Abstract
The stage of progenitor maturation and factors that determine the fate and clonal acquisition of human natural killer (NK) cell receptors during development are unknown. To study human NK cell receptor ontogeny, umbilical cord blood CD34+/Lin−/CD38− cells were cultured with a murine fetal liver line (AFT024) and defined cytokines. In the absence of lymphocyte-stimulating cytokines or when contact with AFT024 was prohibited, NK cell progeny were killer immunoglobulinlike receptor (KIR) and CD94 lectin receptor negative. In contrast, efficient NK cell differentiation and receptor acquisition was dependent on direct contact of progenitors with AFT024 and the addition of interleukin-15 (IL-15) or IL-2 but not IL-7. To address the question of whether receptor acquisition was determined at the stem cell level, single CD34+/Lin−/CD38−progenitors were studied. More than 400 single cell progeny were analyzed from cultures containing IL-15 or IL-2 and NK cells were always polyclonal, suggesting that receptor fate is determined beyond an uncommitted progenitor and that receptor-negative NK cells acquire class I-recognizing receptors after lineage commitment. KIR2DL2/L3/S2 was expressed more than KIR2DL1/S1 or KIR3DL1, and NKG2A was the dominant CD94 receptor, independent of whether the stem cell source contained the respective major histocompatibility complex class I ligand, suggesting a nonrandom sequence of receptor acquisition. The conclusion is that NK receptor fate is determined after NK cell commitment, does not require stromal presentation of human class I alleles, and is clonally stable after expression but dynamic because new receptors are acquired over time.
Introduction
In contrast to T cells, natural killer (NK) cells kill a broad array of targets in an HLA-unrestricted manner. However, current observations counter the notion that NK cell killing is totally major histocompatibility complex (MHC) unrestricted.1-3Although antigen presented by MHC class I molecules activates T cells, “self” class I presentation renders some targets resistant to lysis by NK cells—exactly the opposite. As originally proposed by Ljunggren and Karre2 in the “missing self” hypothesis, it is now well recognized that class I down-regulation by transformation or infection may make cells susceptible to NK lysis. Class I NK receptors were first identified in the mouse as the Ly49 proteins.4,5 In contrast, human class I–recognizing NK receptors are members of the immunoglobulin superfamily designated killer immunoglobulinlike receptors (KIRs).6-10
KIRs are encoded on chromosome 19 and designated by their number of immunoglobulin domains (KIR2D, KIR3D, etc). Their inhibitory and activating function is determined by the length of their associated cytoplasmic tail, long (L) or short (S), respectively.11-13 At least 3 groups of inhibitory receptors (KIR2DL1/S1, KIR2DL2/L3/S2, and KIR3DL1) have class I ligands identified (cw4, cw3, and bw4).14-16 There has been much progress in understanding the determinants of how class I and associated binding peptides may influence receptor ligation17,18 and subsequent signaling.19-22
The lectin-type human NK cell receptors on chromosome 12 initially described by Houchins et al23 and Yabe et al24 were difficult to characterize because they exist as heterodimers, disulfide-linked to CD94.25,26 It is now well characterized that NKG2/CD94 family members are inhibitory (NKG2A, NKG2B) and activating (NKG2C, NKG2E) but recognize nonclassical class 1b HLA-E molecules bound with peptide leader sequences from several classical ABC alleles as well as HLA-G.27-29 The crystal structure of the extracellular domain of CD94 shows a unique c-type lectin fold and a putative ligand binding region for HLA-E with the NKG2/CD94 heterodimer.30 Recently, a unique role of NKG2D, which does not bind CD94, has been elucidated.31 32 NKG2D serves as an activating receptor after interaction with the stress-inducible nonclassical MICA (and possibly MICB) class I homolog. The ontogeny of CD94 lectin and immunoglobulin receptors during human NK cell development and factors that determine NK cell receptor repertoires are unknown.
NK cells are derived from human marrow primitive progenitors.33,34 In the presence of interleukin-2 (IL-2) alone, NK cell differentiation from these progenitors is dependent on direct contact with human stromal ligands.35 The development of NK cells from marrow progenitors has been corroborated in vitro36-38 and in vivo after transplanting fetal sheep with CD34+/Lin−/DR−cells.39 The role of c-kit ligand (KL) in promoting NK progenitor proliferation and the synergistic role of Flt3 ligand (FL) for increasing NK progenitor cloning frequency has been established in vitro40,41 and in vivo.42 The addition of these and other cytokines (eg, IL-7, IL-15) can obviate the absolute requirement for stroma to induce NK cell differentiation.36,38 43
In mice, the ability of stroma to induce differentiation is, at least in part, regulated by the transcription factor interferon-regulatory factor-1 transcription of IL-15.44 In human studies, IL-15 made by stroma and macrophages plays a role in NK development and survival by interaction with components of the IL-2 receptor.38,45 NK cell differentiation from progenitors requires primitive-acting factors like FL, KL, and IL-3,34,40 and the need for lymphoid factors likely occurs later and may coincide with IL-2/IL-15 receptor β (CD122) expression on NK cell progenitors.43
Despite progress in defining the role of cytokines in NK cell ontogeny, differentiation of single human progenitors into NK cells was not possible without murine stromal feeders.46 We have extensively studied a murine cell line (derived from fetal liver) called AFT024 initially described by Moore et al.47 AFT024 and defined cytokines differentiate adult marrow CD34+/Lin−/CD38− progenitors into NK cells, B-lineage cells, and dendritic cells,34 without loss of myeloid-differentiating capacity and while maintaining CD34+ progenitors.48 AFT024 differentiates progenitors into NK cells with a similar phenotype and functional capacity as progenitors that differentiate into NK cells on primary human stroma. In addition, AFT024 is a homogeneous cell line and is more efficient than human stroma on single progenitors; because of its murine origin, the possible confounding variable of human stroma presenting 6 different class I MHC alleles would be avoided. We used AFT024 to induce NK cell differentiation to study CD94 and KIR acquisition during NK cell development from single human stem cells.
Materials and methods
Umbilical cord blood, normal bone marrow, and fetal liver
Umbilical cord blood (UCB) was obtained from full-term mothers from local obstetrical units or from the St Louis Cord Blood Bank (St Louis, MO). Adult bone marrow (BM) was obtained from the posterior iliac crest of normal adult donors. Mononuclear cells were obtained by Ficoll-Hypaque (specific gravity, 1.077; Sigma Diagnostics, St Louis, MO) density gradient centrifugation. Fetal liver hematopoietic progenitors were obtained from 19- to 21-weeks' gestation fetal liver. Fetal liver cells were mechanically disrupted into single cell suspensions as previous described.49 The use of all tissue was approved by the Committee on the Use of Human Subjects in Research at the University of Minnesota.
Purification of primitive progenitors
UCB, fetal liver, or BM mononuclear cells were enriched for CD34+ cells by using the MACS system as recommended by the manufacturer (Miltenyi Biotec, Oberlin, CA). Resultant cells were stained with CD34 allophycocyanin (APC; BD Biosciences, San Diego, CA), and fluorescein isothiocyanate (FITC)-conjugated antibodies against CD2, CD3, CD4, CD5, CD7, CD8, CD10, and CD19 were used for the lineage (Lin) cocktail (all from BD Biosciences). Phycoerythrin (PE)-conjugated anti-CD38 (BD Biosciences) was used as the third fluorescent color for multicolor cell separation and sorting using the fluorescence-activated cell sorter (FACS) Star Plus (Becton Dickinson, San Jose, CA). CD34+/Lin−/38−cells were sorted for limiting dilution experiments or directly into 96-well plates pre-established with AFT024 stroma as described.34
Culture of hematopoietic progenitors
CD34+/Lin−/CD38− cells were plated in 24-well plates (Costar, Cambridge, MA) with a 2:1 (vol/vol) mix of Dulbecco modified Eagle medium (DMEM) high glucose/Ham F12-based medium without stroma (Gibco Laboratories, Grand Island, NY), in direct contact with the AFT024 stromal cell line, or in a Transwell insert separating progenitors from stroma by a 0.4-μm collagen membrane (Costar) as indicated. The DMEM/F12-based medium was developed to maximize NK cell growth50 and is supplemented with 24 μM 2-mercaptoethanol, 50 μM ethanolamine, 20 mg/L ascorbic acid, 50 μg/L sodium selenite (Na2SeO3), 100 U/mL penicillin, and 100 U/mL streptomycin (Gibco). Twenty percent heat-inactivated human AB serum (North American Biologicals, Miami, FL) was used at culture initiation that was reduced to 10% for subsequent media changes. Cytokines were supplemented as indicated with 1000 U/mL IL-2 (a gift from Amgen, Thousand Oaks, CA), 10 ng/mL IL-15 (R&D Systems, Minneapolis, MN), 10 ng/mL FL (a gift from Immunex, Seattle, WA), 20 ng/mL KL (or Stem Cell Factor, a gift from Amgen), and 20 ng/mL IL-7 (R&D Systems). All cytokines were added fresh with weekly media changes except for IL-3 (R&D Systems), which was added at 5 ng/mL only once at culture initiation. Cultures were maintained in a humidified atmosphere at 37°C and 5% CO2.
Antibodies and determination of absolute cell counts
FITC, PE, peridinin chlorophyll protein, and APC coupled control immunoglobulins or specific antibodies directed at APC CD56 (BD Pharmingen, San Diego, CA), CD94 (FITC-conjugated, clone HP-3D9; BD Pharmingen; or PE-conjugated, clone HP-3B1; Beckman Coulter, Miami, FL), NKG2A (clone Z199; Beckman Coulter), CD158a (PE-conjugated, clone EB6; Beckman Coulter; or FITC-conjugated, clone HP-3E4, BD Pharmingen), CD158b (clone GL183; Beckman Coulter), and NKB1 (clone DX9; BD Bioscience) were used to evaluate progeny NK cells from differentiation cultures. Progeny of single cells and bulk cultures were harvested and divided into 3 to 4 aliquots to facilitate evaluation of 9 to 12 surface markers. Absolute cell numbers were determined by addition of 3 × 104 polystyrene microspheres (Polysciences, Warrington, PA) to each aliquot of cultured progeny. After gating out debris, absolute cell numbers were calculated, using the method described by Pribyl et al51 and by Larson and LeBien.52 The absolute number of cells per well was calculated as [(total number of beads added/well)/(number of beads collected) × (number of cells in the phenotype gate of interest) × (the number of times the initial sample was divided)]. All analyses were performed with a FACSCalibur and CELLQuest software (Becton Dickinson).
Class I HLA typing of UCB cells
In some experiments, the CD34− fraction was collected from the CD34+ enrichment step and cryopreserved for HLA typing. HLA typing was performed by a standard complement-dependent cytotoxicity method by using local and commercial serologic typing trays.
Statistics
Results of experimental points obtained from multiple experiments were reported as mean ± 1 SEM. Significance levels were determined by 2-sided Student t test analysis.
Results
UCB CD34+/Lin−/CD38−progenitors efficiently differentiate into NK cells in vitro
We have recently shown that the murine fetal liver cell line, AFT024, and exogenous cytokines support the differentiation of single adult marrow progenitors (CD34+/Lin−/CD38−) along the NK cell lineage.
Primitive-acting cytokines (FL, KL, and IL-3) and lymphocyte-stimulating cytokines (IL-7 and IL-2) were required for NK cell differentiation of adult BM progenitors. Because the cytokine responsiveness of UCB cells may be different from that of adult BM,53 in a first set of experiments, UCB CD34+/Lin−/CD38− cells were cultured in limiting dilution to assess their capacity to differentiate along the NK cell lineage. UCB CD34+/Lin−/CD38− cells cultured on AFT024 with cytokines (FL, KL, IL-3, IL-7, and IL-2) resulted in an NK cell cloning frequency of 15.4% ± 4.6% (n = 4), 3 times higher than that previously found from adult BM progenitors cultured under the same conditions.34 All NK cells throughout this study were phenotypically defined as CD56+/CD3−. After 25 to 36 days of culture, 130 to 160 UCB CD34+/Lin−/CD38−cells resulted in 1.9 ± 0.3 × 105 NK cells (n = 48 replicates from 3 donors) compared with 0.27 ± 0.04 × 105 NK cells from 170 to 240 adult BM CD34+/Lin−/CD38− cells (n = 24 replicates from 2 donors; P < .001). Culture of single UCB CD34+/Lin−/CD38− cells for 50 days followed by 14 days' expansion in IL-2 alone resulted in 5.4 ± 0.9 × 106 [1.92-10.5 × 106] functional NK cells (compared with 8 × 105 from BM) that were cytotoxic against K562 targets (n = 7; data not shown). Therefore, because NK cell differentiation and proliferation from UCB progenitors was more efficient than from adult BM progenitors, UCB progenitors were used to study the ontogeny of class I–recognizing CD94 and immunoglobulinlike receptors on NK cells that develop from human stem cells.
IL-15 or IL-2 and direct contact with AFT024 ligands are optimal for lectin receptor and KIR acquisition
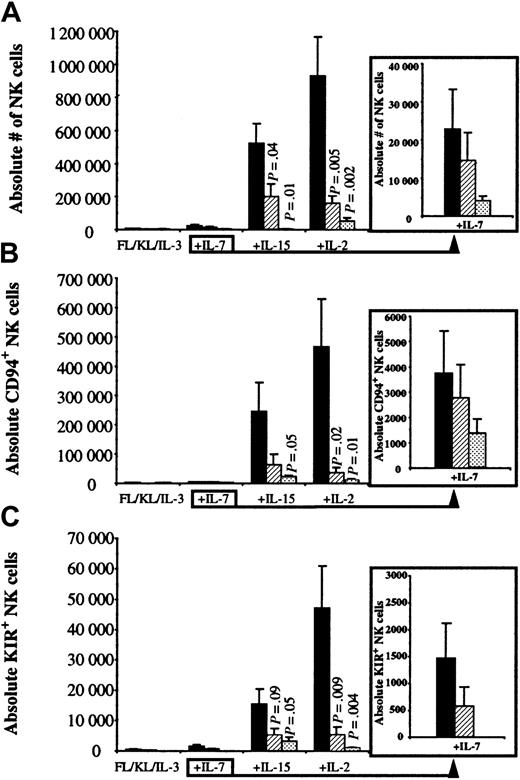
To determine the importance of factors produced by the AFT024 feeder, 300 UCB CD34+/Lin−/CD38−cells were cultured in direct contact with AFT024 (Contact), separated from AFT024 by a microporous Transwell (TW) membrane, or in the absence of AFT024 (No AFT024). All cultures contained FL, KL, and IL-3 with or without the addition of other cytokines (Figure1). In the absence of IL-7, IL-15, or IL-2, the absolute number of NK cells derived from 300 UCB progenitors was less than 6000 and not different for any of the AFT024 conditions (Contact, TW, or No AFT024). Addition of IL-7 to FL, KL, and IL-3 resulted in a small increase in NK cell differentiation. In contrast, addition of IL-15 or IL-2 to FL, KL, and IL-3 resulted in a marked increase in NK cell differentiation, which for each cytokine was greatest when UCB progenitors were cultured in direct contact with AFT024 ligands (Figure 1A).
UCB primitive progenitors differentiate into CD94+/KIR+ NK cells when cultured in direct contact with AFT024.
UCB CD34+/Lin−/CD38− cells (300 per well) were cultured in direct contact with AFT024 (▪), separated from the feeder by a Transwell membrane (▨), or in cytokines alone in the absence of AFT024 (░). All cultures were supplemented with weekly FL in combination with KL alone, or with the addition of a lymphocyte-stimulating cytokine (IL-7, IL-15, or IL-2). IL-3 was only added once at culture initiation. After 30 to 34 days, cell progeny were harvested and evaluated for the absolute number of CD56+, CD56+/CD94+, and CD56+/KIR+ NK cells. (A) The absolute number of NK cells was greatest in the presence of IL-15 or IL-2 when progenitors were cultured in direct contact with AFT024. Under this condition, NK cells accounted for more overall cell growth after the addition of IL-15 (49% ± 5.6%) or IL-2 (56% ± 7.3%) compared with the addition of IL-7 (1.6% ± 0.6%) or in the absence of a lymphocyte-stimulating cytokine (0.6% ± 0.2%). (B) The absolute number of NK cells expressing CD94 (clone HP-3B1) accounted for 38% ± 9.1% of total NK cells for IL-15 and 49% ± 8.5% for IL-2 cultures in direct contact with AFT024. (C) The absolute number of NK cells expressing at least one of the known KIRs (using a PE-cocktail of 3 monoclonal antibodies, DX9, GL183, and EB6) accounted for 2.8% ± 0.8% of total NK cells for IL-15 and 4.9% ± 0.8% for IL-2 cultures in direct contact with AFT024. There were no (FL, KL, IL-3 ± IL-7) or few (+IL-15 or +IL-2) KIR-expressing NK cells in cultures containing cytokines alone in the absence of AFT024.P values are listed for significant differences between progenitors separated from stroma or with cytokines alone compared with NK cell differentiation in contact with AFT024. Each bar represents the mean ± SEM from 6 to 9 UCB progenitor populations sorted for each condition. The insert to the right of the graph shows IL-7 data with a smaller scale to show differences.
UCB primitive progenitors differentiate into CD94+/KIR+ NK cells when cultured in direct contact with AFT024.
UCB CD34+/Lin−/CD38− cells (300 per well) were cultured in direct contact with AFT024 (▪), separated from the feeder by a Transwell membrane (▨), or in cytokines alone in the absence of AFT024 (░). All cultures were supplemented with weekly FL in combination with KL alone, or with the addition of a lymphocyte-stimulating cytokine (IL-7, IL-15, or IL-2). IL-3 was only added once at culture initiation. After 30 to 34 days, cell progeny were harvested and evaluated for the absolute number of CD56+, CD56+/CD94+, and CD56+/KIR+ NK cells. (A) The absolute number of NK cells was greatest in the presence of IL-15 or IL-2 when progenitors were cultured in direct contact with AFT024. Under this condition, NK cells accounted for more overall cell growth after the addition of IL-15 (49% ± 5.6%) or IL-2 (56% ± 7.3%) compared with the addition of IL-7 (1.6% ± 0.6%) or in the absence of a lymphocyte-stimulating cytokine (0.6% ± 0.2%). (B) The absolute number of NK cells expressing CD94 (clone HP-3B1) accounted for 38% ± 9.1% of total NK cells for IL-15 and 49% ± 8.5% for IL-2 cultures in direct contact with AFT024. (C) The absolute number of NK cells expressing at least one of the known KIRs (using a PE-cocktail of 3 monoclonal antibodies, DX9, GL183, and EB6) accounted for 2.8% ± 0.8% of total NK cells for IL-15 and 4.9% ± 0.8% for IL-2 cultures in direct contact with AFT024. There were no (FL, KL, IL-3 ± IL-7) or few (+IL-15 or +IL-2) KIR-expressing NK cells in cultures containing cytokines alone in the absence of AFT024.P values are listed for significant differences between progenitors separated from stroma or with cytokines alone compared with NK cell differentiation in contact with AFT024. Each bar represents the mean ± SEM from 6 to 9 UCB progenitor populations sorted for each condition. The insert to the right of the graph shows IL-7 data with a smaller scale to show differences.
Each of the above culture conditions was then analyzed for the absolute number of NK cells expressing either CD94 (antibody clone HP-3B1) or a KIR (using a PE-combined cocktail of antibodies, DX9, GL183, and EB6). Both CD94- and KIR-expressing NK cells (Table1 for KIR antibody and specificity) were low and independent of AFT024 conditions when 300 UCB CD34+/Lin−/CD38− cells were cultured with FL, KL, and IL-3 alone or with the addition of IL-7. However, addition of IL-15 or IL-2 increased CD94+ (Figure1B) and KIR+ (Figure 1C) NK cells greatest when progenitors were cultured in direct contact with AFT024 ligands compared with when contact was prohibited or the same cytokines were used in the absence of AFT024. These data show that (1) IL-15 or IL-2 in AFT024 cultures is important for CD94 and KIR acquisition on developing NK cells and (2) AFT024 contains contact-dependent ligands important for supporting NK cell differentiation and proliferation as well as CD94 and KIR acquisition.
Antibodies to human natural killer cell receptors
| Clone . | Specificity . | Other designation . |
|---|---|---|
| DX9 | KIR3DL1 | NKB1 |
| GL183 | KIR2DL2/L3/S2 | CD158b |
| EB6 | KIR2DL1/S1 | CD158a |
| HP-3E4 | KIR2DL1/S1/S4 | CD158a |
| Clone . | Specificity . | Other designation . |
|---|---|---|
| DX9 | KIR3DL1 | NKB1 |
| GL183 | KIR2DL2/L3/S2 | CD158b |
| EB6 | KIR2DL1/S1 | CD158a |
| HP-3E4 | KIR2DL1/S1/S4 | CD158a |
IL-15, IL-2, or IL-7 is needed for NK cell long-term proliferation, survival, and receptor acquisition
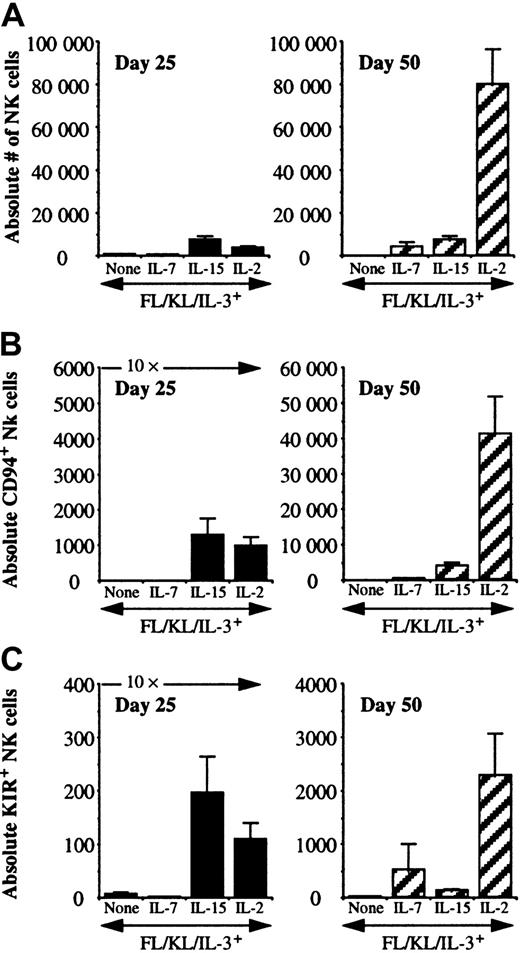
To further define the kinetics of cytokines on NK differentiation from progenitors, UCB CD34+/Lin−/CD38− cells were cultured in direct contact with AFT024 ligands with FL, KL, and IL-3 alone, and with the addition of IL-7, IL-15, or IL-2 for 25 or 50 days. At 25 days, the presence of NK cell differentiation was detected under all conditions, including when FL, KL, and IL-3 were used alone in the absence of a lymphocyte-stimulating cytokine, suggesting that NK cell differentiation can occur on AFT024, albeit with low proliferation (Table 2). These few NK cells that differentiate without IL-7, IL-15, or IL-2 were predominantly CD94− (86% of cultures) and all were KIR−. However, by day 50, NK cells were no longer present in 72% of cultures in the absence of IL-7, IL-15, or IL-2. IL-7 alone was sufficient to induce acquisition of CD94 (75% to 89% of cultures positive) but poorly supported KIR acquisition as shown by the 48% positive cultures at day 25 and only 18% positive cultures at day 50. A majority (> 70%) of cultures containing either IL-15 or IL-2 were positive for both CD94 and at least one KIR at either time point.
Frequency of cultures with natural killer cell growth and receptor expression
| FL, KL, IL-3+ . | NK growth* . | CD94+ NK cells† . | KIR+ NK cells† . | |||
|---|---|---|---|---|---|---|
| Day 25 (25-100 starting cells) (%) . | Day 50 (10-25 starting cells) (%) . | Day 25 (%) . | Day 50 (%) . | Day 25 (%) . | Day 50 (%) . | |
| None | 43/46 (93) | 9/32 (28) | 14 | ND | 0 | ND |
| IL-7 | 45/46 (98) | 28/32 (88) | 75 | 89 | 48 | 18 |
| IL-15 | 29/29 (100) | 28/32 (88) | 100 | 96 | 72 | 89 |
| IL-7 + IL-15 | 29/29 (100) | 26/28 (93) | 100 | 96 | 76 | 85 |
| IL-2 | 45/45 (100) | 35/40 (88) | 100 | 95 | 64 | 91 |
| IL-7 + IL-2 | 46/46 (100) | 29/40 (98) | 98 | 92 | 72 | 82 |
| FL, KL, IL-3+ . | NK growth* . | CD94+ NK cells† . | KIR+ NK cells† . | |||
|---|---|---|---|---|---|---|
| Day 25 (25-100 starting cells) (%) . | Day 50 (10-25 starting cells) (%) . | Day 25 (%) . | Day 50 (%) . | Day 25 (%) . | Day 50 (%) . | |
| None | 43/46 (93) | 9/32 (28) | 14 | ND | 0 | ND |
| IL-7 | 45/46 (98) | 28/32 (88) | 75 | 89 | 48 | 18 |
| IL-15 | 29/29 (100) | 28/32 (88) | 100 | 96 | 72 | 89 |
| IL-7 + IL-15 | 29/29 (100) | 26/28 (93) | 100 | 96 | 76 | 85 |
| IL-2 | 45/45 (100) | 35/40 (88) | 100 | 95 | 64 | 91 |
| IL-7 + IL-2 | 46/46 (100) | 29/40 (98) | 98 | 92 | 72 | 82 |
All cultures were initiated with CD34+/Lin−/CD38− UCB cells, AFT024, FL, KL, IL-3, and the indicated cytokines (n = 3-5 individual UCB for each condition with multiple replicates).
NK indicates natural killer; KIR, killer immuoglobulinlike receptor; FL, Flt3 ligand; KL, c-kit ligand; IL, interleukin; ND, not done; UCB, umbilical cord blood.
The percentage with NK growth is the number of wells with CD56+ cells/replicate wells plated.
The percentage with CD94 or KIR expression is the positive/number of wells with NK cells.
There were distinct patterns of differentiation and proliferation of NK cells between cytokine conditions (Figure2A). At day 18, there were less than 500 absolute NK cells per 10 starting progenitors under all culture conditions (data not shown). Culture of 10 UCB CD34+/Lin−/CD38− cells for 25 days resulted in similar proliferation for IL-15– or IL-2–containing cultures (3500-7500 NK cells), which was greater than when progenitors were cultured in the absence of IL-15, IL-2, and IL-7 or with IL-7 alone (300-500 NK cells). NK cell differences at day 25 were not accounted for simply by cell growth, as there were between 2 to 3 × 104 total cells present under all cytokine conditions. At day 25, 12% to 24% of total cells were NK cells when IL-15 or IL-2 were included in culture and only 2% to 4% in the absence of IL-15 or IL-2, with or without IL-7. NK cell numbers dropped between day 25 and 50 in the absence of any lymphocyte-stimulating cytokine and remained relatively constant in the presence of IL-7 or IL-15. IL-2–containing cultures exhibited substantial proliferation of NK cells between culture day 25 (3611 ± 856 NK cells per 10 starting cells plated; n = 45) and day 50 (79 779 ± 16 450 NK cells per 10 starting cells plated; n = 35; P < .001). Both CD94 and KIR acquisition were cytokine and time dependent (Figure2B,C). In the absence of IL-7, IL-15, or IL-2, essentially no NK cells expressed CD94 or KIR. IL-7 was a weak inducer of CD94 or KIR after short culture intervals. At 25 days, IL-15 and IL-2 were similar in their ability to induce both receptor families (absolute number and percentage of total NK cells). In late cultures (50 days), IL-2 induced the highest absolute number of NK cells, the highest number of CD94+ NK cells, and the highest number of KIR+NK cells. Proliferation correlated with the absolute number of CD94+ and KIR+ NK cells between conditions and individual cultures (data not shown). In IL-2– or IL-15–containing cultures at day 25, an average of 25% (3%-87%) of NK cells expressed CD94 and 2% (0.2%-30%) expressed KIR. Under the same conditions at day 50, CD94 expression significantly increased to 50% (4%-97%) and 4% (0.2%-43%) expressed KIR.
IL-7, IL-15, and IL-2 differ in their capacity to support early and late NK cell differentiation and receptor acquisition.
UCB CD34+/Lin−/CD38− cells were cultured in direct contact with AFT024 and the indicated cytokines. All results are presented normalized to 10 starting cells. Cultures analyzed at day 25 were started with 25 to 100 sorted cells, and cultures analyzed at day 50 were started with 10 to 25 cells to prevent culture overgrowth. (A) There were significantly greater numbers of NK cells at day 50 with IL-2 compared with IL-15 (P < .001). There was no significant difference between these same cytokines at the early time point. (B) The absolute number of CD56+/CD94+ NK cells was determined. The absolute number of CD94-expressing NK cells at day 25 was greater for cultures containing IL-15 or IL-2, but by day 50 cultures containing IL-2 contained significantly greater CD56+/CD94+ NK cells compared with IL-15 (P = .0009). (C) The absolute number of CD56+/KIR+ NK cells was determined by using a KIR cocktail of antibodies as in Figure 1. The absolute number of KIR-expressing NK cells at day 25 was greater for cultures containing IL-15 or IL-2, but by day 50 cultures containing IL-2 contained significantly greater CD56+/KIR+ NK cells compared with IL-15 (P = .009). Each condition represents multiple replicates from 3 to 4 donors for each condition.
IL-7, IL-15, and IL-2 differ in their capacity to support early and late NK cell differentiation and receptor acquisition.
UCB CD34+/Lin−/CD38− cells were cultured in direct contact with AFT024 and the indicated cytokines. All results are presented normalized to 10 starting cells. Cultures analyzed at day 25 were started with 25 to 100 sorted cells, and cultures analyzed at day 50 were started with 10 to 25 cells to prevent culture overgrowth. (A) There were significantly greater numbers of NK cells at day 50 with IL-2 compared with IL-15 (P < .001). There was no significant difference between these same cytokines at the early time point. (B) The absolute number of CD56+/CD94+ NK cells was determined. The absolute number of CD94-expressing NK cells at day 25 was greater for cultures containing IL-15 or IL-2, but by day 50 cultures containing IL-2 contained significantly greater CD56+/CD94+ NK cells compared with IL-15 (P = .0009). (C) The absolute number of CD56+/KIR+ NK cells was determined by using a KIR cocktail of antibodies as in Figure 1. The absolute number of KIR-expressing NK cells at day 25 was greater for cultures containing IL-15 or IL-2, but by day 50 cultures containing IL-2 contained significantly greater CD56+/KIR+ NK cells compared with IL-15 (P = .009). Each condition represents multiple replicates from 3 to 4 donors for each condition.
Single UCB CD34+/Lin−/CD38−progenitors differentiate into polyclonal KIR-expressing NK cells with preferential receptor expression
In the experiments above, each of the individual KIR antibodies (Table 1) contributed to the total KIR expression (data not shown). However, because these were bulk cultures, it was still unclear whether a progenitor resulted in NK cells with clonal or polyclonal KIR- and CD94-receptor expression. To answer this question, single UCB CD34+/Lin−/CD38− cells were cultured from 2 donors with AFT024, FL, KL, IL-3, and single or combinations of lymphocyte-stimulating cytokines. After about 50 days, single cells were capable of NK cell differentiation and proliferation similar to bulk cultures. The progeny of each single cell was then analyzed for individual KIR, either KIR3DL1 (DX9, NKB1), KIR2DL1/S1 (EB6, CD158a), or KIR2DL2/L3/S2 (GL183, CD158b). All single UCB progenitors resulted in NK cells with polyclonal KIR expression, suggesting that KIR fate is determined at a maturational stage beyond a CD34+/Lin−/CD38− cell. Cultures containing IL-2 + IL-7 resulted in the greatest number of KIR-expressing NK cells (data not shown). Therefore, IL-7, IL-7 + IL-15, and IL-7 + IL-2 were studied further to determine the frequency of individual KIR expression from single hematopoietic stem cell populations.
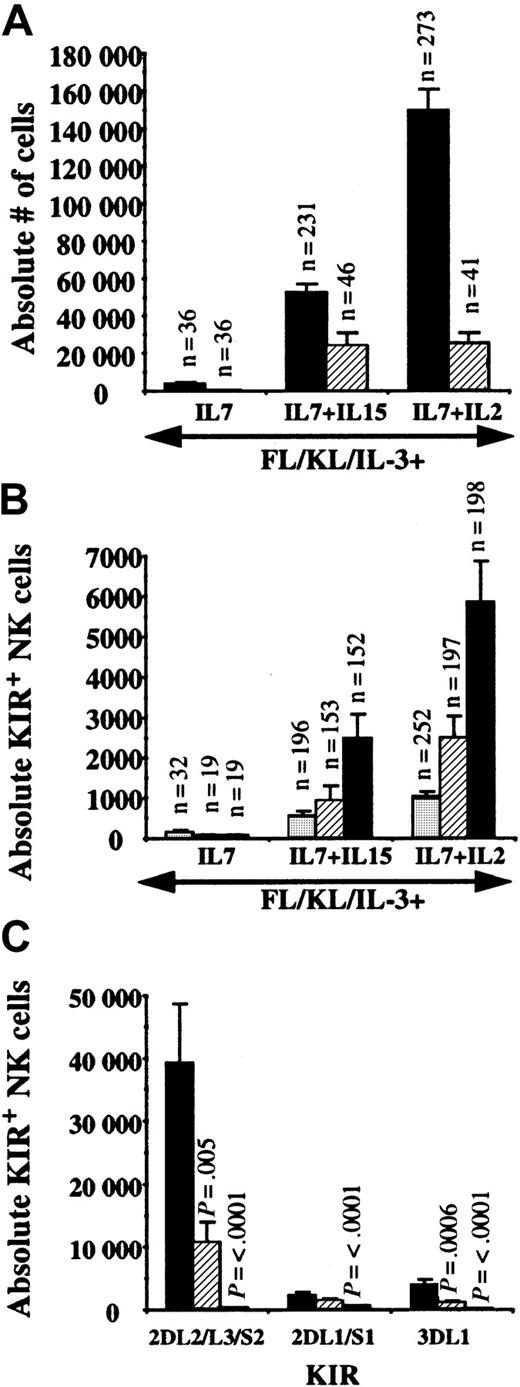
Single UCB CD34+/Lin−/CD38− cells were sorted and progeny were harvested after 26 to 36 days. NK cells were not detected from single cells in the absence of a lymphocyte-stimulating cytokine (data not shown). The cloning frequency from single cells with FL, KL, IL-3, and IL-7 was 7% and increased to 20% when IL-15 or IL-2 was added (Table3). Individual KIRs were found on 25% to 32% of cultured single cell progeny with FL, KL, IL-3, and IL-7 and 51% to 78% of single cell progeny cultured with the addition of IL-15 or IL-2. After analysis of more than 400 clones, no progeny of a single cell was ever found with clonal KIR expression (Figure3A-E). A single UCB progenitor gave rise to 149 020 ± 11 801 NK cells with IL-7 + IL-2 (n = 273) that was greater than the 42 247 ± 4730 NK cells with IL-7 + IL-15 (n = 231; P < .001) (Figure4A). The number of CD94+ NK cells was not different between IL-7 + IL-15 and IL-7 + IL-2. Most CD94+ cells specifically heterodimerized with NKG2A (99.1% ± 0.2% for IL-2– [n = 134] and IL-15–containing [n = 111] cultures) as detected by costaining with the Z199 antibody. However, other CD94+/NKG2A−receptors (0.8% ± 0.2%) were also represented (Figure 3F). Single cells cultured with IL-7 + IL-2 resulted in significantly greater numbers of NK cells expressing individual KIRs (approximately double) than IL-15–containing cultures (Figure 4B), and NK cell progeny expressed an ordered frequency of KIR (KIR2DL2/L3/S2 > KIR2DL1/S1 ≥ KIR3DL1) with either cytokine combination. Although EB6 and DX9 staining were similar in some individual experiments, GL183 always detected a higher percentage of single cell–derived NK progeny than EB6 or DX9. A majority of KIR+ NK cells also expressed CD94 (98% ± 0.2% for IL-2– [n = 17] and 99% ± 0.5% for IL-15–containing [n = 111] cultures) but rare KIR+/CD94− cells were found in some cultures (Figure 3G). To understand the kinetics of KIR expression, CD56+/GL183+/DX9− cells were sorted from primary bulk cultures then replated into secondary culture with fresh AFT024. Essentially all NK cells remained GL183+, suggesting that KIR expression is stable. In addition, a significant number gained DX9, showing that KIR expression is dynamic and new receptors can be acquired at the level of a committed CD56+ cell (Figure 3H).
Frequency of single cells with natural killer cell progeny expressing CD94 or killer immunoglobulinlike receptor
| FL, KL, IL-3+ . | n . | Cloning frequency (%) . | CD94 (%) . | KIR3DL1 (%) . | KIR2DL1/S1 (%) . | KIR2DL2/L3/S2 (%) . |
|---|---|---|---|---|---|---|
| IL-7 | 4 | 7 | 5/12 (42) | 8/32 (25) | 5/19 (26) | 6/19 (32) |
| IL-7 + IL-15 | 10 | 22 | 46/46 (100) | 100/196 (51) | 80/153 (52) | 108/152 (72) |
| IL-7 + IL-2 | 11 | 19 | 38/41 (93) | 144/252 (57) | 141/197 (71) | 155/198 (78) |
| FL, KL, IL-3+ . | n . | Cloning frequency (%) . | CD94 (%) . | KIR3DL1 (%) . | KIR2DL1/S1 (%) . | KIR2DL2/L3/S2 (%) . |
|---|---|---|---|---|---|---|
| IL-7 | 4 | 7 | 5/12 (42) | 8/32 (25) | 5/19 (26) | 6/19 (32) |
| IL-7 + IL-15 | 10 | 22 | 46/46 (100) | 100/196 (51) | 80/153 (52) | 108/152 (72) |
| IL-7 + IL-2 | 11 | 19 | 38/41 (93) | 144/252 (57) | 141/197 (71) | 155/198 (78) |
All cultures were initiated with UCB CD34+Lin−CD38− cells, AFT024, FL, KL, IL-3, and the indicated cytokines (n = the number of individual UCB). Results are from cultures harvested after 26-36 days. The cloning frequency is the number of wells with NK-cell progency divided by the total number of single cells plated. For CD94 and KIR expression, the denominator is the total number of NK-cell populations evaluated.
For abbreviations, see Table 2.
Single UCB primitive progenitors give rise to polyclonal NK cells.
(A-E) These panels (gated on a lymphocyte window) show progeny of single UCB CD34+/Lin−/CD38− cells cultured on AFT024 with FL, KL, IL-3, IL-7, and IL-2 and then stained with antibodies for each of the individual KIR receptors as indicated. Panels A-C were derived from the same single cell with class I HLA type: A1, A2, B8, B62,15 Bw6, Bw6, Cw7, and Cw3 (lacks ligand for NKB1 and CD158a). Panels D and F were from nontyped donors, showing representative NKB1 and CD158a staining patterns. Panel F shows single cell progeny (gated on CD56+ cells), showing the coexpression of CD94 and NKG2A. This example was chosen to show the CD94+/NKG2A− population, although most NK cell progeny derived from a single cell showed less CD94+/NKG2A− cells. Panel G demonstrates single cell progeny (gated on CD56+ cells), showing the coexpression of CD94 and a KIR cocktail of the 3 known antibodies. This example was chosen to highlight the CD94−/KIR+population, although most NK cell progeny derived from a single cell showed less CD94−/KIR+ cells. Panel H shows the progeny of a 4-week secondary culture initiated with CD158b+/NKB1− cells after 42 days of primary culture-initiated UCB CD34+/Lin−/CD38− cells. This example shows that CD158b expression was stable and new KIRs were acquired in secondary culture. Panel I (gated on CD56+cells) shows the expression of KIR3DL1 (NKB1) and KIR2DL2/L3/S2 (CD158b) from a single UCB primitive progenitor. The UCB used for this example showed the following class I HLA type: A1, A3, B35, B35, Bw6, Bw6, and Cw4 (lacks ligand for NKB1 and CD158b). This example also shows the polyclonal nature of KIR acquisition and the expression of one or 2 specific KIRs on each NK cell.
Single UCB primitive progenitors give rise to polyclonal NK cells.
(A-E) These panels (gated on a lymphocyte window) show progeny of single UCB CD34+/Lin−/CD38− cells cultured on AFT024 with FL, KL, IL-3, IL-7, and IL-2 and then stained with antibodies for each of the individual KIR receptors as indicated. Panels A-C were derived from the same single cell with class I HLA type: A1, A2, B8, B62,15 Bw6, Bw6, Cw7, and Cw3 (lacks ligand for NKB1 and CD158a). Panels D and F were from nontyped donors, showing representative NKB1 and CD158a staining patterns. Panel F shows single cell progeny (gated on CD56+ cells), showing the coexpression of CD94 and NKG2A. This example was chosen to show the CD94+/NKG2A− population, although most NK cell progeny derived from a single cell showed less CD94+/NKG2A− cells. Panel G demonstrates single cell progeny (gated on CD56+ cells), showing the coexpression of CD94 and a KIR cocktail of the 3 known antibodies. This example was chosen to highlight the CD94−/KIR+population, although most NK cell progeny derived from a single cell showed less CD94−/KIR+ cells. Panel H shows the progeny of a 4-week secondary culture initiated with CD158b+/NKB1− cells after 42 days of primary culture-initiated UCB CD34+/Lin−/CD38− cells. This example shows that CD158b expression was stable and new KIRs were acquired in secondary culture. Panel I (gated on CD56+cells) shows the expression of KIR3DL1 (NKB1) and KIR2DL2/L3/S2 (CD158b) from a single UCB primitive progenitor. The UCB used for this example showed the following class I HLA type: A1, A3, B35, B35, Bw6, Bw6, and Cw4 (lacks ligand for NKB1 and CD158b). This example also shows the polyclonal nature of KIR acquisition and the expression of one or 2 specific KIRs on each NK cell.
KIR2DL2/L3/S2 is preferentially expressed on NK cells derived from single UBC progenitors.
Single UCB CD34+/Lin−/CD38−cells were cultured on AFT024 and IL-7 (n = 4 UCB donors), IL-7 + IL-15 (n = 10 UCB donors), and IL-7 + IL-2 (n = 11 UCB donors) for 26 to 36 days. The number of replicate single cell progeny analyzed for each condition is listed as the n value above each bar. Progeny of each single cell was split into 3 or 4 aliquots for immunophenotyping. (A) The absolute number of NK cells derived from a single cell was greatest for IL-2 compared with IL-15 (P < .0001). There was no difference in CD94 expression at this early time point. ▪, CD56+, and ▨, CD94+ NK cells. (B) The absolute number of NK cells expressing individual KIR was evaluated. For IL-15– and IL-2–containing cultures, GL183 was expressed greater than EB6 (P = .03 and P = .004, respectively) or DX9 (P = .014 and P = .005, respectively) staining cells. ░, KIR3DL1; ▨, KIR2DL1/S1; ▪, KIR2DL2/L3/S2. (C) Single CD34+/Lin−/CD38− cells were sorted from fetal liver (n = 2), UCB (n = 4), and adult BM (n = 3) on AFT024 with FL, KL, IL-3, IL-7, and IL-2. After 40 to 55 days, replicates from each stem cell source were harvested, split, and stained for each of the individual KIR antibodies as indicated. AllP values listed are for significant differences compared with fetal liver–derived NK cells. Compared with other KIRs, KIR2DL2/L3/S2 expression was significantly greater for fetal liver (P = .003 compared with KIR2DL1/S1; P = .003 compared with KIR3DL1) and UCB (P = .005 compared with KIR2DL1/S1; P = .003 compared with KIR3DL1). ▪, fetal liver (n = 26); ▨, cord blood (n = 69); ■, adult marrow (n = 59).
KIR2DL2/L3/S2 is preferentially expressed on NK cells derived from single UBC progenitors.
Single UCB CD34+/Lin−/CD38−cells were cultured on AFT024 and IL-7 (n = 4 UCB donors), IL-7 + IL-15 (n = 10 UCB donors), and IL-7 + IL-2 (n = 11 UCB donors) for 26 to 36 days. The number of replicate single cell progeny analyzed for each condition is listed as the n value above each bar. Progeny of each single cell was split into 3 or 4 aliquots for immunophenotyping. (A) The absolute number of NK cells derived from a single cell was greatest for IL-2 compared with IL-15 (P < .0001). There was no difference in CD94 expression at this early time point. ▪, CD56+, and ▨, CD94+ NK cells. (B) The absolute number of NK cells expressing individual KIR was evaluated. For IL-15– and IL-2–containing cultures, GL183 was expressed greater than EB6 (P = .03 and P = .004, respectively) or DX9 (P = .014 and P = .005, respectively) staining cells. ░, KIR3DL1; ▨, KIR2DL1/S1; ▪, KIR2DL2/L3/S2. (C) Single CD34+/Lin−/CD38− cells were sorted from fetal liver (n = 2), UCB (n = 4), and adult BM (n = 3) on AFT024 with FL, KL, IL-3, IL-7, and IL-2. After 40 to 55 days, replicates from each stem cell source were harvested, split, and stained for each of the individual KIR antibodies as indicated. AllP values listed are for significant differences compared with fetal liver–derived NK cells. Compared with other KIRs, KIR2DL2/L3/S2 expression was significantly greater for fetal liver (P = .003 compared with KIR2DL1/S1; P = .003 compared with KIR3DL1) and UCB (P = .005 compared with KIR2DL1/S1; P = .003 compared with KIR3DL1). ▪, fetal liver (n = 26); ▨, cord blood (n = 69); ■, adult marrow (n = 59).
NK cells express KIR irrespective of progenitor HLA type
We designed experiments to address whether the HLA type of starting CD34+/Lin−/CD38− cells influenced KIR expression on developing NK cells (eg, would expression of bw4 lead to more or less KIR3DL1 expression). The CD34−fraction of 6 UCB populations was serologically typed for class I MHC to assign the presence or absence of known KIR ligands of corresponding CD34+/Lin−/CD38− cells. Single progenitors were then sorted onto AFT024 and cytokines maximal for inducing NK cell differentiation. Progeny were analyzed for KIR. For these experiments, CD158a was detected by using HP-3E4, which stained similar to EB6. Both recognize KIR2DL1/S1, but recently HP-3E4 has been reported to also recognize KIR2DS4 (Assigned at Sixth International Leukocyte Typing Workshop,www.ncbi.nlm.nih.gov/prow/guide/682_373_991_g.htm). Twenty-nine percent of single progenitors that were heterozygous or homozygous for bw4 gave rise to NK cells expressing KIR3DL1. Similarly, of progenitors that lacked bw4 (those that were homozygous bw6), 28% of NK cell progeny expressed KIR3DL1. There was no difference in the absolute number of KIR3DL1-expressing NK cells from single progenitors with or without bw4. Similar results were seen when comparing starting progenitors with or without a Cw4 or Cw3 group ligand (Table4 and Figure 3I).
The HLA type of the progenitor source does not determine killer immunoglobulinlike receptor expression
| HLA allele4-150 . | KIR mab4-151 . | NK progeny with KIR from HSC with allele‡ (%) . | NK progeny with KIR from HSC without allele‡ (%) . | Absolute number KIR+ NK from HSC with allele4-153 . | Absolute number KIR+ NK from HSC without allele4-153 . |
|---|---|---|---|---|---|
| Bw4 | DX9 | 12/41 (29) | 22/80 (28) | 874 ± 287 | 567 ± 86 |
| Cw4 | HP-3E4 | 12/36 (33) | 57/134 (43) | 488 ± 108 | 1180 ± 197 |
| Cw3 | GL183 | 94/103 (75) | 33/46 (72) | 9671 ± 1929 | 7010 ± 2773 |
| HLA allele4-150 . | KIR mab4-151 . | NK progeny with KIR from HSC with allele‡ (%) . | NK progeny with KIR from HSC without allele‡ (%) . | Absolute number KIR+ NK from HSC with allele4-153 . | Absolute number KIR+ NK from HSC without allele4-153 . |
|---|---|---|---|---|---|
| Bw4 | DX9 | 12/41 (29) | 22/80 (28) | 874 ± 287 | 567 ± 86 |
| Cw4 | HP-3E4 | 12/36 (33) | 57/134 (43) | 488 ± 108 | 1180 ± 197 |
| Cw3 | GL183 | 94/103 (75) | 33/46 (72) | 9671 ± 1929 | 7010 ± 2773 |
mab indicates monoclonal antibody; HSC, hematopoietic stem cell; MHC, major histocompatibility complex; for other abbreviations, see Table 2.
Based on serologic HLA typing of 6 UCB units.
Monoclonal antibody for KIR that recognizes respective class I MHC allele.
Single CD34+/Lin−/CD38− HSCs were sorted from UCB units and data are grouped based on the HLA type of the HSC source. All cultures were initiated with single cells and contained AFT024, FL, KL, IL-3, IL-7, and IL-2. Shown is the number of single-cell NK-cell progeny expressing the KIR that recognize the respective class I ligand (number positive/number analyzed [percent positive]) based on the HSC source with or without the class I MHC KIR ligand of interest (Bw4, Cw4 group, Cw3 group).
The absolute number of NK cells derived from a single HSC expressing the KIR that recognizes the respective class I ligand was calculated. The numerator of the respective previous columns is the number of replicates that contributed to each absolute NK cell number.
CD34+/Lin−/CD38− progenitors from sources earlier in ontogeny differentiate into higher numbers of polyclonal, KIR-expressing NK cells
Last, we questioned the role of the stem cell source and whether hematopoietic cells from earlier in ontogeny would acquire KIR with the same frequency and pattern. Single CD34+/Lin−/CD38− cells were sorted from fetal liver, UCB, and adult BM and cultured on AFT024 with FL, KL, IL-3, IL-7, and IL-2 for 40 to 55 days. Proliferation of NK cells was greatest with starting cells earlier in ontogeny (fetal liver > UCB > adult BM). The absolute number of NK cells derived from a single CD34+/Lin−/CD38− cell was 437 260 ± 54 721 from fetal liver (n = 26), 223 030 ± 20 589 from UCB (n = 144), and 15 584 ± 5923 from adult BM (n = 59). Like UCB, fetal liver–derived NK cells expressed KIR2DL2/L3/S2 with a significantly higher frequency than KIR2DL1/S1 or KIR3DL1 (Figure 4C). The ratio of the average number of GL183+ NK cells over the average number of DX9+NK cells was 10.3 for fetal liver, 9.5 for UCB, and 1.9 for adult BM. The preferential expression of KIR2DL2/L3/S2 on fetal liver–derived NK cells was similar to that seen with UCB, and both were more pronounced than with adult BM.
Discussion
A novel murine fetal liver–derived stromal feeder (AFT024) was used to study NK cell differentiation and the ontogeny of human NK cell receptors. Using this feeder, CD94 and KIR acquisition were optimal when progenitors were in direct contact with AFT024 stromal ligands. Although receptor expression was dependent on exogenous cytokines, cytokines alone in the absence of AFT024 were inefficient in supporting NK differentiation and receptor expression. Soluble factors released by AFT024 only partially restored differentiation along the NK cell lineage and NK cell receptor acquisition. CD94 expression occurred earlier and at a greater frequency than KIR. Both were time dependent with few receptors expressed during short culture intervals. AFT024 supplemented with FL, KL, and IL-3 alone supported a small number of CD56+ NK cells, but these NK cells lacked CD94 and KIR and did not survive long term in the absence of a lymphocyte-stimulating cytokine. Direct comparisons between IL-7, IL-15, and IL-2 show that IL-7 alone is a weak inducer of CD94+ and KIR+NK cells from primitive progenitors. IL-15 and IL-2 are equivalent at short culture intervals and IL-2 is significantly greater at long culture intervals. However, this does not imply that IL-2 has a different capacity to support NK cell differentiation or receptor acquisition but may only reflect the relative potency of these cytokines to induce NK cell proliferation from primitive progenitors in long-term culture. These bulk culture data agree with studies using a mixed CD34+ progenitor population cultured with FL followed by IL-15, resulting in NK cells that were mostly NKG2A+ and expressed some but significantly less KIR.43
The strength of this experimental system is its efficiency to study single primitive cells. The AFT024 murine feeder is unique by efficiently differentiating enough cells to analyze multiple receptors and to calculate cloning efficiencies directly, results we were unable to achieve with primary human stromal layers.40 In addition, NK cells differentiated by AFT024 are similar to those induced by primary human stroma, making this a good model to study NK cell development. The capacity of murine stromal lines to support human NK cell and B-cell differentiation has been reported by others.46,54-56 From single CD34+/Lin−/CD38− cells from multiple cell sources, we provide definitive evidence that NK cell receptor acquisition is polyclonal for both CD94 and KIR. Like bulk cultures, the frequency of CD94+ NK cells was greater than KIR and occurred earlier, suggesting that CD94 expression precedes KIR. Although NKG2A was the dominant receptor of the CD94+ NK cells, distinct populations of CD94+/NKG2A−cells were found. Lectin receptors such as NKG2D, which may not heterodimerize with CD94,31 32 would have been missed in these experiments.
KIR acquisition was polyclonal and more than one receptor could be expressed per cell. All 3 specific KIRs tested were represented in NK cell progeny from single primitive progenitors. These results are distinctly different from IL-15 containing stroma-free cultures using thymic progenitors that differentiated into CD94+/NKG2A+ NK cells but could not express KIR as detected by immunofluorescence or reverse transcriptase–polymerase chain reaction.57 Our data fit with the cumulative and successive expression of Ly49 receptors in a stroma-dependent system in the mouse.58 The capacity to express KIR in vitro increased as the ontogeny of the stem cell source was more immature. Fetal liver CD34+/Lin−/CD38−cells differentiated into a larger number of KIR+ NK cells than UCB, and adult BM progenitors of the same phenotype were the least conducive to KIR acquisition. However, these differences likely reflect the relative proliferation capacities of the respective cell sources rather than inherent differences in mechanisms of NK cell receptor acquisition. Although we hypothesize that CD94 expression generally precedes KIR, distinct small populations of KIR+/CD94− could be found, suggesting that the sequence between CD94 and KIR is not absolute. Alternatively, because receptor expression generally increased over time, we cannot exclude the possibility that CD94 was lost in culture under the conditions tested. CD158b+ (clone GL183) NK cells were always detected at a greater frequency than other receptors tested. Because this antibody recognizes the extracellular surface domain of KIR2DL2, KIR2DL3, and KIR2DS2, one could argue that the high frequency was solely due to the net sum of 3 individual KIR expressed stocastically, at equal frequency. However, GL183+ NK cells were detected on average at a 10-fold higher frequency compared with EB6+ or DX9+ NK cells from UCB and fetal liver stem cell sources in which all KIRs were assessed from each progenitor. In addition, individual NK cell progeny derived from a single cell (example Figure 3A-C) often resulted in a more than 25-fold increase in GL183+ NK cells compared with other KIRs tested, further supporting the nonrandom preferential expression of KIR2DL2/L3/S2 in vitro.
The expression of KIR did not correlate with the HLA type of the progenitor source. This was most definitively shown with class I typing for the mutually exclusive epitopes, Bw4 and Bw6. There was no difference in the number of single cells giving rise to KIR3DL1+ NK cell progeny or the absolute number of KIR3DL1+ NK cells from progenitors that were homozygous Bw6 (lacking Bw4) compared with progenitors containing at least one Bw4-containing allele. The same was true on the basis of serologic typing for the Cw group alleles. This finding agrees with those found in vivo according to a population study of more than 200 normal donors. Normal donors who are homozygous Bw6 still express KIR3DL1 on their circulating NK cells and there was no dose effect based on the presence of one or 2 Bw4 alleles, although studies of family members and twins showed some genetic linkage independent of HLA type.59 Taken together, our data and those published studies show that self alleles do not determine KIR acquisition. However, because all NK cells have at least one receptor recognizing self ligands, it is not yet known how class I alleles educate NK cells to ensure that appropriate receptors are selected.
There has been more progress in the mouse to investigate what determines NK cell receptor repertoires (reviewed in Raulet et al60 and Hoglund et al61). Two major hypotheses have been incorporated into models explaining the education process of NK cells during development. The first hypothesis is that each NK cell must have at least one self-specific NK cell inhibitory receptor to silence auto-aggressive NK cells. The second presumption is that multiple self-specific NK cell receptors are disfavored by the education process so that NK cells will maintain the ability to discriminate between single allelic losses possibly induced by viral infection or malignant transformation. Having too many NK inhibitory receptors would decrease the sensitivity of NK cells to distinguish these single allelic losses. Experiments using class I–deficient mice have been instructive in supporting these hypotheses.62Class I expression is not required for expression of NK cell inhibitory receptors, and β2-microblobulin deficient mice (β2m−) showed no auto-aggressive NK cell compartment. This finding suggests that encountering class I was not required for receptor acquisition. Rather, class I in the environment can induce several changes in Ly49 receptor expression and function. The number of Ly49 receptors per NK cell was higher in β2m− knockout mice compared with β2m+ littermates, suggesting that encountering class I may shut off new receptor acquisition.63 Further evidence comes from transgenic mice that express Ly49A on all NK cells.64 NK cells from these mice exhibit a substantial reduction in usage of endogenous Ly49A and other Ly49 molecules. Other data come from mice expressing H2-Dd in which the frequency of NK cells expressing Ly49A (the receptor for Dd) was not markedly different from control mice. However, the level of Ly49A per cell was lower than in β2m− mice, and the ability to discriminate targets expressing Dd was altered, suggesting that the sensitivity of receptor function may be altered in the education process. These studies support the notion that class I MHC in development may influence both inhibitory receptor usage and receptor function. New studies by Roth et al,58 testing different class I alleles in stroma, show subsequent effects on developing NK cell receptor repertoires in the mouse. The fact that our stromal feeder was murine in origin, therefore, acting as a β2m−feeder for human class I alleles, may be important for the level of NK cell receptor expression shown in our study. Transduction of AFT024 with human β2m and single class I alleles is planned to further test this hypothesis.
The control of NK cell receptor expression may be determined by specific transcription factors that target NK cell receptor genes. Held et al65 have shown that, although the circulating NK cell compartment is normal in TCF-1−/− mice, there are several differences compared with the wild-type mice. Ly49A is a lectin class I–recognizing inhibitory receptor present on 20% of wild-type NK cells in mouse. Strikingly, TCF-1−/− mice develop NK cells in the spleen almost completely lacking Ly49A (1% in TCF-1 knock-out mice) despite normal total NK cell numbers. The near absence of Ly49A+ NK cells in marrow suggests that TCF-1 is affecting an early stage in Ly49A NK cell development. The effect of TCF-1 on Ly49A may be specific to this lectin receptor as no difference was found between TCF-1−/− and TCF-1+/+ mice for expression of Ly49C/I, Ly49C, Ly49D, and Ly49G2.
In summary, NK cell differentiation from single primitive human progenitors requires contact with stroma and defined human cytokines to induce CD94 and KIR on developing NK cells that is independent of progenitor HLA type. The polyclonal nature of this expression shows that receptor fate is determined beyond the maturational stage of an uncommitted CD34+/Lin−/CD38−cell, and KIR acquisition occurs after NK cell commitment (CD56 expression). For KIR, KIR2DL2/L3/S2 is preferentially expressed, which may be genetically programmed because it was seen in all experiments. Even in late cultures, most developing NK cells are still KIR− with a large population of CD94+/KIR− and CD94−/KIR− cells. These immature phenotypes may represent unique maturational stages of NK cell development from primitive, uncommitted progenitors. As the number of reagents for NK cell receptors continues to increase, the precise sequence of CD94 and KIR acquisition will continue to be defined. This model is the first to study NK cell receptor acquisition from single uncommitted hematopoietic cells, allowing for further studies of receptor acquisition in the human system.
We thank Brad Anderson for his help with flow cytometry, Dr Tucker LeBien for his helpful input and support, and Harriet Noreen for facilitating HLA typing and their interpretation.
Supported by grants R01-HL-55417 and PO1-CA-65493 from the National Institutes of Health and supported in part by grant M01-RR00400 from the National Cancer Institute for Research Resources.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Jeffrey S. Miller, University of Minnesota Cancer Center, Box 806, Division of Hematology, Oncology and Transplantation, Harvard Street at East River Road, Minneapolis, MN 55455; e-mail:mille011@tc.umn.edu.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal