The CBA/H mouse model of radiation-induced acute myeloid leukemia (AML) was re-examined using molecular approaches. In addition to the typical promyelocytic AMLs, 34% were reclassified as early pre-B lympho-myeloid leukemias (L-ML) based on leukemic blood cell morphology, immunoglobulin heavy-chain gene re-arrangements (IgHR), or expression of both lymphoid (Vpre-B1 and Rag1) and myeloid (myeloperoxidase and lysozyme M) genes. Allelic loss on chromosome 4 was frequently detected in AMLs (53%) and L-MLs (more than 95%), and the preferential loss of the maternally transmitted allele suggests the locus may be imprinted. A minimally deleted region (MDR) maps to a 3.4-cM interval, which is frequently deleted in radiation-induced thymic lymphomas (TLSR5) and contains a recessive, maternally transmitted genetic locus (Lyr2) that confers resistance to spontaneous and radiation-induced pre-B and T cell lymphomas, suggesting they are one and the same. Thus, theLyr2/TLSR5 locus is frequently implicated in myeloid, lymphoid (B and T), and mixed-lineage mouse leukemias and lymphomas. Epigenetic inactivation of one Lyr2/TLSR5 allele during normal mouse development suggests that only a single hit is required for its inactivation during leukemogenesis, and this may be a significant contributing factor to the efficiency of the leukemogenic process in the mouse.
Introduction
Genetic alterations that interfere with terminal differentiation are key events in the evolution of many leukemias. Most spontaneous human acute leukemias exhibit specific chromosomal translocations that result in either the enforced expression of an oncogene or the fusion of genes normally involved in the control of hemopoietic differentiation.1 Unlike the oncogenic gain-of-function genetic alterations observed in spontaneous leukemias, radiation-induced leukemias in mouse and humans exhibit chromosomal loss,2-11 suggesting that tumor-suppressor gene loss of function has a role in maturation arrest in radiation-induced hemopoietic malignancies.
Although radiation-induced leukemias in humans are predominantly myeloid, both radiation-induced acute myeloid leukemias (AMLs) and lymphomas have been described in the mouse.2-13 Loss of function of the Ikaros gene, which is required for the development of all lymphoid lineages, has been observed in mouse radiation-induced thymic lymphomas8—evidence to support a role of loss-of-gene function in maturation arrest. However, inactivation of the p15INK4b cyclin-dependent kinase inhibitor multitumor-suppressor gene by allelic loss and promoter methylation has also been detected in radiation-induced thymic lymphomas (but not myeloid leukemias), indicating that loss-of-gene function resulting in genetic instability plays a role in lineage-specific multistage radiation lymphomagenesis.9,10Furthermore, despite the fact that an identical or a similar initiating event is responsible for the induction of the different mouse radiation-induced hemopoietic malignancies, allelic loss or tumor-suppressor gene inactivation appears to be largely leukemia/lymphoma-specific: chromosome 2 deletions are characteristic of radiation-induced mouse AMLs, and allelic loss on chromosomes 4, 11, 12, 14,16, and 19 has been reported in radiation-induced thymic lymphomas, though the genetic background of the irradiated mice may influence allelic loss on specific chromosomes.3-11
The diagnosis of radiation-induced leukemia/lymphoma in the mouse does not follow the strict criteria used to diagnose human leukemia. For example, the appearance of metamyelocytes with a thick ring-shaped nucleus in the leukemic blood is characteristic of mouse AML,12 though subclassifications have been proposed.3,13 However, immunophenotype and immunogenotype analyses of leukemias that arose in X-irradiated Eμ–BCL-2 transgenic mice revealed early B lympho-myeloid leukemia (L-ML)14 that might have been diagnosed as AML if leukemic blood cell morphology was the sole criteria used. TheEμ–BCL-2 L-MLs were attributed to the inhibition of apoptosis by BCL-2 in the normally highly radiosensitive pre-B cells, yet they arose in a predominantly CBA/H genetic background,14 and inbred CBA/H mice are susceptible to radiation-induced AML.12 13
To determine whether the generation of radiation-induced L-ML was dependent on the anti-apoptotic effects of the BCL-2transgene in B cell precursor cells and to test the possibility that some radiation-induced AMLs had been misdiagnosed, we re-examined a panel of mouse leukemias diagnosed as AML by blood cell morphology. Two thirds were typical AMLs, and one third were early pre-B lympho-myeloid leukemias similar to the radiation-induced Eμ-BCL-2 L-MLs.14 The radioprotective effect of theBCL-2 transgene is therefore not essential to generate this type of mixed-lineage leukemia in vivo. Allelic loss on chromosome 4 was frequently detected in both L-MLs (more than 95%) and AMLs (approximately 50%), as it was in the Eμ-BCL-2 L-MLs.14 A 3.4-cM minimally deleted region (MDR) on chromosome 4 maps to an interval frequently deleted in radiation-induced thymic lymphomas (thymic lymphoma-suppressor region 5, TLSR5)7 and also contains the recessive maternally transmitted lymphoma resistance 2 (Lyr2) locus.Lyr2 confers resistance to spontaneous and radiation-induced pre-B and T cell lymphomas/lymphocytic leukemias,15-17suggesting that the imprinted locus is involved in most mouse spontaneous and radiation-induced hemopoietic malignancies, either as a tumor-suppressor gene or a susceptibility–resistance locus.
Materials and methods
Mouse irradiations
CBA/H and C57BL/6 mice were from the Harwell colony. Eight- to 12-week-old CBA/H (n = 138), (CBA/H × C57BL/6)F1 (n = 89), F1 × CBA/H (n = 827), F1 × C57BL/6 (n = 114), and F1 × F1 (n = 142) mice were exposed to a single acute dose of 3.0 Gy x-radiation at 0.5 Gy/min (constant potential, 250 kV; HVL, 1.2 mm3). Animal studies were carried out according to the guidelines, Responsibility in the Use of Animals for Medical Research, issued by the MRC in (July 1993) and Home Office Project license numbers PPL 30/689 and 30/1272. Mouse leukemias were initially diagnosed by microscopic examination of blood, bone marrow, spleen, and thymus. Leukemic spleen and thymus was snap-frozen in dry ice.
Molecular studies
DNA was prepared from control tail and leukemic spleen.4,10,14 Approximately 15 μg restriction enzyme-digested DNA was resolved by 1% (wt/vol) agarose gel electrophoresis and transferred to Genescreen (NEN Life Science Products, Boston, MA) nylon membranes. HindIII DNA digests were probed with the pTcrb-J2 probe to screen for T cell receptor β (TCR-β) gene rearrangements,18 andEcoRI and BamHI DNA double digests were probed with p5′(JH) to screen for immunoglobulin heavy chain (IgH) gene rearrangements.19 Loss of heterozygosity analyses using specific microsatellite markers on chromosomes 2 and 4 with tail and leukemic DNA from the same mouse have already been described.4 Genetic map positions are from the Mouse Genome Database,20 and, except for MP-15-3′ on chromosome 4, microsatellite primer sequences were from the Whitehead Institute.21
Depending on the degree of splenomegaly, total cellular RNA was also prepared from approximately half of the leukemic spleen and from control adult spleen, bone marrow, thymus, and kidney, for Northern blot analyses.22 RNA (approximately 10-20 μg) was resolved by 1.0% (wt/vol) denaturing gel electrophoresis, transferred to Genescreen membranes, and probed with myeloperoxidase (MPO),22 lysozyme M (LysM),22CD19,22 PU1,23Vpre-B1 (nucleotides 320-866),24 and Rag1 (nucleotides 207-1896)25 cDNA probes. Normal bone marrow and thymus were used as positive controls for Vpre-B1 and Rag1,respectively (data not shown). Equivalent RNA loading was confirmed by ethidium bromide staining or hybridizations with glutathione peroxidase.26 Probes were labeled with [α-32P]-dATP (3000 Ci/mmol; Amersham Pharmacia Biotech, Amersham, United Kingdom) using the Random Prime Labeling Kit (Life Technologies, Paisley, United Kingdom). Blots were quantitated using a Bio-Rad Molecular Imager FX (Bio-Rad Laboratories, Hemel Hempstead, Bucks, United Kingdom).
Results
Leukemia presentation
AML diagnosis in the mouse relies predominantly on an increased white blood cell count and the appearance of immature myeloid cells in the peripheral blood.12,13,27 Leukemic cells infiltrate the spleen, resulting in splenomegaly, and may be detected in the bone marrow, though bone marrow failure is common. Healthy mouse peripheral blood predominantly contains neutrophils and lymphoid cells (Table1), and the relative proportion of these cells decreases significantly in myeloid leukemias. As illustrated in Table 1, the distribution of the different immature myeloid cells in the mouse leukemias allows them to be further subclassified (A-D)3 13 in a manner that in some respects is analogous to the French–American–British (FAB) classification of human leukemic bone marrow. In a study involving 1310 X-irradiated mice, 195 myeloid leukemias were diagnosed, and the relative incidence of each leukemia subclass is shown in Table 1.
Leukocyte distribution in the peripheral blood of normal and leukemic mice
| Cell type . | White blood cell count (%) . | ||||
|---|---|---|---|---|---|
| Normal blood . | Leukemia subclass . | ||||
| A . | B . | C . | D . | ||
| Neutrophil | ∼ 25 | — | < 2 | < 2 | — |
| Metamyelocyte | — | 0-3 | ∼ 60-70 | 15-20 | — |
| Myelocyte | — | < 2 | ∼ 30-40 | ∼ 10 | < 2-3 |
| Myeloblast | — | 20-40 | ∼5 | ∼2 | < 1 |
| Blast | — | 60-70 | < 5 | ∼ 2 | < 1 |
| Eosinophil | ∼ 0.5 | — | — | — | — |
| Monocyte/macrophage | ∼ 2 | — | < 1 | 50-60 | 80-90 |
| Lymphocytic | 60-66 | — | ∼ 5 | ∼ 5 | ∼ 5 |
| Leukemia subclass incidence (%) (no. leukemias diagnosed) | 19.9 | 43.7 | 27.8 | 8.6 | |
| (n = 39) | (n = 85) | (n = 54) | (n = 17) | ||
| Cell type . | White blood cell count (%) . | ||||
|---|---|---|---|---|---|
| Normal blood . | Leukemia subclass . | ||||
| A . | B . | C . | D . | ||
| Neutrophil | ∼ 25 | — | < 2 | < 2 | — |
| Metamyelocyte | — | 0-3 | ∼ 60-70 | 15-20 | — |
| Myelocyte | — | < 2 | ∼ 30-40 | ∼ 10 | < 2-3 |
| Myeloblast | — | 20-40 | ∼5 | ∼2 | < 1 |
| Blast | — | 60-70 | < 5 | ∼ 2 | < 1 |
| Eosinophil | ∼ 0.5 | — | — | — | — |
| Monocyte/macrophage | ∼ 2 | — | < 1 | 50-60 | 80-90 |
| Lymphocytic | 60-66 | — | ∼ 5 | ∼ 5 | ∼ 5 |
| Leukemia subclass incidence (%) (no. leukemias diagnosed) | 19.9 | 43.7 | 27.8 | 8.6 | |
| (n = 39) | (n = 85) | (n = 54) | (n = 17) | ||
Leukemia immunogenotype
Leukemic spleen DNA samples were immunogenotyped forIgH and TCR-β gene rearrangements. Although all the leukemias had a germline TCR-β gene configuration (TCR-βG; data not shown), 34% hadIgH gene rearrangements (IgHR; Figure1 and data not shown). The relative proportions of IgHG andIgHR leukemias in each of the A-D leukemia subclasses (Table 2) indicate that although subclasses B and C are most typical of IgHGleukemias and A and D are most typical of theIgHR leukemias, there is significant overlap.
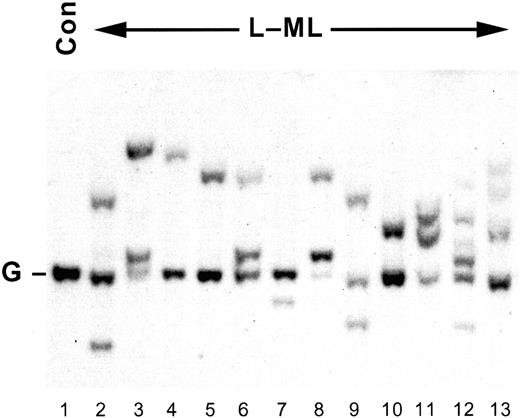
IgH gene rearrangements.
Representative Southern blot analysis of EcoRI- andBamHI-digested genomic DNA (approximately 15 μg) prepared from control spleen (lane 1) or L-MLs (lanes 2-13). DNA was resolved by 1% (wt/vol) agarose gel electrophoresis, blotted, and probed with p5′(JH).14 19 G, IgH gene germline configuration; Con, control spleen.
IgH gene rearrangements.
Representative Southern blot analysis of EcoRI- andBamHI-digested genomic DNA (approximately 15 μg) prepared from control spleen (lane 1) or L-MLs (lanes 2-13). DNA was resolved by 1% (wt/vol) agarose gel electrophoresis, blotted, and probed with p5′(JH).14 19 G, IgH gene germline configuration; Con, control spleen.
Immunogenotype and leukemia subclasses
| Genotype . | Leukemia subclass (%) . | |||
|---|---|---|---|---|
| A . | B . | C . | D . | |
| IgHG | 23.3 | 83.3 | 66.7 | 30.8 |
| (n = 9) | (n = 71) | (n = 36) | (n = 5) | |
| IgHR | 76.7 | 16.7 | 33.3 | 69.2 |
| (n = 30) | (n = 14) | (n = 18) | (n = 12) | |
| Genotype . | Leukemia subclass (%) . | |||
|---|---|---|---|---|
| A . | B . | C . | D . | |
| IgHG | 23.3 | 83.3 | 66.7 | 30.8 |
| (n = 9) | (n = 71) | (n = 36) | (n = 5) | |
| IgHR | 76.7 | 16.7 | 33.3 | 69.2 |
| (n = 30) | (n = 14) | (n = 18) | (n = 12) | |
As shown in Figure 1, though mono-allelic (lanes 4, 5, 7, 10) and bi-allelic (lanes 3 and 8) IgH gene rearrangements were detected, 2 rearranged alleles in addition to the germline configuration allele (lanes 2, 6, and 11) were also frequently observed. Although this might be attributed to the presence of contaminating normal cells in the spleen, the hybridization signal is variable. In many cases (Figure 1, lanes 2-11 and 13), loss of heterozygosity (LOH) on chromosome 4 was detectable by polymerase chain reaction in the same DNA samples, indicating that less than 20% contaminating normal cells were present in the leukemic spleen (see below). Furthermore, 3 or more rearranged alleles were also detected in 5 leukemias (Figure 1, lanes 12 and 13, and data not shown), suggesting either the presence of subclonal variants within a clonal leukemia, or the presence of more than one independently arising leukemia in the same mouse. Given that the mean latency of the IgHGand IgHR leukemias is approximately 18 months and that the lifetime incidence of theIgHR leukemias in irradiated CBA/H (8.7%), (CBA/H × C57BL/6)F1 (10.1%), F1 × CBA/H (7.9%), F1 × C57BL/6 (7.9%), and F1 × F1 (6.3%) is low, the probability that 2 or more independent IgHR leukemias arose in the same mouse at the same (or at a similar) time is negligible. Multiple IgHR alleles within individual leukemias (Figure 1, lanes 2, 6, 9, 11-13) most likely represent subclonal variants of a clonal proB cell leukemia with a leaky proB(IgHG) → pre-B(IgHR) cell differentiation block. Furthermore, because there was no statistically significant difference in the incidence of IgHR leukemias in the inbred CBA/H and the hybrid mice analyzed, the C57BL/6 genetic background in the backcross and intercross mice is not a confounding factor.
Leukemia phenotype
To further characterize the IgHG andIgHR leukemias, total cellular RNA was prepared from 71 IgHG and 38 IgHRleukemic spleens and analyzed for the expression of lineage-specific/restricted markers by Northern blot (Figure2A-B). Because the source of leukemic cells is the leukemic spleen and leukemic cell infiltration into the spleen is variable, only significant differences in the mRNA profile of a leukemic spleen compared to a normal spleen can be attributed to the leukemia cells. Hence, enrichment (+) of an mRNA species in an individual leukemia is defined as more than 2-fold and depletion (−) as less than 0.5-fold, compared to its level in control spleen (MPO, CD19, LysM, PU1), or as the presence (+) or absence (−) when that mRNA is not detectable in the spleen (Vpre-B1, Rag1). Only large differences in mRNA levels are considered informative and are larger than those that might be attributed to gel loading. Leukemias that contained mRNA levels less than 2 and greater than 0.5 compared to control spleen are, therefore, not considered informative and are not scored in Table 3.
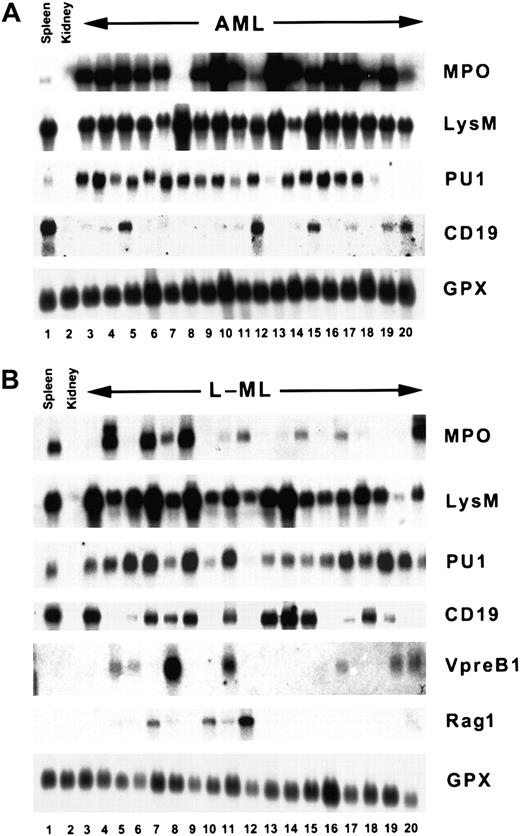
AML and L-ML mRNA expression profiles.
Representative Northern blots containing total cellular RNA (approximately 20 μg) prepared from control spleen or kidney. (A)IgHG leukemias. (B) IgHRleukemias. Separate blots were probed with myeloperoxidase (MPO), lysozyme M (LysM), CD19, PU1, Vpre-B1, Rag1, or glutathione peroxidase cDNA probes. Quantitation of the Northern blots is summarized in Table 3.
AML and L-ML mRNA expression profiles.
Representative Northern blots containing total cellular RNA (approximately 20 μg) prepared from control spleen or kidney. (A)IgHG leukemias. (B) IgHRleukemias. Separate blots were probed with myeloperoxidase (MPO), lysozyme M (LysM), CD19, PU1, Vpre-B1, Rag1, or glutathione peroxidase cDNA probes. Quantitation of the Northern blots is summarized in Table 3.
Leukemia mRNA expression profile compared to control spleen
| mRNA . | Leukemia . | |||
|---|---|---|---|---|
| IgHG (AML) (%) . | IgHR (L-ML) (%) . | |||
| + . | − . | + . | − . | |
| MPO | 92 (61 of 66) | 5 (3 of 60) | 21 (8 of 38) | 68 (26 of 38) |
| LysM | 6 (4 of 65) | 26 (17 of 65) | 22 (7 of 32) | 53 (17 of 32) |
| PU1 | 47 (15 of 32) | 6 (4 of 65) | 47 (15 of 32) | 9 (3 of 32) |
| Vpre-B | 0 (0 of 71) | 100 (71 of 71) | 47 (18 of 38) | 53 (20 of 38) |
| CD19 | 0 (0 of 51) | 96 (49 of 51) | 0 (0 of 32) | 75 (24 of 32) |
| Rag1 | 0 (0 of 54) | 100 (0 of 54) | 36 (12 of 33) | 64 (21 of 33) |
| mRNA . | Leukemia . | |||
|---|---|---|---|---|
| IgHG (AML) (%) . | IgHR (L-ML) (%) . | |||
| + . | − . | + . | − . | |
| MPO | 92 (61 of 66) | 5 (3 of 60) | 21 (8 of 38) | 68 (26 of 38) |
| LysM | 6 (4 of 65) | 26 (17 of 65) | 22 (7 of 32) | 53 (17 of 32) |
| PU1 | 47 (15 of 32) | 6 (4 of 65) | 47 (15 of 32) | 9 (3 of 32) |
| Vpre-B | 0 (0 of 71) | 100 (71 of 71) | 47 (18 of 38) | 53 (20 of 38) |
| CD19 | 0 (0 of 51) | 96 (49 of 51) | 0 (0 of 32) | 75 (24 of 32) |
| Rag1 | 0 (0 of 54) | 100 (0 of 54) | 36 (12 of 33) | 64 (21 of 33) |
+, > 2-fold control spleen; −, < 0.5-fold control spleen.
As shown in Figure 2A and Table 3 and compared to control spleen, the typical IgHG leukemia is MPO+ (mean expression, 54.3 ± 76.8), PU1+ (mean expression, 4.35 ± 3.89), LysM− (mean expression, 1.0 ± 1.0), CD19− (mean expression, 0.16 ± 0.196),Vpre-B1−, and Rag1−.They are clearly AMLs and resemble a human M2 AML.28,29 In contrast, the IgHR leukemias exhibit a much more heterogeneous mRNA expression profile (Figure 2B). No leukemia exhibited greater than 2-fold enrichment of CD19 mRNA compared to control spleen (mean expression, 0.4 ± 0.43), and on average they expressed lower levels of PU1 mRNA (mean expression, 1.72 ± 1.03) than the AMLs, though a similar proportion (47%, Table 3) did show some enrichment (mean expression, 2.6 ± 0.78) of the myeloid and B cell PU1 mRNA marker.33 Because 36% to 47% of the leukemias express Vpre-B1 and Rag1, this is consistent with the ProB/early pre-B differentiation block inferred from the IgH gene rearrangements.14,24,25,30Although the IgHR leukemias were generally depleted for the myeloid MPO and LysM mRNAs, 8 of 38 exhibited enrichment of MPO mRNA (Figure 2B, lanes 4, 6, 8, and 20; mean expression, 4 ± 2.1), and 7 of 32 exhibited enrichment of LysM mRNA (Figure 2B, lanes 3, 6, 8, and 13; mean expression, 3.9 ± 1.1), consistent with the detection of immature myeloid cells in the leukemic peripheral blood (Table 1). These IgHR, CD19−, MPO+/−, LysM+/−, PU1+/−, Vpre-B1+/−, Rag1+/− leukemias are, therefore, mixed-lineage early B L-MLs and are similar to the IgHRCD19−, Mac-1+, and B220+ L-MLs that arise in irradiated Eμ–BCL-2 transgenic mice in a predominantly CBA/H genetic background.14 Thus, theEμ–BCL-2 transgene is not essential for the generation of this type of leukemia.
Loss of heterozygosity
Allelic loss on chromosome 2 is detected in more than 90% of radiation-induced AMLs in CBA/H mice,3,4,27,31,32 and allelic loss on chromosome 4 is frequently detected in radiation-induced thymic lymphomas and Eμ–BCL-2L-MLs.5,7,9,11,14,33 Leukemias in this study were screened for LOH on chromosomes 2 and 4. Because most of the leukemias arose in F1 backcross or intercross mice, there is a 50% probability of homozygosity at any given microsatellite marker. Similarly, only those leukemic DNA samples that contain less than 20% contaminating normal cells reveal LOH.4,31 32
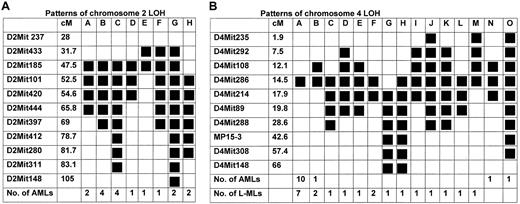
Thirty AMLs were informative for chromosome 2 LOH (Chr.2LOH), and the LOH patterns for the 17 AMLs that were informative (heterozygous) for both proximal and distal breakpoints are shown in Figure 3A. A 5-cM MDR (47.5-52.5 cM) is defined by LOH patterns E and H (Figure 3A) and is consistent with the recently defined 0.6-cM MDR (47.5-48.1 cM).32There was no differential loss of the CBA/H or C57BL/6 allele (18:12, respectively), consistent with findings of other studies.4,31 32 In contrast, only 2 L-MLs (less than 10%) had detectable Chr.2LOH, indicating that allelic loss on chromosome 2 is specific for radiation-induced AML.
Deletion mapping by LOH in informative leukemias that have heterozygous proximal and distal breakpoints.
(A) Chromosome 2 (AML) (B) Chromosome 4 (AML and L-ML). Polymorphic microsatellite markers used and their relative genetic positions (cM)20 are shown, as are the number (No.) of AMLs or L-MLs that exhibited a particular LOH pattern (A-O). Solid boxes show regions of LOH.
Deletion mapping by LOH in informative leukemias that have heterozygous proximal and distal breakpoints.
(A) Chromosome 2 (AML) (B) Chromosome 4 (AML and L-ML). Polymorphic microsatellite markers used and their relative genetic positions (cM)20 are shown, as are the number (No.) of AMLs or L-MLs that exhibited a particular LOH pattern (A-O). Solid boxes show regions of LOH.
A similar analysis for chromosome 4 LOH (Chr.4LOH) revealed 15 AML and 25 L-ML informative leukemias. Chr.4LOHpatterns for the 34 leukemias heterozygous at both proximal and distal breakpoints are illustrated in Figure 3B. The maternally transmitted CBA/H allele was preferentially lost in both AMLs (9 of 10) and L-MLs (16 of 18) that arose in irradiated CBA/H × (C57BL/6 × CBA/H)F1 backcross mice, and 19 of 28 of these leukemias arose in female mice. However, the D4Mit286 genotype of CBA/H × F1 backcross mice affected with L-ML (homozygous–heterozygous ratio, 19:23) or AML (homozygous–heterozygous ratio, 25:34) revealed no significant excess homozygosity or heterozygosity (P > .5), indicating that there is no genetic linkage between genotype and phenotype at this locus in the CBA/H and C57BL/6 inbred mouse strains.
Seventeen AMLs were informative for LOH and were heterozygous on chromosomes 2 and 4. Eight of 17 exhibited Chr.2LOH and an apparently normal chromosome 4; 8 of 17 exhibited Chr.2LOHand Chr.4LOH; and 1 of 17 exhibited Chr.4LOHand an apparently normal chromosome 2. At the level of detection defined by the microsatellite markers used, Chr.2LOH was detected in 94% of AMLs and can be described as a primary genetic lesion, whereas Chr.4LOH was a recurrent (53%) secondary chromosomal abnormality.
AML Chr.4LOH patterns define a 5.8-cM (12.1-17.9 cM) MDR (pattern A, Figure 3B), and this is reduced to a 3.4-cM interval (14.5 cM-17.9 cM), when the L-ML LOH patterns (A, B, and G) are included. TLSR5 on chromosome 4 has been mapped to an approximately 20-cM interval centered on D4Mit21 (15.6 cM),7 and, because most of the leukemias in this study exhibit LOH in this region (Figure 3B), allelic loss at TLSR5 is associated with all 3 types of radiation-induced hemopoietic malignancies.
Discussion
The diagnosis of mouse radiation-induced AMLs has traditionally relied on an increased white blood cell count and the appearance of immature myeloid cells in the blood, bone marrow and spleen,12,13,27 and the subclassification of the AMLs using FAB criteria has been suggested.3 13 Our molecular analyses of 195 leukemias that arose in irradiated mice and that were originally diagnosed as AMLs using leukemic blood cell morphology has revealed 2 distinct malignancies. Sixty-six percent were typical AMLs, and 34% were mixed-lineage early B L-MLs. A screen for LOH on chromosomes 2 and 4 revealed that chromosome 2 allelic loss is specific to the AMLs (more than 90%), but chromosome 4 allelic loss is frequently observed in both AMLs (approximately 50%) and L-MLs (more than 90%).
We have mapped a 3.4-cM MDR on chromosome 4 that is common to both radiation-induced AMLs and L-MLs. Allelic loss on chromosome 4 is also frequently observed in radiation-induced thymic lymphomas (a T cell malignancy), and one tumor-suppressor gene locus, TLSR5,7maps to (or very near) the MDR identified in our study. The lymphoma resistance 2 (Lyr2, 14.5 cM)20 locus that confers a resistance to spontaneous or radiation-induced (pre-B cell) lymphocytic leukemias/lymphomas was fine mapped to this interval by genetic linkage analyses using SL/Kh mice that are highly susceptible to spontaneous pre-B lymphomas and SL/Ni mice that are resistant. Resistance to spontaneous pre-B lymphomas in SL/Ni mice is conferred by a recessive maternally transmitted resistance locus,15-17 34 but there is no linkage between theLyr2 genotype and phenotype (radiation-induced AML or L-ML) in the irradiated CBA/H × F1 mice in our study.
The preferential loss of the maternally transmitted CBA/H allele (25 of 28 leukemias) in the radiation-induced L-MLs and AMLs that arose in our study is consistent with the maternal effects observed at theLyr2 locus in spontaneous pre-B lymphomas.17Loss of gene function in the radiation-induced L-MLs and AMLs presumably occurs by the deletion of the active maternally derived allele and the inactivation of the paternal allele by imprinting, and this single hit inactivation of a tumor-suppressor gene may be a contributing factor to the relative efficiency of radiation leukemogenesis in the mouse. The preferential loss of the maternal allele at a different chromosome 4 locus (42.6-55.6 cM) has also been observed in methylene chloride-induced lung carcinoma.35However, genetic background effects are observed in LOH analyses of radiation-induced thymic lymphomas. The preferential loss of the paternal RF/J allele was observed at the TLSR1 locus (42.6-44.5 cM) in radiation-induced thymic lymphomas that arose in a (C57BL/6 × RF/J)F1 genetic background,5 but not in those that arose in a (C57BL/6 × BALB/c)F1 genetic background.7
The observation that approximately 50% of the mouse radiation-induced AMLs exhibit allelic loss at TLSR5/Lyr2, in addition to the characteristic (more than 95%) allelic loss on chromosome 2, is unexpected given that chromosome 4 aberrations have been infrequently detected (less than 20%) in cytogenetic studies.3,27 As illustrated in Figure 3A, most of the chromosome 2 deletions are large (greater than 20 cM), consistent with the terminal and interstitial deletions involving the loss of approximately 33% to 66% of the approximately 120-cM chromosome observed by cytogenetics.3,27,31,32 In contrast, most (11 of 13) of the AML chromosome 4 deletions are relatively small (less than 10 cM; Figure 3B), representing less than 10% of the whole chromosome, and are presumably too small to detect reliably using conventional cytogenetic methods. There is increasing evidence that gene deletions directly induced by ionizing radiation tend to be considerably larger than those that occur either as an indirect consequence of irradiation or as a consequence of cytoplasmic irradiation.36-38 This raises the possibility that radiation-induced allelic loss on chromosome 2 is the initiating event in mouse myeloid radiation leukemogenesis and that allelic loss at Lyr2/TLSR5 is a recurrent secondary event that arises as an indirect or a delayed effect of the initiating exposure to radiation.
The presence of IgH gene rearrangements in approximately 10% of human AMLs39-44 has been attributed to either lineage infidelity, involving an aberrant differentiation program in the leukemic cell, or lineage promiscuity of a lympho-myeloid progenitor cell and the corresponding leukemia.14,43 Pre-B progenitor cells that can undergo myeloid differentiation under specific conditions have been detected in vitro,45 46supporting evidence for the lineage promiscuity model. If the choice of commitment to a particular lineage is intrinsically flexible and the differentiation block in the corresponding leukemia is incomplete, subclonal variants generated by the different maturation options and external microenvironmental influences available in vivo should be detected within a clonal leukemia. The detection of more than 2 rearranged IgH alleles within individual leukemias (Figure1) is consistent with a leaky ProB (IgHG)⇒ pre-B (IgHR) differentiation block.
The involvement of the apparently imprinted Lyr2/TLSR5 locus in spontaneous and induced myeloid, lymphoid, and mixed-lineage leukemias/lymphomas suggests it may play a key role in the control of differentiation of multipotential lympho-myeloid progenitor cells. In many respects, the Pax5 gene is an excellent candidate. B cell development in Pax5−/− cells is arrested at the Pre-BI (IgHR) stage in vivo, but they can be induced to undergo terminal myeloid, T-lymphoid, or B-lymphoid differentiation in vitro depending on the presence or absence of cytokines or stromal support cells.46 Although thePax5 gene maps (20.7 cM)20 near theLyr2/TLSR5 locus (14.5-17.9 cM, Figure 3B), our mapping studies excluded it as a candidate, and no Pax5 gene rearrangements or homozygous deletions have been detected in our leukemias (M.P., unpublished results). Given that we have evidence that maturation arrest can occur at the ProB to early pre-B stage, the Lyr2/TLSR5 locus may act upstream ofPax5. One prediction from this model is that (pre)-leukemic L-ML cells will occasionally infiltrate the thymus, where the microenvironment will induce TCR-β gene rearrangements, resulting in mixed-lineageIgHR-TCR-βR thymic lymphomas/T cell leukemias.
A second candidate gene that maps to the Lyr2/TLSR5 locus is the type 1 receptor for transforming growth factor TGF-β (Tgfbr1) because its inactivation may contribute to the transformation process by rendering leukemic cells insensitive to growth inhibition by TGF-β. The human orthologue (TGFBR1) maps to chromosome 9q22, and a tumor-suppressor gene locus identified in this region in bladder cancer47 and TGFBR1has been implicated directly as a tumor susceptibility allele in human cancer48 and cutaneous T cell lymphoma.49
We thank T. Ford, J. Neil, M. Cross, N. Kerr, P. Harrison, and T. Tedder for the cDNA probes, and M. Greaves and E. Wright for help in the course of these studies.
Supported by the Medical Research Council and by the Leukaemia Research Fund as part of a Specialist Programme in Radiation Leukaemogenesis. H.C. was supported by an MRC Research Studentship.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Mark Plumb, Department of Genetics, University of Leicester, University Road, Leicester LE1 7RH, United Kingdom; e-mail: map12@le.ac.uk.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal