cac agt g | |
| Thymus | c att gtg CCT TCC TAC | N region | T GGG GGA TAC G cac agt g |
| c att gtg CCT TCC TA | ACT GGG GGA TAC G cac agt g | ||
| c att gtg | cac agt g | ||
| Dδ2-Dδ3 excision circles‡ | |||
| Germline2-154 | c act gtg | cac aca g | |
| E12/RR transfectant | c act gtg ACT GGG GGA | T TCC TAC cac aca g | |
| E47/RR transfectant | c act gtg ACT GGG GGA TAC G | C TAC cac aca g | |
| HEB/RR transfectant | c act gtg ACT GGG GGA | TAC cac aca g | |
| c act gtg ACT GGG GGA TAC | cac aca g | ||
| Thymus | c act gtg | cac aca g | |
| c act gtg | N region | cac aca g | |
| c act gtg | N region | AC cac aca g | |
| c act | N region | cac aca g |
| Sample . | 5′ gene segment* . | P nucleotides . | 3′ gene segment* . |
|---|---|---|---|
| Vδ2-Dδ3 rearrangements | |||
| Germline† | TGT GCC TGT GAC ACC | ACT GGG GGA TAC G cac agt g | |
| E47/RR transfectant | 0 | −3 | |
| 0 | −4 | ||
| HEB/RR transfectant | −2 | −3 | |
| −1 | −2 | ||
| −1 | −3 | ||
| −2 | T | 0 | |
| 0 | −3 | ||
| Dδ2-Dδ3 rearrangements‡ | |||
| Germline2-153 | c att gtg CCT TCC TAC | ACT GGG GGA TAC G cac agt g | |
| E12/RR transfectant (4/4)# | c att gtg | cac agt g | |
| E47/RR transfectant (10/11)# | c att gtg | cac agt g | |
| E47/RR transfectant (1/11)# | c att gtg CCT TCC TAC | CT GGG GGA TAC G cac agt g | |
| HEB/RR transfectant (7/7)# | c att gtg | cac agt g | |
| Thymus | c att gtg CCT TCC TAC | N region | T GGG GGA TAC G cac agt g |
| c att gtg CCT TCC TA | ACT GGG GGA TAC G cac agt g | ||
| c att gtg | cac agt g | ||
| Dδ2-Dδ3 excision circles‡ | |||
| Germline2-154 | c act gtg | cac aca g | |
| E12/RR transfectant | c act gtg ACT GGG GGA | T TCC TAC cac aca g | |
| E47/RR transfectant | c act gtg ACT GGG GGA TAC G | C TAC cac aca g | |
| HEB/RR transfectant | c act gtg ACT GGG GGA | TAC cac aca g | |
| c act gtg ACT GGG GGA TAC | cac aca g | ||
| Thymus | c act gtg | cac aca g | |
| c act gtg | N region | cac aca g | |
| c act gtg | N region | AC cac aca g | |
| c act | N region | cac aca g |
RSS indicates recombination signal sequence; RR, RAG1 plus RAG2.
Indicated are the number of nucleotides deleted at the 5′ and 3′ gene segments. Lower case represents RSS nucleotides; upper case, nucleotides of gene segments.
For 5′ gene segment, this is Vδ2 (coding); for 3′ gene segment, this is Dδ3 (coding + 3′ RSS).
See Figure 2 for the Dδ2-Dδ3 rearrangement and excision circle composition, such as they occur in the classical and alternative recombination mechanisms.
For 5′ gene segment, this is Dδ2 (5′ RSS + coding); for 3′ gene segment, this is Dδ3 (coding + 3′ RSS).
For 5′ gene segment, this is Dδ3 (5′ RSS); for 3′ gene segment, this is Dδ2 (3′ RSS).
#Indicated are the number of PCR products of the total number of sequenced PCR products.
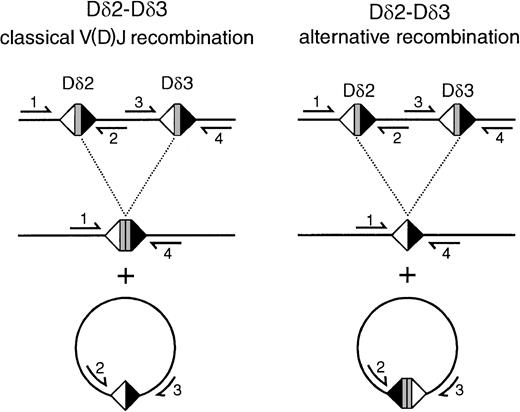
Scheme of classical V(D)J recombination and alternative recombination mechanisms resulting in Dδ2-Dδ3 rearrangement products and circular excision products.
In classical recombination, Dδ2 and Dδ3 coding sequences are coupled to a coding joint, whereas the intervening sequence is coupled via the downstream Dδ2 RSS and the upstream Dδ3 RSS to a signal joint on the circular excision product. In the alternative mechanism that was observed in the transfected BOSC 23 cells on the basis of the sequencing of the rearrangements, a signal joint of the upstream Dδ2 RSS and the downstream Dδ3 RSS is formed, whereas the Dδ2 and Dδ3 coding sequences are deleted out on the excision circles. Primers mentioned are as follows: 1, Dδ2-5′ RO2; 2, Dδ2-lower; 3, Dδ3-upper; 4, Dδ3-3′ N.
Scheme of classical V(D)J recombination and alternative recombination mechanisms resulting in Dδ2-Dδ3 rearrangement products and circular excision products.
In classical recombination, Dδ2 and Dδ3 coding sequences are coupled to a coding joint, whereas the intervening sequence is coupled via the downstream Dδ2 RSS and the upstream Dδ3 RSS to a signal joint on the circular excision product. In the alternative mechanism that was observed in the transfected BOSC 23 cells on the basis of the sequencing of the rearrangements, a signal joint of the upstream Dδ2 RSS and the downstream Dδ3 RSS is formed, whereas the Dδ2 and Dδ3 coding sequences are deleted out on the excision circles. Primers mentioned are as follows: 1, Dδ2-5′ RO2; 2, Dδ2-lower; 3, Dδ3-upper; 4, Dδ3-3′ N.
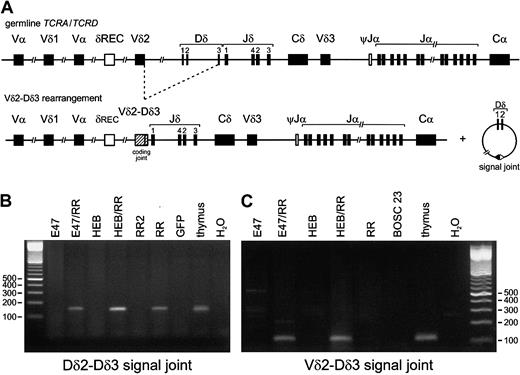
In contrast to the Dδ2-Dδ3 and Vδ2-Dδ3 rearrangements, we did not detect the other 4 frequently occurring types ofTCRD rearrangements, which all involve the Jδ1 gene segment (Table 3). Dδ2-Dδ3 recombinations can be formed only by direct rearrangement of the 2 neighboring Dδ2 and Dδ3 segments. The intermediate circular excision products or signal joints that are formed as a side product during this process (Figure 3A) were indeed detected in the same transfection combinations (E2A or HEB plus RAG) in which the corresponding Dδ2-Dδ3 coding joints were found (Figure 3B). Sequencing of these excision circle products from transfected cells showed a coding joint–like configuration, confirming the less common mechanism of Dδ2-Dδ3 recombination (Table 2; Figure2); similar products were occasionally found in thymocytes as well. Vδ2-Dδ3 rearrangements can be formed either by direct coupling or via a 2-step mechanism involving the Dδ2-Dδ3 joint as an intermediate. In the former case, Vδ2-Dδ3 signal joints should be detectable, whereas in the latter option both the Dδ2-Dδ3 signal joints as well as Vδ2-Dδ2 signal joints should be observed. PCR analysis did show Vδ2-Dδ3 signal joints in the E2A or HEB plus RAG transfectants (Figure 3A,C), whereas Vδ2-Dδ2 signal joints were not found, indicating that the observed Vδ2-Dδ3 recombination takes place via direct coupling of the 2 gene segments, rather than as a 2-step process in which Dδ2-Dδ3 coding joints act as intermediates.
TCRD rearrangements in various E2A- or HEB-transfected BOSC 23 cells
| Rearrangement . | E47 . | E47/RR . | E12 . | E12/RR . | HEB . | HEB/RR . | RR . | BOSC-233-150 . | Thymus . |
|---|---|---|---|---|---|---|---|---|---|
| Dδ2-Dδ3 | − | + | − | + | − | + | +/− | − | ++ |
| Vδ2-Dδ3 | − | + | − | + | − | + | − | − | ++ |
| Dδ2-Jδ1 | − | − | − | − | − | − | − | − | ++ |
| Vδ1-Jδ1 | − | − | − | − | − | − | − | − | ++ |
| Vδ2-Jδ1 | − | − | − | − | − | − | − | − | ++ |
| Vδ3-Jδ1 | − | − | − | − | − | − | − | − | ++ |
| Dδ2-Dδ3 signal joint | − | + | − | + | − | + | +/− | − | ++ |
| Vδ2-Dδ2 signal joint | − | − | − | − | − | − | − | − | ++ |
| Vδ2-Dδ3 signal joint | − | + | − | + | − | + | − | − | ++ |
| Dδ3-Jδ1 | − | − | − | − | − | − | − | − | ++ |
| Dδ3-Jδ3 | − | − | − | − | − | − | − | − | ++ |
| Dδ3-Jδ1 signal joint | − | − | − | − | − | − | − | − | ++ |
| Dδ3-Jδ3 signal joint | − | − | − | − | − | − | − | − | ++ |
| Rearrangement . | E47 . | E47/RR . | E12 . | E12/RR . | HEB . | HEB/RR . | RR . | BOSC-233-150 . | Thymus . |
|---|---|---|---|---|---|---|---|---|---|
| Dδ2-Dδ3 | − | + | − | + | − | + | +/− | − | ++ |
| Vδ2-Dδ3 | − | + | − | + | − | + | − | − | ++ |
| Dδ2-Jδ1 | − | − | − | − | − | − | − | − | ++ |
| Vδ1-Jδ1 | − | − | − | − | − | − | − | − | ++ |
| Vδ2-Jδ1 | − | − | − | − | − | − | − | − | ++ |
| Vδ3-Jδ1 | − | − | − | − | − | − | − | − | ++ |
| Dδ2-Dδ3 signal joint | − | + | − | + | − | + | +/− | − | ++ |
| Vδ2-Dδ2 signal joint | − | − | − | − | − | − | − | − | ++ |
| Vδ2-Dδ3 signal joint | − | + | − | + | − | + | − | − | ++ |
| Dδ3-Jδ1 | − | − | − | − | − | − | − | − | ++ |
| Dδ3-Jδ3 | − | − | − | − | − | − | − | − | ++ |
| Dδ3-Jδ1 signal joint | − | − | − | − | − | − | − | − | ++ |
| Dδ3-Jδ3 signal joint | − | − | − | − | − | − | − | − | ++ |
RR indicates RAG1 plus RAG2.
In some experiments, GFP-transfected cells were used as nonspecific template control.
Analysis of intermediate excision circles or signal joints.
Analysis of intermediate excision circles or signal joints illustrates that E2A and HEB induce single-step TCRD recombinations in nonlymphoid cells. (A) In the example of a Vδ2-Dδ3 rearrangement, the coding and signal joints formed during direct coupling of the Vδ2 and Dδ3 segments are shown. (B,C) Analysis of signal joints in BOSC 23 cells transfected with E47, HEB, RAG1, and RAG2 expression vectors. Genomic DNA (200 ng) of the various transfectants was used for PCR amplification by means of Dδ2-lower plus Dδ3-upper (panel B) or Vδ2-sj 3′ plus Dδ3-5′ S (panel C) primers that specifically detect Dδ2-Dδ3 and Vδ2-Dδ3 signal joints, respectively. Thymus DNA was used as positive control, and GFP- or mock-transfected BOSC 23 DNA as nonspecific template control. PCR products were run on a 2% agarose gel and stained with ethidium bromide. Similar to the coding joints shown in Figure 1, Dδ2-Dδ3 and Vδ2-Dδ3 signal joints were observed upon combined transfection of either E47 or HEB together with RR. In line with the data on Dδ2-Dδ3 PCR products, Dδ2-Dδ3 signal joints were also detectable in genomic DNA derived from transfectants expressing RR only (lane RR). Importantly, transfection with 2 μg RAG expression vectors (lane RR2) did not result in detectable signal joints; this amount of RAG1/RAG2 vector leads to RAG activity levels that are more in line with the RAG activity levels in those cases in which 6 μg RAG1/RAG2 is cotransfected with E2A or HEB (W. J. Romanow, personal communication, 2001).
Analysis of intermediate excision circles or signal joints.
Analysis of intermediate excision circles or signal joints illustrates that E2A and HEB induce single-step TCRD recombinations in nonlymphoid cells. (A) In the example of a Vδ2-Dδ3 rearrangement, the coding and signal joints formed during direct coupling of the Vδ2 and Dδ3 segments are shown. (B,C) Analysis of signal joints in BOSC 23 cells transfected with E47, HEB, RAG1, and RAG2 expression vectors. Genomic DNA (200 ng) of the various transfectants was used for PCR amplification by means of Dδ2-lower plus Dδ3-upper (panel B) or Vδ2-sj 3′ plus Dδ3-5′ S (panel C) primers that specifically detect Dδ2-Dδ3 and Vδ2-Dδ3 signal joints, respectively. Thymus DNA was used as positive control, and GFP- or mock-transfected BOSC 23 DNA as nonspecific template control. PCR products were run on a 2% agarose gel and stained with ethidium bromide. Similar to the coding joints shown in Figure 1, Dδ2-Dδ3 and Vδ2-Dδ3 signal joints were observed upon combined transfection of either E47 or HEB together with RR. In line with the data on Dδ2-Dδ3 PCR products, Dδ2-Dδ3 signal joints were also detectable in genomic DNA derived from transfectants expressing RR only (lane RR). Importantly, transfection with 2 μg RAG expression vectors (lane RR2) did not result in detectable signal joints; this amount of RAG1/RAG2 vector leads to RAG activity levels that are more in line with the RAG activity levels in those cases in which 6 μg RAG1/RAG2 is cotransfected with E2A or HEB (W. J. Romanow, personal communication, 2001).
TCRD recombination involving Jδ gene segments cannot be induced by E2A and HEB
From various studies, it is known that Vδ-Jδ rearrangements are formed via multiple (consecutive) couplings involving Dδ segments, rather than as a direct joining of Vδ to Jδ gene segments. This even applies to Dδ2-Jδ1 joints that are known to be formed in 2 steps, given the presence of identifiable Dδ3 segment sequences in virtually all of these coding joints.40 41 To determine whether E2A or HEB can induce recombination to Jδ1, we studied a rarer type of TCRD rearrangement, Dδ3-Jδ1, which can occur only as a direct coupling and which is known to be present in human thymocytes (T. M. Breit et al, unpublished observations, 2001). Although readily detectable in thymocytes, this rearranged Dδ3-Jδ1 product could not be detected in any of the transfected cell populations; the only PCR product apparent was the larger germline fragment encompassing the nonrearranged Dδ3 and Jδ1 segments that lie within 1 kb (Table 3). Even after nested PCR, no clear signs of Dδ3-Jδ1 rearrangements were found (data not shown), and also no Dδ3-Jδ1 signal joints were detectable (Table 3). To fully exclude recombination to Jδ gene segments, we studied another type of TCRD rearrangement, Dδ3-Jδ3, which is also formed in a 1-step reaction and occurs in thymocytes as well. Also this type of coupling (either coding joint or signal joint) could not be observed in any of the transfected combinations (Table 3). Collectively, these data illustrate that E2A and HEB have the ability to induce recombination in the TCRDlocus, but that this concerns only 1-step rearrangements in the Vδ2-Dδ region, and not in the more downstream Jδ region.
V-J rearrangements in the TCRG locus are induced by E2A and HEB in cooperation with the RAG proteins
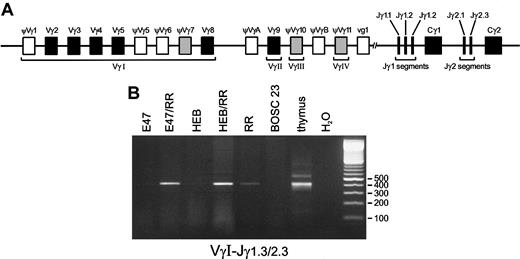
Although the TCRD locus is generally believed to be the first TCR locus that is rearranged during T-cell differentiation, we wished to ascertain whether in the transfected BOSC 23 cells recombination events would also be detectable in theTCRG locus, which starts rearranging later thanTCRD during thymocyte differentiation, but earlier than theTCRB and TCRA genes.9-11 The humanTCRG locus is composed of a limited set of Vγ gene segments that are grouped in Vγ families and 5 Jγ gene segments clustered in the homologous Jγ1 and Jγ2 regions (Figure4A). To analyze Vγ-Jγ recombinations, we employed 4 Vγ family primers in combination with 3 primers known to recognize the Jγ1.1/2.1, Jγ1.2, and Jγ1.3/2.3 segments. VγI-Jγ1.3/2.3 products were found to be induced by the basic HLH (bHLH) proteins E2A or HEB in the presence of RAG proteins as compared with RAG proteins only (Figure 4B). Similar to Dδ2-Dδ3 recombination levels, TaqMan RQ-PCR revealed an inducing effect (3- to 5-fold) in the E2A or HEB plus RAG transfectants as compared with transfections with the RAG genes alone. Heteroduplex analysis of the Vγ-Jγ PCR products to discriminate between polyclonal and clonal recombination products revealed some level of heterogeneity in the various transfection combinations (data not shown). Since the VγI gene family consists of many distinct Vγ gene segments that can rearrange, we sequenced these VγI-Jγ1.3/2.3 recombinations to study the diversity of V gene segment usage (Table4). All (approximately 20) sequenced products were found to contain the Jγ2.3 gene segment, which is discernible from Jγ1.3 at a single nucleotide position. At the V side, 2 gene segments were identified: ψVγ7 and Vγ8. Interestingly, within the VγI cluster, these 2 segments are most proximal to the Jγ segments (Figure 4A), which might explain their predominance. Rearrangements between VγIV (Vγ11) and Jγ1.3/2.3 gene segments were also observed in the E2A or HEB plus RAG transfectants, but they could be found at similar levels in the RAG1/RAG2 alone transfectants (data not shown). Apparently, the VγIV and Jγ2.3 gene segments are relatively easily accessible to the action of the RAG proteins, even without the presence of lymphoid transcription factors. The position of the VγIV gene segment just proximal to the Jγ gene segments might explain the finding of the otherwise rare VγIV-Jγ1.3/2.3 recombination. Nevertheless, rearrangements between the VγIV segment and the more proximal Jγ segments (Jγ1.1 and Jγ2.1) were not seen. Moreover, recombination products between any of the other Vγ gene segments and these Jγ1.1 and Jγ2.1 segments were not detectable at all in the BOSC 23 transfectants. The same was true for Vγ9-Jγ1.2 rearrangements, which are frequently found in peripheral blood TCRγδ+ T cells. The E2A and HEB bHLH proteins thus not only induceTCRD recombination, but also direct the RAG proteins to rearrange particular V and J gene segments within theTCRG locus.
Induction of Vγ-Jγ rearrangements by E2A or HEB in the presence of RAG1 and RAG2.
(A) Schematic diagram of the human TCRG locus. The humanTCRG locus comprises 2 constant region gene segments (Cγ) preceded by 2 or 3 joining gene segments (Jγ) and at least 14 variable gene segments (Vγ) belonging to 4 subgroups. Nine Vγ gene segments (5 functional genes and 4 pseudogenes) belong to subgroup I, whereas subgroups II, III, and IV each consist of a single gene segment designated Vγ9, Vγ10, and Vγ11, respectively. Two pseudogenes, VγA and VγB, located upstream of Vγ9 and Vγ11, respectively, belong to none of these subgroups. Five joining segments have been identified: Jγ1.1, Jγ1.2, and Jγ1.3 upstream of Cγ1; and Jγ2.1 and Jγ2.3 upstream of Cγ2. In addition to the 6 functional Vγ gene segments (solid blocks), 3 pseudogenes (shaded blocks) can also rearrange to Jγ gene segments. (B) Following transfection of BOSC 23 cells, PCR amplification with 200 ng genomic DNA of the transfected cells was performed by means of VγI-3′– and Jγ1.3/2.3-3′–specific primers. Thymus DNA and BOSC 23 DNA were used as positive and nonspecific template controls, respectively. Electrophoresis of PCR products on 2% agarose gels shows that E47 or HEB and RAG1 plus RAG2 proteins can promote VγI-Jγ1.3/2.3 rearrangements at high levels as compared with transfectants expressing RR only (lane RR) as quantitated by RQ TaqMan PCR analysis.
Induction of Vγ-Jγ rearrangements by E2A or HEB in the presence of RAG1 and RAG2.
(A) Schematic diagram of the human TCRG locus. The humanTCRG locus comprises 2 constant region gene segments (Cγ) preceded by 2 or 3 joining gene segments (Jγ) and at least 14 variable gene segments (Vγ) belonging to 4 subgroups. Nine Vγ gene segments (5 functional genes and 4 pseudogenes) belong to subgroup I, whereas subgroups II, III, and IV each consist of a single gene segment designated Vγ9, Vγ10, and Vγ11, respectively. Two pseudogenes, VγA and VγB, located upstream of Vγ9 and Vγ11, respectively, belong to none of these subgroups. Five joining segments have been identified: Jγ1.1, Jγ1.2, and Jγ1.3 upstream of Cγ1; and Jγ2.1 and Jγ2.3 upstream of Cγ2. In addition to the 6 functional Vγ gene segments (solid blocks), 3 pseudogenes (shaded blocks) can also rearrange to Jγ gene segments. (B) Following transfection of BOSC 23 cells, PCR amplification with 200 ng genomic DNA of the transfected cells was performed by means of VγI-3′– and Jγ1.3/2.3-3′–specific primers. Thymus DNA and BOSC 23 DNA were used as positive and nonspecific template controls, respectively. Electrophoresis of PCR products on 2% agarose gels shows that E47 or HEB and RAG1 plus RAG2 proteins can promote VγI-Jγ1.3/2.3 rearrangements at high levels as compared with transfectants expressing RR only (lane RR) as quantitated by RQ TaqMan PCR analysis.
Sequences of cloned TCRG recombination products
| . | E12/RR . | E47/RR . | HEB/RR . | ||||||
|---|---|---|---|---|---|---|---|---|---|
| . | 5′ del . | P region . | 3′ del . | 5′ del . | P region . | 3′ del . | 5′ del . | P region . | 3′ del . |
| Vγ7-Jγ2.3 rearrangement | |||||||||
| 0 | C | −4 | −2 | 0 | 0 | CC | −7 | ||
| −2 | 0 | −9 | −4 | ||||||
| Vγ8-Jγ2.3 rearrangement | |||||||||
| −3 | −15 | −1 | 0 | 0 | C | −7 | |||
| −5 | −2 | −2 | C | 0 | −2 | −4 | |||
| −5 | −3 | −4 | −4 | ||||||
| −4 | −2 | ||||||||
| −6 | 0 | ||||||||
| . | E12/RR . | E47/RR . | HEB/RR . | ||||||
|---|---|---|---|---|---|---|---|---|---|
| . | 5′ del . | P region . | 3′ del . | 5′ del . | P region . | 3′ del . | 5′ del . | P region . | 3′ del . |
| Vγ7-Jγ2.3 rearrangement | |||||||||
| 0 | C | −4 | −2 | 0 | 0 | CC | −7 | ||
| −2 | 0 | −9 | −4 | ||||||
| Vγ8-Jγ2.3 rearrangement | |||||||||
| −3 | −15 | −1 | 0 | 0 | C | −7 | |||
| −5 | −2 | −2 | C | 0 | −2 | −4 | |||
| −5 | −3 | −4 | −4 | ||||||
| −4 | −2 | ||||||||
| −6 | 0 | ||||||||
RR indicates RAG1 plus RAG2.
Indicated are the number of nucleotides deleted at the 5′ and 3′ gene segments.
Absence of TCRB rearrangements andTCRD deletions in E2A and HEB transfectants
To further analyze potential effects of E2A and/or HEB on V(D)J recombination in the TCR loci that are normally rearranged in later stages of T-cell differentiation (late double-negative and immature single-positive stages), we first studied rearrangements in the humanTCRB locus. Unlike the TCRG locus, theTCRB locus is built up of a large number of distinct Vβ segments, clustered in approximately 25 Vβ families, and 2 Dβ segments each lying upstream of a cluster of 6 or 7 Jβ gene segments. The presence of V, D, and J segments in the TCRB locus implies that both incomplete and complete rearrangements can occur. Incomplete TCRB rearrangements almost exclusively concern Dβ-Jβ recombinations, as incomplete Vβ-Dβ joints are rarely found in T cells. For this reason, we studied Dβ1-Jβ1, Dβ1-Jβ2, and Dβ2-Jβ2 rearrangements, which can readily be detected in thymocytes. However, we could not find either one of these products in any of the transfected cell populations, even after nested PCR reactions (data not shown). Given these results and also the results from the TCRD analysis concerning 2-step recombinations, we anticipated that Vβ-Jβ joints, which normally include Dβ sequences, would not be detectable either. Employing Jβ1 or Jβ2 primers in combination with specific primers for the frequently used Vβ2 and Vβ5 families,42 we indeed could not observe complete Vβ-Jβ joints (data not shown).
To further substantiate the absence of other mature TCR gene rearrangements, we studied TCRD deletional rearrangements mediated by the nonfunctional δREC and ψJα segments, which flank the coding elements of the TCRD locus (Figure 1). TheseTCRD deletions precede rearrangements in the humanTCRA locus, and they are known to occur at a relatively late stage in T-cell differentiation. However, no δREC-ψJα rearrangements were identified in transfected BOSC 23 cells (data not shown). Taken together, these data do not provide any evidence for a role for E2A or HEB in inducing TCRB rearrangements orTCRD deletional rearrangements.
Discussion
In this study, we provide evidence that the bHLH transcription factors E2A and HEB play a role in the induction of V(D)J recombination in human TCRD and TCRG loci, employing a transfection-based model of nonlymphoid cells. E2A and HEB were found to induce immature Dδ2-Dδ3 and Vδ2-Dδ3 rearrangements upon cotransfection with RAG proteins in nonlymphoid cells. Rearrangements of VγI family (especially ψVγ7 and Vγ8) gene segments to Jγ2.3 were also easily induced by the action of both transcription factors. All except the Dδ2-Dδ3 rearrangements were very similar to the majority of rearrangements found in lymphoid cells, with junctional regions characterized by nucleotide deletion and occasionally P nucleotides. The lack of N regions can be explained by the absence of terminal deoxynucleotidyl transferase, which mediates nontemplated nucleotide insertion. Remarkably, the junctions of virtually all Dδ2-Dδ3 rearrangements from the transfected cells were found to be identical, because of an unusual direct coupling of the intact upstream RSS of the Dδ2 segment to the intact downstream RSS of the Dδ3 segment. This unusual type of rearrangement shares similarity to atypical IGK gene recombinations between Jκ segments and an isolated RSS-like sequence in the J-Cκ intron.43 In fact, both types of recombinations represent signal joints, which normally are not found on the genome, but rather on the excision circles that are formed during V(D)J recombination (see Figure2).
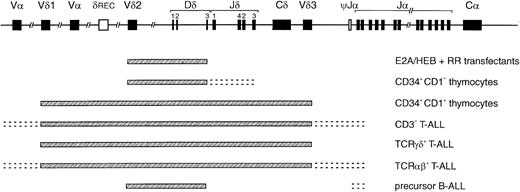
The virtually identical patterns of TCR gene recombinations in both the E2A plus RAG– and the HEB plus RAG–transfected cells showed a striking similarity to the most immature types of rearrangements seen in human thymocyte subpopulations (Table5). Dδ2-Dδ3 and Vδ2-Dδ3 rearrangements represent the earliest TCRD rearrangements in thymocytes, being clearly detectable via Southern blot analysis next to Dδ2-Jδ1 rearrangements in sorted cells of the most immature CD34+/CD1a− subset.11 In the CD34+/CD1a+ fraction, complete Vδ1-Jδ1 and Vδ2-Jδ1 rearrangements become more prominent. However, rearrangements to Jδ segments, even simple single-step couplings like Dδ3-Jδ1 and Dδ3-Jδ3 as also seen in thymus, could not be induced in the transfected cells. TCRG rearrangements involving Jγ1.3/2.3 segments and TCRB rearrangements (most probably, incomplete Dβ-Jβ joinings) were clearly detectable only in CD34+/CD1a+ cells (Table 5), although low levels of (other) TCRD, TCRG, and/or TCRBrecombinations in the CD34+/CD1a− subset cannot be fully excluded. The TCRD rearrangement pattern of the transfectants was also compared with TCR recombination data in human T-ALL (CD3−, TCRαβ+, and TCRγδ+) and precursor B-ALL samples, as investigated in our laboratory (Table 5).6,44-47 In T-ALL, Jδ recombinations are predominant, especially in CD3+ T-ALL, whereas Dδ2-Dδ3 and Vδ2-Dδ3 rearrangements are found only at low frequencies in immature CD3-T-ALL.6,41,44Interestingly, these latter 2 TCRD rearrangements are very characteristic in precursor B-ALL, where they constitute approximately 80% of the identified (cross-lineage) TCRDrecombinations.6,44 Rearrangements involving Jδ have not been found in precursor B-ALL.6,44 48 The data thus suggest that E2A or HEB can induce the very early TCRDrecombinations that carry a less T-cell–specific character, as they are also readily observed in precursor B-ALL (Figure5).
Detection of cloned TCRD, clonedTCRG, and cloned TCRB gene rearrangements in E2A/HEB-transfected cells as compared with human thymocytes and acute lymphoblastic leukemias
| Rearrangement . | E2A/HEB transfectants . | Thymocytes . | T-ALL, %5-150 . | Precursor B-ALL, %5-150 . | ||||
|---|---|---|---|---|---|---|---|---|
| CD34+ 1a− . | CD34+1a+ . | Total . | CD3− . | TCRγδ+ . | TCRαβ+ . | |||
| TCRD | ||||||||
| Dδ2-Dδ3 | + | ++ | ++ | + | 4 | — | — | 13 |
| Vδ2-Dδ3 | + | + | + | + | 6 | 4 | — | 67 |
| Dδ2-Jδ1 | − | + | ++ | + | 10 | 15 | 7 | — |
| Vδ1-Jδ1 | − | − | + | + | 24 | 56 | 13 | — |
| Vδ2-Jδ1 | − | − | + | + | 10 | 6 | 20 | — |
| Vδ3-Jδ1 | − | − | + | + | 5 | 8 | 7 | — |
| Other | NT | NT | NT | NT | 40 | 10 | 53 | 20 |
| TCRG | ||||||||
| VγI-Jγ1.1/2.1 | − | NT | NT | + | 8 | 9 | 20 | 18 |
| VγII-Jγ1.1/2.1 | − | NT | NT | + | — | — | — | 1 |
| VγIII-Jγ1.1/2.1 | − | NT | NT | + | 1 | — | 1 | — |
| VγIV-Jγ1.1/2.1 | − | NT | NT | + | 1 | 4 | — | 1 |
| VγI-Jγ1.2 | − | NT | NT | + | — | — | — | 1 |
| VγII-Jγ1.2 | − | NT | NT | + | 2 | — | — | — |
| VγIII-Jγ1.2 | NT | NT | NT | + | — | — | — | — |
| VγIV-Jγ1.2 | NT | NT | NT | + | — | — | — | — |
| VγI-Jγ1.3/2.3 | + | − | + | + | 69 | 52 | 58 | 56 |
| VγII-Jγ1.3/2.3 | − | − | + | + | 7 | 24 | 6 | 18 |
| VγIII-Jγ1.3/2.3 | − | − | + | + | 4 | 7 | 9 | — |
| VγIV-Jγ1.3/2.3 | +/− | − | + | + | 4 | 4 | 5 | 5 |
| Other | NT | NT | NT | NT | 4 | 1 | — | — |
| TCRB | ||||||||
| Dβ1-Jβ1 | − | − | + | + | 12 | 16 | 9 | — |
| Dβ1-Jβ2 | − | − | + | + | 4 | 14 | 3 | 16 |
| Dβ2-Jβ2 | − | − | + | + | 11 | 5 | 17 | 17 |
| Vβ-Jβ1 | − | − | − | + | 18 | 13 | 28 | — |
| Vβ-Jβ2 | − | − | − | + | 47 | 45 | 30 | 53 |
| Other | NT | NT | NT | NT | 9 | 7 | 13 | 14 |
| Rearrangement . | E2A/HEB transfectants . | Thymocytes . | T-ALL, %5-150 . | Precursor B-ALL, %5-150 . | ||||
|---|---|---|---|---|---|---|---|---|
| CD34+ 1a− . | CD34+1a+ . | Total . | CD3− . | TCRγδ+ . | TCRαβ+ . | |||
| TCRD | ||||||||
| Dδ2-Dδ3 | + | ++ | ++ | + | 4 | — | — | 13 |
| Vδ2-Dδ3 | + | + | + | + | 6 | 4 | — | 67 |
| Dδ2-Jδ1 | − | + | ++ | + | 10 | 15 | 7 | — |
| Vδ1-Jδ1 | − | − | + | + | 24 | 56 | 13 | — |
| Vδ2-Jδ1 | − | − | + | + | 10 | 6 | 20 | — |
| Vδ3-Jδ1 | − | − | + | + | 5 | 8 | 7 | — |
| Other | NT | NT | NT | NT | 40 | 10 | 53 | 20 |
| TCRG | ||||||||
| VγI-Jγ1.1/2.1 | − | NT | NT | + | 8 | 9 | 20 | 18 |
| VγII-Jγ1.1/2.1 | − | NT | NT | + | — | — | — | 1 |
| VγIII-Jγ1.1/2.1 | − | NT | NT | + | 1 | — | 1 | — |
| VγIV-Jγ1.1/2.1 | − | NT | NT | + | 1 | 4 | — | 1 |
| VγI-Jγ1.2 | − | NT | NT | + | — | — | — | 1 |
| VγII-Jγ1.2 | − | NT | NT | + | 2 | — | — | — |
| VγIII-Jγ1.2 | NT | NT | NT | + | — | — | — | — |
| VγIV-Jγ1.2 | NT | NT | NT | + | — | — | — | — |
| VγI-Jγ1.3/2.3 | + | − | + | + | 69 | 52 | 58 | 56 |
| VγII-Jγ1.3/2.3 | − | − | + | + | 7 | 24 | 6 | 18 |
| VγIII-Jγ1.3/2.3 | − | − | + | + | 4 | 7 | 9 | — |
| VγIV-Jγ1.3/2.3 | +/− | − | + | + | 4 | 4 | 5 | 5 |
| Other | NT | NT | NT | NT | 4 | 1 | — | — |
| TCRB | ||||||||
| Dβ1-Jβ1 | − | − | + | + | 12 | 16 | 9 | — |
| Dβ1-Jβ2 | − | − | + | + | 4 | 14 | 3 | 16 |
| Dβ2-Jβ2 | − | − | + | + | 11 | 5 | 17 | 17 |
| Vβ-Jβ1 | − | − | − | + | 18 | 13 | 28 | — |
| Vβ-Jβ2 | − | − | − | + | 47 | 45 | 30 | 53 |
| Other | NT | NT | NT | NT | 9 | 7 | 13 | 14 |
Data on thymocytes and acute lymphoblastic leukemia compiled from Southern blot and/or polymerase chain reaction data from this study and earlier reports from our laboratory.6,11,44,45 47
T-ALL indicates T-lineage acute lymphoblastic leukemia; B-ALL, B-lineage ALL; TCR, T-cell receptor; TCRD, TCRδ;TCRG, TCRγ; TCRB, TCRβ; NT, not tested.
Frequencies defined as percentage of detectedTCRD rearrangements (on the basis of TCRDJ1 probe hybridization), percentage of detected TCRG rearrangements, and percentage of TCRB rearrangements (involving both the Jβ1 and Jβ2 regions).
Compilation of TCRD recombination patterns.
Indicated are the parts of the TCRD locus that can rearrange in the E2A and HEB transfectants described in this study, CD34+/CD1a− and CD34+/CD1a+ thymocyte subsets, CD3− and TCRγδ and TCRαβ T-ALL, and precursor B-ALL. The pattern observed in the E2A/HEB-plus-RR–transfected cells is strikingly similar to that of the most immature thymocytes and the cross-lineage pattern of precursor B-ALL, illustrating that E2A and HEB can induce the immature and less T-cell–specific rearrangements in theTCRD locus.
Compilation of TCRD recombination patterns.
Indicated are the parts of the TCRD locus that can rearrange in the E2A and HEB transfectants described in this study, CD34+/CD1a− and CD34+/CD1a+ thymocyte subsets, CD3− and TCRγδ and TCRαβ T-ALL, and precursor B-ALL. The pattern observed in the E2A/HEB-plus-RR–transfected cells is strikingly similar to that of the most immature thymocytes and the cross-lineage pattern of precursor B-ALL, illustrating that E2A and HEB can induce the immature and less T-cell–specific rearrangements in theTCRD locus.
In the transfected cells, VγI-Jγ1.3/2.3 rearrangements were readily observed; such recombinations are also frequent in both T-ALL and precursor B-ALL, albeit in the latter Jγ2.3 is less frequently involved than Jγ1.3; VγIV-Jγ1.3/2.3 couplings are less prominent in both ALL subsets. Finally, incomplete Dβ-Jβ and complete Vβ-Jβ recombinations were not detected in the transfectants, but occur in all 3 T-ALL subtypes and at a lower frequency in human precursor B-cell leukemias (approximately 35% of cases).6Together, these data illustrate that the TCR gene recombination pattern as observed in the E2A or HEB plus RAG–transfected cells concerns the more immature types of TCR recombinations, as observed in CD34+/CD1a− thymocytes and also in precursor B-ALL, with the exception of the lack of TCRB rearrangements (Figure 5).
Although recombination can be induced relatively easily in this transfection system, the efficiency seems to be too low to allow detection of recombination events that occur in 2 consecutive steps. This is best illustrated by our study on circular intermediate products that are formed during recombination. Upon study of Vδ2-Dδ3 recombinations, we exclusively found Vδ2-Dδ3 signal joints and no Vδ2-Dδ2 plus Dδ2-Dδ3 circular intermediates. Although we cannot formally exclude that, eg, 2-step Vδ1-Jδ1 or Vδ2-Jδ1 rearrangements can be induced at very low levels, the fact that the single-step Dδ3-Jδ1 and Dδ3-Jδ3 recombinations are also undetectable strengthens our view that Jδ1 rearrangements are truly absent in E2A- or HEB-transfected cells rather than being an artifact in the model system. The same seems to be true for Dβ-Jβ and Vβ-Jβ rearrangements, as well as δREC-ψJα TCRDdeletional rearrangements. The TCRD recombination data thus suggest that E2A and HEB are able to regulate recombination of, especially, the Dδ3 gene segment, but that other factors are required to open up the downstream region of the Jδ segments to the V(D)J recombinase. The area between Dδ3 and the Jδ region might harbor important regulatory elements that can bind factors mediating the truly T-cell–specific types of TCRDrecombinations.49 Interestingly, Vδ-Dδ intermediates are lacking in E2A−/− mice, but Dδ-Jδ intermediates do still occur,15 which indeed suggests the involvement of one or more yet unknown transcription factors. From E2A−/− mice, it is further deduced that E2A plays a role in regulation of Vγ/Vδ subsets during ontogeny by activating and repressing V segments.15 These data fit perfectly with our model system, in which E2A can induce rearrangements of specific Vγ gene segments and Vδ-Dδ rearrangements, but not incomplete Dδ-Jδ or complete Vδ-Jδ rearrangements.
From the initial V(D)J recombination study in this nonlymphoid model system, it is known that induction of recombination by, eg, E2A is critically dependent on activation domains.23 This has led to the hypothesis that E-box proteins such as E2A are involved in regulating chromatin accessibility by relieving the repressive effect of nucleosomes, most probably through recruitment of complexes containing histone acetyl transferase activity.23 50 We therefore hypothesize that the interaction of E2A and HEB with chromatin leads to accessibility of RSS sequences of TCR loci as well, which is supported by the observation of VγI and Vδ2 germline transcripts upon E2A or HEB transfection (J. K. Ghosh et al, personal communication, 2001). The proximity of gene segments, as in the case of Dδ2 and Dδ3 and also ψVγ7 and Vγ8 within the VγI family, might be another relevant factor. Finally, the exact nucleotide sequence of the RSS might also influence recombination, although the RSSs of, eg, the various VγI family gene segments are equally perfect in that respect.
The observations in this study raise the issue of how E2A and HEB are involved in regulating TCR recombination during in vivo T-cell differentiation. Both E2A and HEB knock-out mice show a block, though not complete, in the early double-negative stages of T-cell differentiation. The phenotype of these mice thus illustrates the important role of both E2A and HEB in the earliest phases of T-cell differentiation, although the incompleteness of the differentiation arrest indicates some degree of redundancy by other regulatory factors. Using the in vitro model, we show that some types of immature TCR rearrangements can be induced by E2A or HEB, whereas the absence of these rearrangements in E2A knock-out mice shows that E2A and HEB are probably also involved in stimulating the immature TCR recombination in vivo. For induction of further (more mature) rearrangements, additional cofactors or transcription factors might be required or negative regulatory elements might have to be downregulated at certain differentiation stages. Moreover, we cannot fully exclude a cooperative action of both E2A and HEB in particular recombinations in vivo, although we have not seen a synergistic effect of combined transfection of E2A and HEB with RAG proteins (data not shown).
Besides mediating physiological immunoglobulin/TCR gene rearrangements, V(D)J recombination is also thought to be involved in the formation of particular chromosome aberrations in human leukemias.25,26 This especially concerns chromosome aberrations in T-ALL, in which TCRB or TCRD gene segments and their regulatory elements are translocated to oncogenes on partner chromosomes, resulting in activation of the oncogenes. Examples include translocations t(1;14) and t(11;14), involving the genes encoding the TAL1 and LMO1/LMO2 transcription factors, respectively.25,51 As the breakpoint regions on these chromosomes have been found to be located near RSS or RSS-like sequences,25-27 it is suggestive that accessibility of the involved oncogenes to RAG protein activity might be a critical step in the formation of these chromosome aberrations as well. We therefore also studied TAL1 deletions, which are V(D)J-like rearrangements that are found exclusively in T-ALL, particularly in T-ALL with TCRD deletions.52 However, such RSS-mediated TAL1 deletions were not induced upon E2A or HEB transfection (data not shown). Given the relatively low frequency of V(D)J recombinations in nonlymphoid cells, it can be argued that detection of TAL1 deletions in this model system would be very difficult. However, it might also be that transcription factors other than E2A and HEB regulate accessibility of theTAL1 gene and that an aberrant combination of regulatory factors is required to induce these unwanted oncogenic TAL1 deletions.
In summary, our data demonstrate a role for the bHLH proteins E2A and HEB in induction of TCR rearrangements. Comparison with the TCR-rearrangement patterns found in thymocyte subsets and in T- and B-lineage leukemias indicates that the identified rearrangements in, especially, the TCRD locus are immature and carry a less T-cell–specific character (Figure 5). A challenge for future studies remains the identification of factors that are involved in induction of more T-cell–specific TCR rearrangements, such as (complete)TCRD recombinations involving Jδ couplings and/orTCRB recombinations. Furthermore, it will be interesting to see whether the here-described model system will be helpful in dissecting the molecular processes regulating RSS-mediated chromosome aberrations in human lymphoid leukemias.
Dr R. Benner for continuous support and Drs F. J. T. Staal and T. Szczepanski for critical reading and valuable comments.
Supported by grant EUR 95-1015 from the Dutch Cancer Society (Koningin Wilhelmina Fonds); and additional financial support (to A.W.L.) from the Haak Bastiaanse Kuneman Foundation.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Anton W. Langerak, Department of Immunology, Erasmus University Rotterdam/University Hospital Rotterdam, PO Box 1738, 3000 DR Rotterdam, The Netherlands; e-mail:langerak@immu.fgg.eur.nl.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal