Activating point mutations in codons 12, 13, or 61 of the K-ras and N-ras genes have been reported to occur in up to 40% of patients with multiple myeloma at presentation. In a study of 34 presentation myeloma cases using a sensitive polymerase chain reaction-restriction fragment length polymorphism strategy on enriched tumor cell populations, the present study detected N-ras codon 61 mutation-positive cells in all patients. Quantitative plaque hybridization using allele-specific oligonucleotide probes showed that in the majority of patients, ras mutation-positive cells comprise only a subpopulation of the total malignant plasma cell compartment (range, 12%-100%). Using clonospecific point mutations in the 5′ untranslated region of the BCL6 gene to quantitate clonal B cells in FACS-sorted bone marrow populations from 2 patients, the representation of ras mutation-positive cells was independent of immunophenotype. These observations imply that mutational activation of N-ras codon 61 is a mandatory event in the pathogenesis of multiple myeloma; such mutations provide a marker of intraclonal heterogeneity that may originate at an earlier ontologic stage than immunophenotypic diversification of the malignant B cell clone.
Introduction
Multiple myeloma accounts for about 10% of all hemopoietic malignancies in the Western world. The disease is characterized by a monoclonal expansion of postgerminal center B-lymphocytes whose descendants infiltrate bone marrow as malignant plasma cells. Following high-dose chemotherapy, myeloablative therapy, and subsequent hemopoietic stem cell “rescue,” patients enter a remission phase of highly variable duration that ultimately culminates in relapse, which is typified by more aggressive disease.1-3
Although the phenotype of the predominant tumor cell population in multiple myeloma closely resembles that of a plasma cell, a number of studies have reported evidence for intraclonal heterogeneity within the clonal B-cell population.4-6 This phenomenon is highly variable among individual patients and is reflected by the expression of, for example, CD10, CD45, and CD126 on a minor subpopulation of the malignant B-cell population (reviewed in San-Miguel et al5).
A number of onco/tumor suppressor genes such as ras7-11FGFR3,12c-maf,13MUM1,14cyclin D1,15p53, andpRb16 have been implicated in the pathogenesis of multiple myeloma (reviewed in Hallek et al15 and Berenson et al17). Of these, “activating” point mutations affecting codons 12, 13, and 61 of the ras family of oncogenes represent the most common, single, nonconstitutional genetic lesion reported in this disease.7-11,15 As in other hemopoietic malignancies, these mutations affect the N-ras and K-ras genes but rarely, if ever, involve Ha-ras.10 However, in contrast to ras mutations documented in acute leukemias,18-22 which commonly involve N-ras codons 12/13, those in myeloma are predominantly of N-ras codon 61.9-11 15
The reported frequency of myeloma cases harboring mutant ras varies from 10% to 40% at presentation, rising to 70% at relapse (reference 11 and references therein) implying a possible role for this oncogene in disease progression. However, malignant plasma cells usually comprise less than 25% of the bone marrow mononuclear cells in multiple myeloma and most of these earlier studies used strategies for detecting mutations that have limited sensitivity. Therefore, the frequency of ras mutation-positive cases may have been underestimated in these earlier studies. This problem could be further exacerbated if, as reported in some cases of acute leukemia, mutations affecting the N-ras gene (but not apparently those in the K-rasgene) occur in only a subpopulation of tumor cells.21-23
We report here that N-ras 61 mutation-positive cells can be detected in subpopulations of tumor cells in all (34 of 34) cases of multiple myeloma at presentation. Such ras mutation-positive cells are equally distributed among different immunophenotypic compartments of the clonal B-cell population, suggesting that mutation in the N-rasgene arises as an ontogenically early event in disease pathogenesis.32 33
Patients, materials, and methods
Patient material and CD138+ cell enrichment
Thirty-four consecutive patients recruited from the northwest of England presenting with multiple myeloma, by standard criteria, were enrolled in the study. All but 3 patients were white. Twenty-one were men; 13 were women. Six patients had stage I disease, with the remainder in stage II/III. The subtypes were reflective of an unselected myeloma population being predominantly IgG (18 cases), followed by IgA (10 cases) and light-chain disease only (5 cases); one patient had nonsecretory myeloma. An aliquot of the diagnostic bone marrow aspirate was used. Bone marrow infiltration by plasma cells ranged from 4% to 82%. Aspirates were layered on Lymphoprep (Nycomed, Buckinghamshire, United Kingdom) and enriched for mononuclear cells. Cells were then labeled with CD138 microbeads and purified using the miniMACS immunomagnetic system (Miltenyi Biotech, Surrey, United Kingdom). Enriched fractions were assessed for purity by CD138-fluorescein isothiocyanate (FITC) monoclonal antibody (mAb; Serotec, Littlehampton, United Kingdom) labeling and FACScan analysis (Becton Dickinson, San Jose, CA) and morphologic examination. Cells were suspended in 50 μL lysate buffer (45 μL 0.5% Tween 20 and 5 μL buffer containing 100 mM Tris HCl, 15 mM MgCl2, and 500 mM KCl), incubated with 1 μL proteinase K (20 mg/mL) at 56°C for 1 hour and heat inactivated at 96°C for 10 minutes.
Polymerase chain reaction (PCR)-restriction fragment length polymorphism, enrichment PCR, and restriction enzyme digests
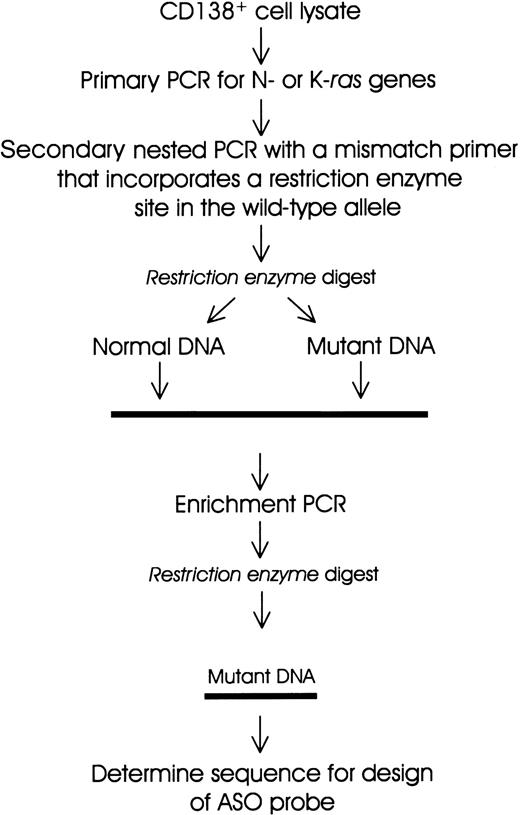
A polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) strategy22,24 was adapted for screening of activating mutations in N-ras and K-ras genes (Figure 1 and Table 1). Cell lysate DNA (from a minimum of 5000 cell equivalents) was used for primary PCR amplification (20 cycles) of sequences encompassing codons 12, 13, and 61 of N-ras and K-ras genes using previously described primers with some modifications24 (Table 1). One microliter of a 1:50 dilution was subjected to secondary nested PCR (30 cycles) to incorporate a restriction enzyme site in the wild-type, but not in the mutant, allele (Figure 1). The PCR product (20 μL) was digested with the appropriate restriction enzyme (New England Biolabs, Hertfordshire, United Kingdom). An enrichment PCR stage (10 cycles) was carried out on an aliquot of the restriction enzyme digest using the same primers as in secondary PCR (see above). The reaction products were then subjected to further restriction enzyme digests. All PCR reactions were performed using a “hot-start, touchdown” protocol (96°C for 5 minutes; 96°C for 1 minute, 65°C to 55°C for 1 minute, and 68°C for 1 minute; final extension 68°C for 10 minutes) in a 100-μL reaction mix containing 10 mM Tris HCl, 1.5 mM MgCl2, 50 mM KCl, 100 μM dNTPs, 100 ng of each primer, and 2.5 U high-fidelity Taq polymerase (Boehringer Mannheim, Sussex, United Kingdom). DNAs from a panel of cell lines with knownras gene mutations were used as positive controls. Digests of PCR products were resolved on 3% agarose gels containing ethidium bromide. DNA bands that were resistant to digestion were excised and the DNA recovered and sequenced using an ABI-PRISM 377 analyzer.
Schematic representation of the PCR-RFLP and enrichment PCR strategy for the detection of mutations in N-ras and K-ras genes.
Schematic representation of the PCR-RFLP and enrichment PCR strategy for the detection of mutations in N-ras and K-ras genes.
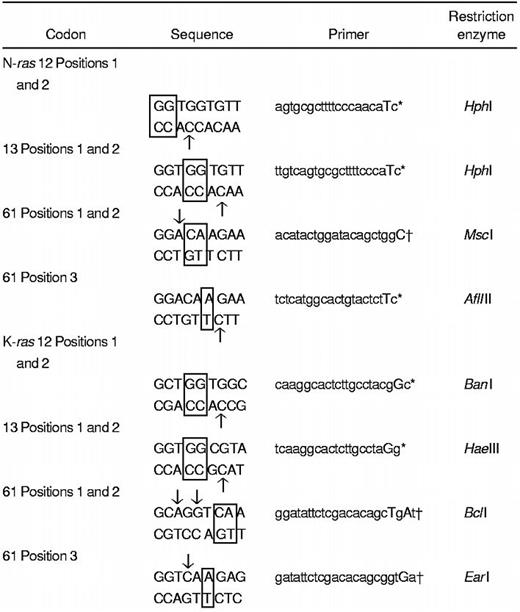
Primers and restriction enzyme sites for detection of hot-spot mutations in N- and K-ras genes
Nucleotide positions in codon 12, 13, or 61 targeted for screening are depicted in boxes.
Arrows show the nucleotide targeted for design of the mismatch primer. Uppercase nucleotide in the primer represents the mismatch.
Antisense primer.
Sense primer.
Detection of mutations in the 5′ untranslated region of theBCL6 gene
A 790-bp region in the 5′ noncoding region of theBCL6 gene was PCR amplified from CD138+-enriched cell lysate DNA using published primers and conditions described elsewhere.25 PCR products were cloned into pCR-Blunt II-TOPO (Invitrogen, Groningen, Netherlands). DNA sequence was determined using an ABI-Prism 377 sequencer in 6 independent clones from each PCR reaction.
Quantitative plaque hybridization analysis
The PCR products were generated for N-ras exon 2 and BCL-6 5′ untranslated region (UTR) by using primers that incorporatedEcoRI sites at the ends to facilitate cloning into dephosphorylated Lambda ZAP II bacteriophage vector (Stratagene). The primer combinations for N-ras and BCL-6 were 5′-AGGAATTCAGTGGTTTATAGATGGTGAAAC-3′ and 5′-ATGAATTCGTACCTGTAGAGGTTAATATC-3′; 5′-GGAATTCCGCTGCTCATGATCATTATTT-3′ and GGAATTCAGACACGATACTTCATCTCAT-3′, respectively. Ligated inserts were packaged into Gigapack II Gold extract (Stratagene) and phage libraries were obtained on an XL1-Blue MRF′ bacterial lawn. Duplicate plaque lifts on Hybond-N membranes (Amersham, Buckinghamshire, UK) were denatured, neutralized, and cross-linked (Stratagene UV autocross-linker). Filters were then hybridized with 32P γ-ATP–labeled allele-specific oligonucleotide probes and washed stringently26 to allow discrimination of mutant and wild-type alleles. The percentage of cells that were positive for N-ras and BCL6 gene mutations was calculated assuming that mutations were present on only one allele.
FACS analysis of myeloma cells
Frozen aliquots of presentation bone marrow mononuclear cells were dual labeled with CD138-FITC (Serotec)/CD10-phycoerythrin (PE), CD45RO-PE (Becton Dickinson), or CD126-PE (Immunotech). Cells were sorted on a Vantage FACS (Becton Dickinson). At least 1000 cells were sorted from each immunophenotypic subset. Cells were collected into PCR buffer for subsequent quantitative analysis of N-ras and 5′ UTR BCL6 gene mutations.
Results
Mutations of codon 61 of the N-ras gene are universal at presentation in multiple myeloma
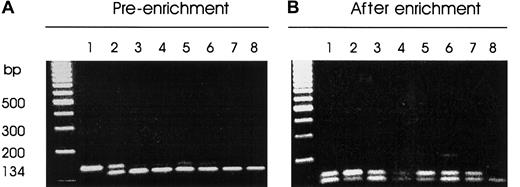
Thirty-four consecutive patients with multiple myeloma were screened by a sensitive PCR-RFLP strategy for the presence of mutations in codons 12, 13, and 61 of the N-ras and K-rasgenes (Table 1 and Figure 1). Bone marrow aspirates from 8 normal subjects served as negative controls for the study. Two patients referred with polyclonal elevation of IgG levels whose bone marrow aspirates were enriched for CD138-expressing cells served as further negative controls (data not shown). Figure2 illustrates an agarose gel electrophoretic analysis of Msc I digests of a 134-bp PCR fragment encompassing codon 61 of the N-ras gene from a representative cohort of myeloma patients. The patient samples (lanes 3-7) generated faint bands resistant to digestion of varying intensity (Figure 2A), which were all enhanced after enrichment (Figure 2B). All of the 34 patients had activating mutations of codon 61 of the N-ras gene as determined from direct sequencing of DNA excised from the agarose gels (Table 2). One patient also had an additional mutation in codon 12 of the K-ras gene (data not shown). No further mutations affecting either the N-ras or K-ras oncogene were found in the patient series. None of the control groups displayed detectable mutations in either N-ras or K-ras oncogenes. As a further control, we also verified the absence of mutations in CD138− bone marrow populations from 10 patients analyzed (data not shown).
Screening for N-ras codon 61 mutations by mismatch PCR-RFLP.
MscI restriction enzyme digests of PCR-DNA resolved on 3% metaphor agarose gel containing ethidium bromide. PCR products harboring codon 61 of the N-ras gene were generated by nested PCR of CD138+-enriched cell lysate DNA. The mutant allele is resistant to enzyme digest. (A) Pre-enrichment: lane 1, uncut PCR-DNA; lane 2, HL60 DNA-positive control; lanes 3 to 7, representative patient samples; lane 8, normal DNA. (B) After-enrichment PCR: lane 1, an aliquot of the enzyme digest product of HL60 PCR-DNA from panel A; lane 2, HL60 PCR-DNA showing enrichment of the mutant allele; lanes 3 to 7, patient samples; lane 8, normal DNA.
Screening for N-ras codon 61 mutations by mismatch PCR-RFLP.
MscI restriction enzyme digests of PCR-DNA resolved on 3% metaphor agarose gel containing ethidium bromide. PCR products harboring codon 61 of the N-ras gene were generated by nested PCR of CD138+-enriched cell lysate DNA. The mutant allele is resistant to enzyme digest. (A) Pre-enrichment: lane 1, uncut PCR-DNA; lane 2, HL60 DNA-positive control; lanes 3 to 7, representative patient samples; lane 8, normal DNA. (B) After-enrichment PCR: lane 1, an aliquot of the enzyme digest product of HL60 PCR-DNA from panel A; lane 2, HL60 PCR-DNA showing enrichment of the mutant allele; lanes 3 to 7, patient samples; lane 8, normal DNA.
Characteristics of N-ras codon 61 mutations in myeloma
| Patient no. . | Nucleotide sequence* . | Amino acid substitution† . | % Tumor cells with mutant allele‡ . |
|---|---|---|---|
| 1 | CcA | Proline | 12 |
| 2 | CtA | Leucine | 15 |
| 3 | CgA | Arginine | 16 |
| 4 | CgA | Arginine | 18 |
| 5 | CcA | Proline | 18 |
| 6 | aAA | Lysine | 20 |
| 7 | CcA | Proline | 20 |
| 8 | CcA | Proline | 22 |
| 9 | CtA | Leucine | 22 |
| 10 | aAA | Lysine | 24 |
| 11 | CcA | Proline | 26 |
| 12 | CcA | Proline | 28 |
| 13 | Cat | Histidine | 30 |
| 14 | CgA | Arginine | 32 |
| 15 | CgA | Arginine | 32 |
| 16 | CcA | Proline | 34 |
| 17 | aAA | Lysine | 36 |
| 18 | CtA | Leucine | 42 |
| 19 | CgA | Arginine | 44 |
| 20 | CtA | Leucine | 46 |
| 21 | CAt | Histidine | 52 |
| 22 | CcA | Proline | 54 |
| 23 | CtA | Leucine | 64 |
| 24 | aAA | Lysine | 66 |
| 25 | CtA | Leucine | 68 |
| 26 | CtA | Leucine | 70 |
| 27 | aAA | Lysine | 72 |
| 28 | aAA | Lysine | 76 |
| 29 | CgA | Arginine | 82 |
| 30 | CtA | Leucine | 90 |
| 31 | aAA | Lysine | 100 |
| 32 | CgA | Arginine | 100 |
| 33 | CcA | Proline | 100 |
| 34 | CcA | Proline | 100 |
| Patient no. . | Nucleotide sequence* . | Amino acid substitution† . | % Tumor cells with mutant allele‡ . |
|---|---|---|---|
| 1 | CcA | Proline | 12 |
| 2 | CtA | Leucine | 15 |
| 3 | CgA | Arginine | 16 |
| 4 | CgA | Arginine | 18 |
| 5 | CcA | Proline | 18 |
| 6 | aAA | Lysine | 20 |
| 7 | CcA | Proline | 20 |
| 8 | CcA | Proline | 22 |
| 9 | CtA | Leucine | 22 |
| 10 | aAA | Lysine | 24 |
| 11 | CcA | Proline | 26 |
| 12 | CcA | Proline | 28 |
| 13 | Cat | Histidine | 30 |
| 14 | CgA | Arginine | 32 |
| 15 | CgA | Arginine | 32 |
| 16 | CcA | Proline | 34 |
| 17 | aAA | Lysine | 36 |
| 18 | CtA | Leucine | 42 |
| 19 | CgA | Arginine | 44 |
| 20 | CtA | Leucine | 46 |
| 21 | CAt | Histidine | 52 |
| 22 | CcA | Proline | 54 |
| 23 | CtA | Leucine | 64 |
| 24 | aAA | Lysine | 66 |
| 25 | CtA | Leucine | 68 |
| 26 | CtA | Leucine | 70 |
| 27 | aAA | Lysine | 72 |
| 28 | aAA | Lysine | 76 |
| 29 | CgA | Arginine | 82 |
| 30 | CtA | Leucine | 90 |
| 31 | aAA | Lysine | 100 |
| 32 | CgA | Arginine | 100 |
| 33 | CcA | Proline | 100 |
| 34 | CcA | Proline | 100 |
Wild-type N-ras codon 61 nucleotide sequence-CAA.
Wild-type amino acid-Gln.
CD138+ cells.
Lowercase and bold letters denote nucleotide substitution.
Cells harboring N-ras mutations constitute a variable subset of CD138+tumor cells
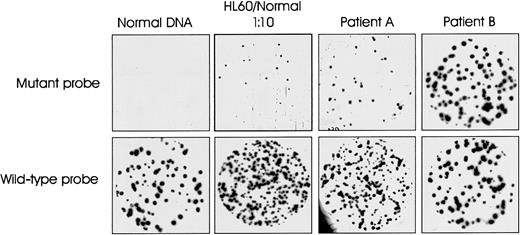
To determine the representation of N-ras codon 61 mutation-positive cells in CD138+ tumor cell populations, quantitative, allele-specific oligonucleotide plaque hybridization was performed on phage libraries generated from cloning PCR-amplified DNA from each patient. Figure 3 illustrates an example of the results obtained. Plaques from normal DNA hybridized to wild-type but not to the mutant-specific probe. A 1:10 mix of HL60 (harboring an N-ras 61 mutation) and normal DNA was used as positive control for the experiment. The 2 patient samples illustrate the variation in quantitative levels of ras mutation-positive cells in the tumor cell population. The mutation was essentially clonal (100%) in patient 32 but constituted only 26% of the CD138+ tumor cell population in patient 11. Table 2 summarizes the quantitative data for all 34 patients. The ras mutation-positive cells as a percentage of the CD138+ tumor cell population ranged from 12% to 100%. The mutation was essentially clonal in just 4 patients.
Representative allele-specific oligonucleotide plaque hybridization for quantitation of N-ras gene mutations.
Duplicate filters hybridized with allele-specific mutant and wild-type oligonucleotide probes for N-ras codon 61. Samples: 1, normal DNA; 2, positive control—10% HL60 in normal DNA; 3 and 4, representative patient samples A and B (patients 11 and 32, Table 2).
Representative allele-specific oligonucleotide plaque hybridization for quantitation of N-ras gene mutations.
Duplicate filters hybridized with allele-specific mutant and wild-type oligonucleotide probes for N-ras codon 61. Samples: 1, normal DNA; 2, positive control—10% HL60 in normal DNA; 3 and 4, representative patient samples A and B (patients 11 and 32, Table 2).
Ras-mutation positive cells are evenly distributed among immunophenotypic subpopulations of myeloma cells
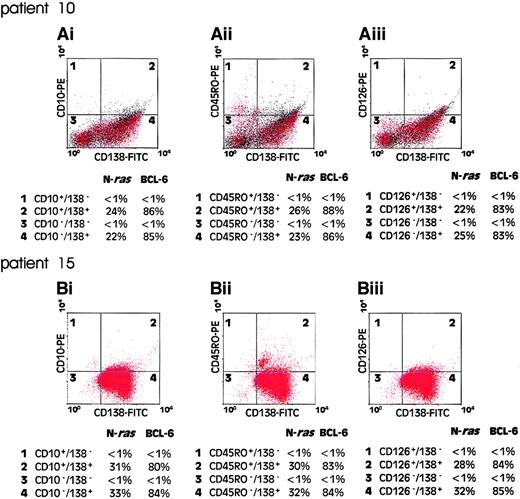
Intraclonal immunophenotypic diversity is a well-described characteristic of malignant cells in multiple myeloma and has, in some studies, been shown to correlate with disease behavior.4-6To determine whether the mutational activation of ras genes occurs before or after the acquisition of variant phenotypes in myeloma, we examined the representation of ras mutation-positive cells among immunophenotypic subsets of FACS sorted myeloma cells. Mononuclear cells from presentation bone marrow aspirates were FACS sorted for coexpression of CD138 with either CD10, CD45RO, or CD126. As a marker of B-cell clonality for these experiments we exploited the occurrence of clonal mutations in the 5′ UTR of the BCL6gene in B-cell malignancies of postgerminal center phenotype.25,27 Previously published reports have shown that about 33% of patients with multiple myeloma harbor one or more mutations in the 5′ noncoding region of the BCL6gene.27 28 Figure 4 shows the results of flow cytometric analysis and corresponding quantitative plaque hybridization data in 2 patients analyzed (patients 10 and 15, Table 2). In both cases, a clonal BCL-6 5′ UTR mutation was present in more than 80% of the CD138+ cells, but not in CD138− cell populations. In patient 10, the ras mutation-positive cells comprised approximately 24% of the CD138+ tumor cell population (Table 2). As shown in Figure4, a comparable value for the representation of ras mutation-positive cells was obtained using BCL-6 5′ UTR mutation as a clonal marker. However, when cells were sorted for expression of either CD10, CD45RO, or CD126, the representation of ras mutation-positive cells, as a fraction of the sorted clonal B-cell populations, remained unchanged. Essentially similar results were obtained in the analysis of patient 15 (Figure 4). These data demonstrate that ras mutation-positive cells are evenly distributed among different immunophenotypic compartments of the malignant clone and suggest that ras mutational events have occurred as an ontogenically earlier event than immunophenotypic diversification of the malignant clone.
Quantitation of mutant N-ras gene in clonal B-cell populations.
Bone marrow mononuclear cells from patient presentation samples were dual labeled with CD138-FITC/CD10-PE (Ai) and (Bi), CD138-FITC/CD45RO-PE (Aii) and (Bii), or CD138-FITC/CD126-PE (Aiii) and (Biii), FACS analyzed, and flow sorted for immunophenotypic subsets. One thousand cells from each quadrant 1 to 4 were subjected to PCR to amplify exon 2 of the N-ras gene and an 850-bp fragment of the 5′ UTR of the BCL6 gene. Products were cloned into a bacteriophage vector for quantitative plaque hybridization to determine the relative levels of mutant and wild-type alleles as shown. The quantitative data are presented as percentage of each cell population positive for an allelic mutation of the N-ras or BCL6 gene.
Quantitation of mutant N-ras gene in clonal B-cell populations.
Bone marrow mononuclear cells from patient presentation samples were dual labeled with CD138-FITC/CD10-PE (Ai) and (Bi), CD138-FITC/CD45RO-PE (Aii) and (Bii), or CD138-FITC/CD126-PE (Aiii) and (Biii), FACS analyzed, and flow sorted for immunophenotypic subsets. One thousand cells from each quadrant 1 to 4 were subjected to PCR to amplify exon 2 of the N-ras gene and an 850-bp fragment of the 5′ UTR of the BCL6 gene. Products were cloned into a bacteriophage vector for quantitative plaque hybridization to determine the relative levels of mutant and wild-type alleles as shown. The quantitative data are presented as percentage of each cell population positive for an allelic mutation of the N-ras or BCL6 gene.
Discussion
Studies in experimental model systems have provided evidence supporting a causative role for mutant ras genes in the pathogenesis of multiple myeloma. For example, transduction of lymphoblastoid cells with constructs expressing mutant N-rasdrives the cells into a plasmacytoid differentiation program with accompanying secretion of monoclonal immunoglobulin, reminiscent of the phenotype in multiple myeloma.29 In more recent studies, mutant ras has been shown to confer a growth advantage and to suppress apoptosis in the interleukin-6–dependent, ANBL-6 myeloma cell line model.30,31 Interestingly, this effect was observed with an N-ras codon 61 mutant, but not with a K-ras codon 12 mutant,31 an observation that may well attest to the functional significance of the predominance of N-ras codon 61 mutations in multiple myeloma.
By using bone marrow preparations that were highly enriched for tumor plasma cells in combination with a sensitive PCR-RFLP strategy, we found that all 34 patients in our series harbored mutations in N-ras codon 61. In addition, we were able to accurately quantitate the representation of mutant ras-containing cells by plaque hybridization in conjunction with ASO probes. This revealed that in the majority of patients, such mutations were present in only a subset of the malignant, clonal B cell population. For this reason it seems likely that earlier studies that used less sensitive screening strategies on nonenriched bone marrow cells7-9,11would have underestimated the true frequency of this mutational event in multiple myeloma. In particular, N-ras gene mutations that are present in only a subpopulation of malignant cells may not have been detected in earlier studies in cases reportedly harboring mutations in other ras genes/N-ras codons. Intriguingly, we did not observe any increased frequency of detection of mutations affecting the K-ras gene; indeed, only one patient in our series was found to harbor a K-ras codon 12 mutation. This finding reinforces the inference alluded to in earlier studies on human leukemia, that only mutations in the N-ras gene manifest in “subclonal” ras mutation-positive tumor cells.21-23
Our finding that mutation of N-ras codon 61 is essentially universal at presentation in multiple myeloma has a number of implications, not least that mutational activation of this oncogene is a mandatory event in the pathogenesis of this disease. This is in contrast to other mutational changes affecting onco/tumor suppressor genes that occur more sporadically in multiple myeloma (reviewed in Hallek et al15). In acute leukemia, disease progression and subsequent relapse in some patients is accompanied by loss of ras mutation-positive blasts.19,22 By contrast, in multiple myeloma, the previously reported incidence of ras mutation-positive cases in disease relapse is notably higher that at presentation (70% versus 10% to 40%) (reviewed in Corradini et al,10 Liu et al,11 and Hallek et al15 and references cited therein). This may well reflect a higher representation of ras mutation-bearing cells in relapsed disease, which would facilitate a higher detection rate in these earlier studies. It may therefore be relevant that, of the 4 patients in our study who presented with more than 90% representation of ras mutation-positive cells, 3 have since succumbed to early relapse (within 6 months). It remains to be determined whether the population dynamics of ras mutation-positive cells in multiple myeloma has prognostic relevance.
Immunophenotypic studies in several laboratories have documented intraclonal heterogeneity in the malignant cell population in multiple myeloma.4-6 This phenomenon and its precise ontologic origin have been the subject of much controversy.6 Our data show that the mutational status of N-ras codon 61 provides a marker for distinguishing 2 distinct subpopulations of tumor cells in most myeloma patients. In further exploring the origin of this “genotypic” intraclonal heterogeneity, we exploited clonotypic point mutations in the BCL-6 5′ UTR to quantitate and track the distribution of ras mutation-positive and ras mutation-negative cells within the clonal B-cell compartment among FACS sorted subpopulations from patients' bone marrow. Using 3 independent immunophenotypic markers in 2 different patients, we found that the representation of ras mutation-positive clonal B cells was independent of immunophenotype of the malignant cells. Such mutational activation of N-ras codon 61 may therefore arise at an ontologically earlier stage in disease pathogenesis than immunophenotypic diversification of the malignant clone. Serial studies on individual patients using ras and BCL-6 5′ UTR-based gene markers to track tumor cell population dynamics should provide further insight into the pathobiology of multiple myeloma.
We thank Dr Jim Heighway for helpful discussions during the initial phase of the work.
Supported by The Gardner Bequest Fund, the Christie Hospital Endowment Fund, and the United Kingdom Cancer Research Campaign.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
John D. Norton, Department of Biological Sciences, University of Essex, Wivenhoe Park, Colchester, Essex, CO4 3SQ, United Kingdom; e-mail: jnorton@essex.ac.uk.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal