Abstract
Fanconi anemia (FA) is an autosomal recessive disease with congenital anomalies, bone marrow failure, and susceptibility to leukemia. Patient cells show chromosome instability and hypersensitivity to DNA cross-linking agents. At least 8 complementation groups (A-G) have been identified and 6 FAgenes (for subtypes A, C, D2, E, F, and G) have been cloned. Increasing evidence indicates that a protein complex assembly of multiple FA proteins, including FANCA and FANCG, plays a crucial role in the FA pathway. Previously, it was reported that FANCA was phosphorylated in lymphoblasts from normal controls, whereas the phosphorylation was defective in those derived from patients with FA of multiple complementation groups. The present study examined phosphorylation of FANCA ectopically expressed in FANCA− cells. Several patient-derived mutations abrogated in vivo phosphorylation of FANCA in this system, suggesting that FANCA phosphorylation is associated with its function. In vitro phosphorylation studies indicated that a physiologic protein kinase for FANCA (FANCA-PK) forms a complex with the substrate. Furthermore, at least a part of FANCA-PK as well as phosphorylated FANCA were included in the FANCA/FANCG complex. Thus, FANCA-PK appears to be another component of the FA protein complex and may regulate function of FANCA. FANCA-PK was characterized as a cytoplasmic serine kinase sensitive to wortmannin. Identification of the protein kinase is expected to elucidate regulatory mechanisms that control the FA pathway.
Introduction
Fanconi anemia (FA) is an autosomal recessive disease characterized by congenital anomalies, progressive bone marrow failure, and susceptibility to leukemia (reviewed by Buchwald and Moustacchi1 and Garcia-Higuera and colleagues2). Cells derived from patients with FA show chromosomal instability, hypersensitivity to DNA cross-linking agents such as mitomycin C (MMC), abnormal cell cycle progression, and reduced cell survival.1,2 FA has at least 8 different complementation groups (FA-A to FA-G) as defined by cell fusion analysis.3-7 To date, 6 of the FA genes (FANCA, FANCC, FANCD2, FANCE, FANCF, and FANCG) have been cloned.7-13
The FA proteins have no functional domains, such as protein kinase (PK) and DNA helicase motifs, and no homology to each other or other known proteins. The FANCC gene, cloned by functional complementation of FA-C cells, encodes a protein of 558 amino acids with a molecular mass of 63 kd.8 The cellular localization of FANCC protein is mostly cytoplasmic,14,15 but a fraction of this protein does translocate into nuclei.16-19 The FANCA gene encodes a protein of 1455 amino acids with a molecular mass of approximately 163 kd.9,10 The protein contains a bipartite nuclear localization signal (NLS) at its N-terminus and a partial leucine zipper between amino acids 1069 and 1090. This protein is detected both in the nuclei and in the cytoplasm.16-19 TheFANCG gene cloned by its potential to complement FA-G cells is identical to the previously identified human XRCC9 gene, cloned by its potential to partially complement sensitivity to DNA-damaging agents of a mutant Chinese hamster ovary (CHO) cell line.11,20 This gene encodes a 68-kd protein with an internal leucine zipper. The FANCF and FANCEgenes, cloned by functional complementation, encode a nuclear 42-kd protein and a 60-kd protein with 2 NLS motifs, respectively.12,13 The FANCD2 gene, cloned positionally, encodes a 163-kd nuclear protein.7This protein is monoubiquitinated in response to DNA damage and interacts with the breast cancer susceptibility protein, BRCA1, in nuclear foci.21
A number of studies from our group16,18,22-25 and others26-29 indicate that the protein complex assembly of FANCA, FANCC, and FANCG in the cytoplasm and nuclear accumulation of the protein complex play a crucial role in the FA pathway. FANCF and FANCE are likely to assemble in the protein complex in nuclei.11,12,29 Defects of either of these FA proteins abrogate monoubiquitination and targeting to nuclear foci of FANCD2.21 These findings support a proposal of the FA pathway that the multiple FA proteins sequentially assemble and finally activate FANCD2, a downstream nuclear component of the pathway.18,21,29 Thus, this pathway appears to regulate nuclear functions such as DNA repair, replication, and transcription. On the other hand, significant amounts of FANCA, FANCC, and FANCG proteins localize in the cytoplasm and these proteins interact with various molecules in the cytoplasm and in the membrane.30-34 However, little is known regarding regulatory mechanisms of the FA protein interaction in the cytoplasm or nuclear translocation of the FA protein complex. We found that FANCA is phosphorylated in normal lymphoblasts and that this phosphorylation is defective in lymphoblasts from multiple FA patients, thus raising the possibility that FANCA phosphorylation is involved in regulation of the pathway.18 22 We now report further evidence supporting the functional significance of FANCA phosphorylation and biochemical characteristics of a PK for FANCA (FANCA-PK). FANCA-PK may be another component of the FA protein complex.
Materials and methods
Cell culture
The SV40-immortalized fibroblasts GM6914 were maintained in Dulbecco modified Eagle medium (DMEM) containing 10% fetal calf serum (FCS), as described.24,25 The Epstein-Barr virus (EBV)–immortalized FA-A lymphoblast, HSC72, and FA-G lymphoblasts, EUFA316 and EUFA143, were cultured in RPMI 1640 containing 15% FCS.25 HeLa cells, Jurkat cells, and Mo-7e cells were maintained in DMEM containing 7.5% FCS, RPMI 1640 containing 10% FCS, and RPMI 1640 containing 10% FCS and recombinant human interleukin (IL)-3 (10 ng/mL), respectively. Mononuclear cells from adult peripheral blood and cord blood were cultured for 4 days in RPMI 1640 with 10% FCS in the presence of phytohemagglutinin (5 μg/mL) and recombinant human IL-2 (20 U/mL).
Generation of human complementary DNA mutant constructs and retroviral infection of FA cell lines
Complementary DNAs (cDNAs) of wild-type FANCA (wt-FANCA), with or without a Flag epitope at its N-terminus, and mutant FANCA cDNAs were generated by polymerase chain reaction with Pfu polymerase (Stratagene, La Jolla, CA). cDNA inserts, verified by DNA sequencing, were subcloned into the retroviral expression vector, pMMP, as described.24 The indicated pMMP constructs were transfected by lipofection into 293 producer cells (human embryonic kidney cells) expressing the VSV-G envelope protein. Retroviral supernatants, collected on day 5 following lipofection, contained 4.6 × 106 infectious units/mL, as estimated by Southern blot analysis of infected NIH-3T3 cells (data not shown). Infection of fibroblasts and lymphoblasts was done as described previously.24
Cell survival assay
The cell survival assay was performed as described previously.24
In vivo phosphorylation of FANCA
In vivo phosphorylation of FANCA was examined, as described.18 Briefly, cells (approximately 107cells/sample) were incubated in medium containing 1 mCi (37 MBq)/mL [32P]orthphosphate for 2 hours and lysed in 1 mL lysis buffer. Immunoprecipitates with affinity-purified anti-FANCA antibody (2 μg) raised against the N-terminus16 or affinity-purified anti-FANCG antibody25 (2 μg) were analyzed on sodium dodecyl sulfate (SDS)-polyacrylamide gels. To examine effects of inhibitors, cells were pretreated with inhibitors for 30 minutes and metabolically labeled with 32P in the presence of inhibitors.
In vitro phosphorylation of FANCA
Cell lysates (approximately 2 mg protein) were subjected to immunoprecipitation, using the indicated antibodies. Immune complexes on beads were washed 3 times with the lysis buffer containing 1 mM dithiothreitol (DTT), then 3 times with kinase buffer (50 mM HEPES, pH 7.5, and 10 mM MgCl2, 1 mM DTT), followed by incubation in the kinase buffer containing 10 μCi (0.37 MBq) [γ-32P] ATP at 30°C, for the indicated time. The samples were denatured and loaded on SDS-polyacrylamide gels. To examine the effects of inhibitors, immunoprecipitates were incubated in kinase buffer–containing inhibitors, then in vitro kinase reactions were observed in the presence of inhibitors.
In vitro phosphorylation of glutathione S-transferase-calmodulin–dependent protein kinase I
Glutathione S-transferase-calmodulin–dependent protein kinase I (GST-CaM-KI; 1-293; Lys49Glu) was incubated with GST-CaM–dependent kinase kinase (CaM-KK; 84-434) in a kinase buffer containing 10 μCi (0.37 MBq) [γ-32P] ATP at 30°C for 20 minutes as described.35 In this reaction, GST-CaM-KI is phosphorylated on threonine residues.35These GST-fusion proteins were kindly provided by Dr H. Tokumitsu.
Monoubiquitination of FANCD2
A monoubiquitinated form of FANCD2 was detected as a protein band showing a slower mobility on Western blotting using anti-FANCD2 monoclonal antibody.21
Effects of a dominant-negative IκB kinase
The GM6914 cells were transiently transfected using Fugene (Roche, Mannheim, Germany) with eukaryotic expression constructs, pRc-HA epitope-tagged IκB kinase (IKK)-2(Lys44Ala) (kindly provided by Dr M. Karin),36 pIRES-FANCA, and a reporter gene construct pNF-κB-Luc (Stratagene). The total DNA transfected was kept constant by supplementation with the control vector. At 48 hours after transfection, cells were used for various experiments. Reporter gene activity was determined as described.37 Other details are described in the legend to Figure 8.
Detection of phosphorylation and Western blotting
Proteins separated on SDS-polyacrylamide gels were blotted onto a polyvinylidene difluoride (PVDF) membrane. Phosphorylation of proteins on the membrane was visualized autoradiographically. The same membrane was probed with indicated antibodies, as described.25
Phosphoamino acid analysis
Phosphorylated proteins transferred onto a PVDF membrane were excised and subjected to partial acid hydrolysis. Phosphoamino acids liberated were separated by a 2-dimensional electrophoresis on cellulose thin layer plates (Merck, Whitehouse Station, NJ), according to the method of Cooper and coworkers.38Phosphoamino acids were visualized autoradiographically.
In situ phosphopeptide mapping
Cell fractionation
Fractionation of lymphoblasts was done as described.14 Anti–c-abl antibody (Ab-3) and anti–β-tubulin antibody were from Oncogene Science (Cambridge, MA) and Roche, respectively.
Coimmunoprecipitation of PKs with FANCA
Anti-FANCA immune complexes from lysates of GM6914/wt-FANCA were analyzed on Western blotting with anti-HA monoclonal antibody (12CA5, Roche) for HA-IKK-2 (Lys44Ala) and antibodies against IKK-2 (I87320, Transduction Laboratories, Franklin Lakes, NJ), a p85 subunit of phosphoinositide-3 kinase (PI3K) (P13020, Transduction Laboratories), and a catalytic subunit of DNA-dependent PK (SC-1552, Santa Cruz, Santa Cruz, CA).
Reagents
Phorbol-12-myristate-13-acetate, KT5823, autocamitide-2–related inhibitory protein, calfostin, hypercin, and cyclosporine A were purchased from Calbiochem-Novabiochem (San Diego, CA). Wortmannin and okadaic acid were from Sigma.
Results
We earlier reported that FANCA protein is phosphorylated in EBV-transformed lymphoblasts from normal controls.18 In the present study, we observed FANCA phosphorylation in mitogen-stimulated normal lymphocytes from cord and peripheral blood (Figure 1A). FANCA was also phosphorylated in various human cell lines, including GM6914, an SV40-transformed FANCA− fibroblast, transduced with wt-FANCA (GM6914/wt-FANCA), HeLa, a fibroblastoid cell line derived from cervical carcinoma, Jurkat, a T-cell leukemia cell line, and Mo-7e, an IL-3–dependent myeloid cell line (Figure 1B).
In vivo phosphorylation of FANCA protein in various cells and patient-derived mutant FANCA proteins.
(A) In vivo phosphorylation of FANCA in normal lymphocytes. Lymphocytes from cord blood (CB Ly; lanes 1 and 2) and peripheral blood (PB Ly; lane 3), and lymphoblastic cells (LCL; lane 4) from a normal control were metabolically labeled with 32P, lysed, and subjected to immunoprecipitation (IP) with preimmune serum (Pre; lane 1) or anti-FANCA antibody (lanes 2-4). Western blots of FANCA are shown at the bottom. (B) In vivo phosphorylation of FANCA in GM6914/wt-FANCA, HeLa, Jurkat, and Mo-7e cells. Western blots of FANCA are shown at the bottom. (C) MMC sensitivities of GM6914 cells transduced with wt-FANCA, FANCA(Arg435Cys), and FANCA(1239-43del). Cells were seeded in the presence of various concentrations of MMC and cell survival was assayed 4 days after seeding. (D) Monoubiquitination of FANCD2 in GM6914 cells transduced with wt-FANCA, FANCA(His1110Pro), FANCA(Arg1117Gly), FANCA(Arg435Cys), and FANCA(1239-43del). (E) In vivo phosphorylation of wt-FANCA, FANCA(His1110Pro), FANCA(Arg1117Gly), FANCA(Arg435Cys), and FANCA(1239-43del) expressed in GM6914 cells. Western blots of FANCA are shown at the bottom.
In vivo phosphorylation of FANCA protein in various cells and patient-derived mutant FANCA proteins.
(A) In vivo phosphorylation of FANCA in normal lymphocytes. Lymphocytes from cord blood (CB Ly; lanes 1 and 2) and peripheral blood (PB Ly; lane 3), and lymphoblastic cells (LCL; lane 4) from a normal control were metabolically labeled with 32P, lysed, and subjected to immunoprecipitation (IP) with preimmune serum (Pre; lane 1) or anti-FANCA antibody (lanes 2-4). Western blots of FANCA are shown at the bottom. (B) In vivo phosphorylation of FANCA in GM6914/wt-FANCA, HeLa, Jurkat, and Mo-7e cells. Western blots of FANCA are shown at the bottom. (C) MMC sensitivities of GM6914 cells transduced with wt-FANCA, FANCA(Arg435Cys), and FANCA(1239-43del). Cells were seeded in the presence of various concentrations of MMC and cell survival was assayed 4 days after seeding. (D) Monoubiquitination of FANCD2 in GM6914 cells transduced with wt-FANCA, FANCA(His1110Pro), FANCA(Arg1117Gly), FANCA(Arg435Cys), and FANCA(1239-43del). (E) In vivo phosphorylation of wt-FANCA, FANCA(His1110Pro), FANCA(Arg1117Gly), FANCA(Arg435Cys), and FANCA(1239-43del) expressed in GM6914 cells. Western blots of FANCA are shown at the bottom.
We next examined phosphorylation of patient-derived FANCA mutant proteins in GM6914 cells. FANCA mutant proteins (His1110Pro and Arg1117Gly) failed to correct MMC hypersensitivity in GM6914 cells.25 In addition, we prepared stable transformants of GM6914 expressing other patient-derived mutants (Arg435Cys and 1239-43del).41 These transformants are also hypersensitive to MMC (Figure 1C). Furthermore, all the FANCA mutants failed to induce monoubiquitination of FANCD2 (Figure 1D). The in vivo phosphorylation of all these 4 mutants was markedly reduced (Figure1E). Western blotting showed that the mutant proteins were expressed at comparable levels with wt-FANCA. These results suggest that phosphorylation of the exogenously expressed FANCA proteins is associated with their function. Thus, use of this system should be appropriate to study FANCA-PK.
We attempted to develop an in vitro phosphorylation system of this protein. Flag-tagged wt-FANCA was immunoprecipitated from lysates of GM6914/Flag-wt-FANCA and incubated in kinase buffer containing Mg++ and [γ-32P] ATP. In immunoprecipitation with either of anti-FANCA or anti-Flag antibody (M2), FANCA was phosphorylated (Figure2A, left and middle). Of note, Flag-wt-FANCA was similarly phosphorylated even after a 2-step immunopurification (anti-Flag → anti-FANCA), suggesting that a PK specifically associates with FANCA (Figure 2A, right). The in vitro phosphorylation occurred in a time-dependent manner (2-30 minutes; Figure 2B). Thus, we performed standard in vitro kinase assays at 15 to 20 minutes.
In vitro phosphorylation of FANCA.
(A) In vitro phosphorylation of Flag-wt-FANCA in GM6914 cells. Cell lysates from GM6914 and GM6914/Flag-wt-FANCA were immunoprecipitated (IP) with anti-Flag antibody (M2) or anti-FANCA antibody. For a 2-step immunoprecipitation, Flag-wt-FANCA was absorbed to anti-Flag antibody-conjugated Sepharose, eluted with Flag peptide (100 μg/mL), then immunoprecipitated with anti-FANCA antibody. Immunoprecipitates were incubated in a kinase buffer containing [γ-32P] ATP (10 μCi [0.37 MBq]) for 20 minutes. (B) A time course of in vitro phosphorylation of Flag-wt-FANCA expressed in GM6914 cells. Anti-FANCA immunoprecipitates from GM6914/Flag-wt-FANCA were incubated in a kinase buffer containing [γ-32P] ATP (10 μCi [0.37 MBq]) for the indicated times. Western blots of FANCA were shown at the bottom.
In vitro phosphorylation of FANCA.
(A) In vitro phosphorylation of Flag-wt-FANCA in GM6914 cells. Cell lysates from GM6914 and GM6914/Flag-wt-FANCA were immunoprecipitated (IP) with anti-Flag antibody (M2) or anti-FANCA antibody. For a 2-step immunoprecipitation, Flag-wt-FANCA was absorbed to anti-Flag antibody-conjugated Sepharose, eluted with Flag peptide (100 μg/mL), then immunoprecipitated with anti-FANCA antibody. Immunoprecipitates were incubated in a kinase buffer containing [γ-32P] ATP (10 μCi [0.37 MBq]) for 20 minutes. (B) A time course of in vitro phosphorylation of Flag-wt-FANCA expressed in GM6914 cells. Anti-FANCA immunoprecipitates from GM6914/Flag-wt-FANCA were incubated in a kinase buffer containing [γ-32P] ATP (10 μCi [0.37 MBq]) for the indicated times. Western blots of FANCA were shown at the bottom.
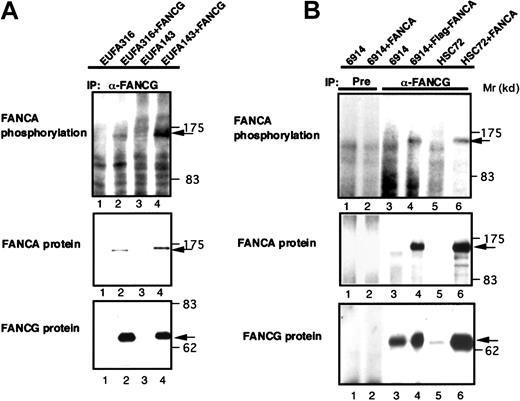
To determine if FANCA-PK binds and phosphorylates FANCA in the FA protein complex, we examined in vivo and in vitro phosphorylation of FANCA coimmunoprecipitated with FANCG. Analyses of anti-FANCG immune complexes from 32P-labeled lysates of FANCG−cells, EUFA316 and EUFA143, and the cells corrected by introduction of wt-FANCG (EUFA316/wt-FANCG and EUFA143/wt-FANCG) showed that phosphorylated FANCA specifically coimmunoprecipitated with FANCG (Figure 3A). When anti-FANCG immune complexes from cell lysates of HSC72/wt-FANCA and GM6914/wt-FANCA were incubated with [γ-32P] ATP, FANCA was phosphorylated in vitro (Figure 3B).
In vivo and in vitro phosphorylation of FANCA coimmunoprecipitated with FANCG.
(A) Cell lysates from 32P-labeled EUFA316, EUFA316/wt-FANCG, EUFA143, and EUFA143/wt-FANCG cells were subjected to immunoprecipitation (IP) with anti-FANCG antibody. An arrow indicates phosphorylated FANCA. The lower panels show Western blots of FANCA and FANCG. (B) Cell lysates from HSC72, HSC72/wt-FANCA, GM6914, and GM6914/wt-FANCA were subjected to immunoprecipitation (IP) with preimmune serum (Pre) or anti-FANCG antibody; then in vitro phosphorylation of the immunoprecipitates was analyzed. An arrow indicates phosphorylated FANCA. The lower panels show Western blots of FANCA and FANCG.
In vivo and in vitro phosphorylation of FANCA coimmunoprecipitated with FANCG.
(A) Cell lysates from 32P-labeled EUFA316, EUFA316/wt-FANCG, EUFA143, and EUFA143/wt-FANCG cells were subjected to immunoprecipitation (IP) with anti-FANCG antibody. An arrow indicates phosphorylated FANCA. The lower panels show Western blots of FANCA and FANCG. (B) Cell lysates from HSC72, HSC72/wt-FANCA, GM6914, and GM6914/wt-FANCA were subjected to immunoprecipitation (IP) with preimmune serum (Pre) or anti-FANCG antibody; then in vitro phosphorylation of the immunoprecipitates was analyzed. An arrow indicates phosphorylated FANCA. The lower panels show Western blots of FANCA and FANCG.
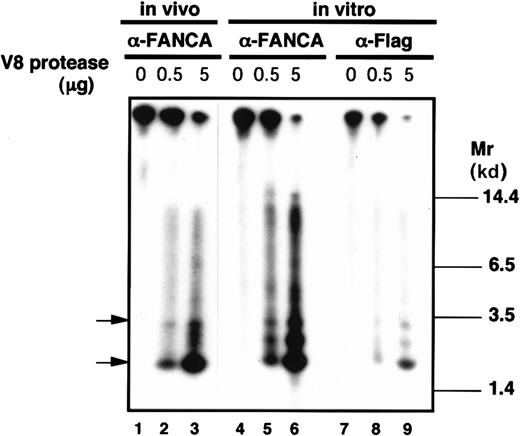
To compare phosphorylation sites between in vivo phosphorylated FANCA and in vitro phosphorylated FANCA, in situ phosphopeptide mapping was done (Figure 4). The32P-labeled FANCA proteins were digested in a gel by V8 protease and the phosphopeptides were separated on a polyacrylamide gel. Flag-tagged FANCA protein, immunoprecipitated with either of anti-FANCA polyclonal antibody or anti-Flag monoclonal antibody and phosphorylated in vitro, showed identical phosphopeptide mapping (Figure 4, lanes 6 and 9). More importantly, a comparison of phosphopeptide mapping patterns between in vivo and in vitro phopshorylated FANCA proteins showed that at least 2 major phosphopeptides (indicated by arrows) showed the same mobilities (compare lanes 3, 6, and 9 in Figure 4), although the 2 patterns were not identical. Thus, FANCA proteins phosphorylated in vivo and in vitro appear to share, at least in part, common phosphorylation sites.
In situ phosphopeptide mapping of FANCA.
Flag-tagged wt-FANCA expressed in GM6914 cells was phosphorylated in vivo or immunoprecipitated with either anti-FANCA antibody or anti-Flag monoclonal antibody and phosphorylated in vitro. Phosphorylated FANCA proteins were treated with V8 protease (0.5 or 5 μg) and separated on an SDS-polyacrylamide gel.
In situ phosphopeptide mapping of FANCA.
Flag-tagged wt-FANCA expressed in GM6914 cells was phosphorylated in vivo or immunoprecipitated with either anti-FANCA antibody or anti-Flag monoclonal antibody and phosphorylated in vitro. Phosphorylated FANCA proteins were treated with V8 protease (0.5 or 5 μg) and separated on an SDS-polyacrylamide gel.
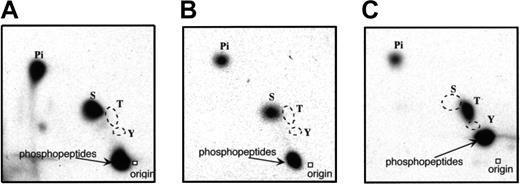
To characterize FANCA-PK, we first did a phosphoamino acid analysis, using in vivo and in vitro phosphorylated FANCA proteins in GM6914/wt-FANCA. The proteins were subjected to acid hydrolysis and phosphoamino acid analysis by 2-dimensional chromatography. In both, only phosphoserine was detected (Figure5A,B). As a control for threonine phosphorylation, GST-CaM-KI (1-293, Lys49Glu) was phosphorylated by GST-CaM-KK (84-434) in vitro and subjected to phosphoamino acid analysis (Figure 5C).
Phosphoamino acid analysis of FANCA.
FANCA protein phosphorylated in vivo (A) and in vitro (B) were subjected to phosphoamino acid analysis. As a control of threonine phosphorylation, GST-CaM-KI(1-293, Lys49Glu), phosphorylated by GST-CaM-KK(84-434) in vitro, was subjected to phosphoamino acid analysis (C). The positions of phosphoamino acid standards, as determined by ninhydrin staining, are shown.
Phosphoamino acid analysis of FANCA.
FANCA protein phosphorylated in vivo (A) and in vitro (B) were subjected to phosphoamino acid analysis. As a control of threonine phosphorylation, GST-CaM-KI(1-293, Lys49Glu), phosphorylated by GST-CaM-KK(84-434) in vitro, was subjected to phosphoamino acid analysis (C). The positions of phosphoamino acid standards, as determined by ninhydrin staining, are shown.
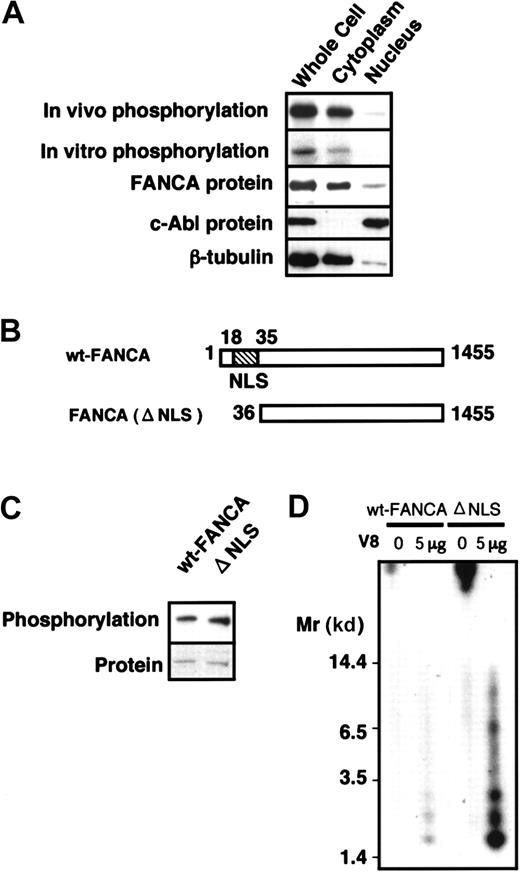
Next, to determine the subcellular localization of FANCA-PK and phosphorylated FANCA protein, we fractionated HSC72/wt-FANCA. Both in vivo phosphorylated FANCA and in vitro kinase activity were detected mainly in the cytoplasm (Figure 6A). To further examine whether or not cytoplasmic FANCA-PK phosphorylates FANCA in vivo, we examined the phosphorylation of a mutant FANCA protein, FANCA(del NLS), which lacks an N-terminal portion containing NLS (Figure 6B) and fails to translocate into the nuclei in GM6914 cells.24 FANCA(del NLS) was phosphorylated equally to wt-FANCA in vivo (Figure 6C). V8 protease–digested phosphopeptides derived from in vitro phosphorylated protein were identical between wt-FANCA and FANCA(delNLS) (Figure 6D). Taken together, these results are interpreted to mean that FANCA-PK binds and phosphorylates FANCA in the cytoplasm.
Cytoplasmic localization of FANCA phosphorylation.
(A) Cytoplasmic localization of phosphorylated FANCA and FANCA-PK activities. HSC72/wt-FANCA cells (approximately 3 × 107cells) were metabolically labeled by 32P and fractionated to cytoplasmic and nuclear fractions; then each fraction was subjected to immunoprecipitation with anti-FANCA antibody. In vitro FANCA phosphorylation was examined using each fraction from HSC72/wt-FANCA cells (approximately 3 × 107 cells). The middle panel shows Western blots of FANCA. In parallel experiments, each fraction from the cells (approximately 5 × 105 cells) was subjected to Western blotting with antibodies against c-Abl and β-tubulin, nuclear and cytoplasmic markers, respectively (lower panels). (B) Structure of FANCA(delNLS). (C) In vivo phosphorylation of FANCA(delNLS) in GM6914. Western blots of FANCA are shown at the bottom. (D) Phosphopeptide mapping of wt-FANCA and FANCA(delNLS) phosphorylated in vitro.
Cytoplasmic localization of FANCA phosphorylation.
(A) Cytoplasmic localization of phosphorylated FANCA and FANCA-PK activities. HSC72/wt-FANCA cells (approximately 3 × 107cells) were metabolically labeled by 32P and fractionated to cytoplasmic and nuclear fractions; then each fraction was subjected to immunoprecipitation with anti-FANCA antibody. In vitro FANCA phosphorylation was examined using each fraction from HSC72/wt-FANCA cells (approximately 3 × 107 cells). The middle panel shows Western blots of FANCA. In parallel experiments, each fraction from the cells (approximately 5 × 105 cells) was subjected to Western blotting with antibodies against c-Abl and β-tubulin, nuclear and cytoplasmic markers, respectively (lower panels). (B) Structure of FANCA(delNLS). (C) In vivo phosphorylation of FANCA(delNLS) in GM6914. Western blots of FANCA are shown at the bottom. (D) Phosphopeptide mapping of wt-FANCA and FANCA(delNLS) phosphorylated in vitro.
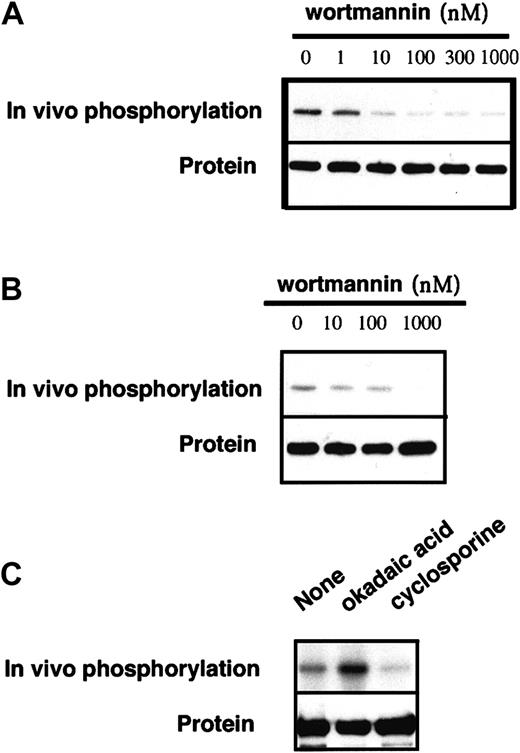
We screened effects of various PK stimulators and inhibitors on FANCA phosphorylation in vitro and in vivo (Table1). Although most of these compounds were without effect, wortmannin, a potent inhibitor of phosphoinositide (PI) kinases (reviewed by Fruman and colleagues42), did suppress FANCA phosphorylation at 10 nM to 1 μM (Figure7A). This compound also suppressed in vivo phosphorylation of FANCA (Figure 7B). When we examined effects of okadaic acid, an inhibitor of serine/threonine protein phosphatases 1 and 2A, and cyclosporine A, an inhibitor of a protein phosphatase 2B (calcineurine), okadaic acid increased the in vivo phosphorylation of FANCA, thereby suggesting that FANCA phosphorylation levels are regulated by a serine/threonine protein phosphatase (Figure 7C).
Effects of PK stimulators and inhibitors on FANCA phosphorylation
| Agent . | Major target . | Assay . | Effect . |
|---|---|---|---|
| PK stimulator | |||
| cAMP (4 μM) | cAMP-dependent PK | In vitro | − |
| cGMP (4 μM) | cGMP-dependent PK | In vitro | − |
| Calcium (100 μM) | Calmodulin-dependent PK | In vitro | − |
| PMA (100 nM) | Protein kinase C | In vivo† | − |
| PK inhibitor | |||
| cAMP inhibitor protein* (10 U/assay) | cAMP-dependent PK | In vitro | − |
| KT5823 (1 μM) | cGMP-dependent PK | In vitro | − |
| AIP (1 μM) | Calmodulin-dependent PK | In vitro | − |
| Calfostin (500 nM) | Protein kinase C | In vitro | − |
| Hypericin (50 nM) | MAPK, casein kinase II | In vitro | − |
| PD98059 (50 μM) | MAPK kinase | In vivo | − |
| SB203580 (10 μM) | p38 MAPK | In vivo | − |
| Rapamycin (20 nM) | FRAP | In vivo | − |
| Aspirin (5 mM) | IKK-2 | In vivo | − |
| (1 mM) | In vitro | − |
| Agent . | Major target . | Assay . | Effect . |
|---|---|---|---|
| PK stimulator | |||
| cAMP (4 μM) | cAMP-dependent PK | In vitro | − |
| cGMP (4 μM) | cGMP-dependent PK | In vitro | − |
| Calcium (100 μM) | Calmodulin-dependent PK | In vitro | − |
| PMA (100 nM) | Protein kinase C | In vivo† | − |
| PK inhibitor | |||
| cAMP inhibitor protein* (10 U/assay) | cAMP-dependent PK | In vitro | − |
| KT5823 (1 μM) | cGMP-dependent PK | In vitro | − |
| AIP (1 μM) | Calmodulin-dependent PK | In vitro | − |
| Calfostin (500 nM) | Protein kinase C | In vitro | − |
| Hypericin (50 nM) | MAPK, casein kinase II | In vitro | − |
| PD98059 (50 μM) | MAPK kinase | In vivo | − |
| SB203580 (10 μM) | p38 MAPK | In vivo | − |
| Rapamycin (20 nM) | FRAP | In vivo | − |
| Aspirin (5 mM) | IKK-2 | In vivo | − |
| (1 mM) | In vitro | − |
PMA indicates phorbol-12-myristate-13-acetate; AIP, autocamitide-2–related inhibitory protein; and MAPK, mitogen-activated PK.
Recombinant PK A heat-stable inhibitor, isoform α.
32P-labeled cells were treated with PMA for 10 minutes and in vivo phosphorylation of FANCA was estimated.
Effects of wortmannin and okadaic acid on FANCA phosphorylation.
(A) Wt-FANCA in GM6914 cells was immunoprecipitated and phosphorylated in vitro, in the presence of various concentrations of wortmannin. (B) GM6914/wt-FANCA was metabolically labeled with32P for 2 hours in the presence of various concentrations of wortmannin, then FANCA phosphorylation was examined. (C) GM6914 cells/wt-FANCA was labeled with 32P for 2 hours in the presence of okadaic acid (1 μM) or cyclosporine (10 μg/mL), lysed, and subjected to immunoprecipitation with anti-FANCA antibody. Western blots of FANCA are shown at the bottom.
Effects of wortmannin and okadaic acid on FANCA phosphorylation.
(A) Wt-FANCA in GM6914 cells was immunoprecipitated and phosphorylated in vitro, in the presence of various concentrations of wortmannin. (B) GM6914/wt-FANCA was metabolically labeled with32P for 2 hours in the presence of various concentrations of wortmannin, then FANCA phosphorylation was examined. (C) GM6914 cells/wt-FANCA was labeled with 32P for 2 hours in the presence of okadaic acid (1 μM) or cyclosporine (10 μg/mL), lysed, and subjected to immunoprecipitation with anti-FANCA antibody. Western blots of FANCA are shown at the bottom.
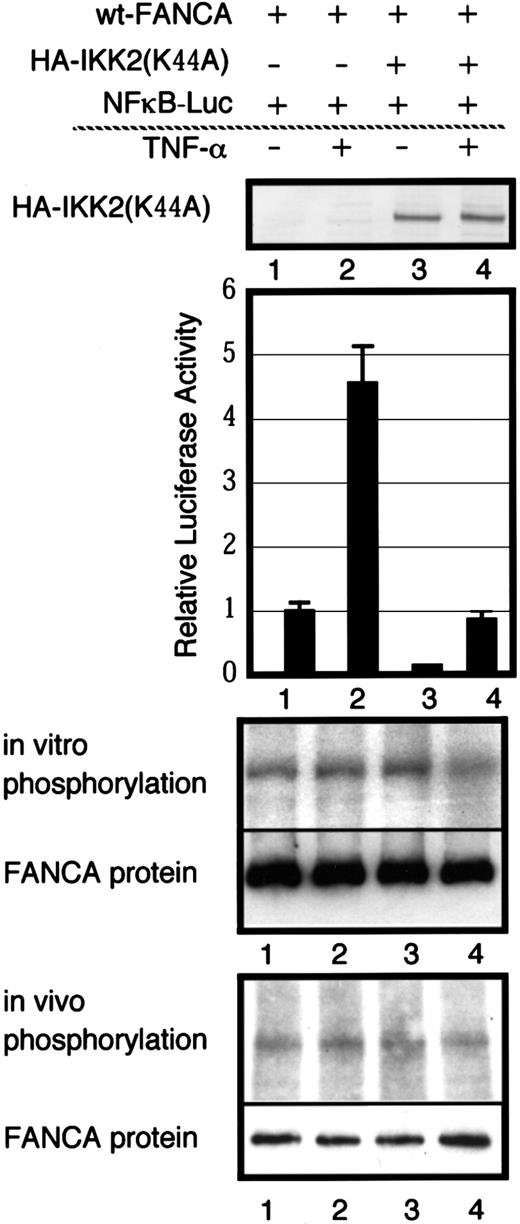
Otsuki and coworkers found IKK-2 to be a FANCA-binding protein, using a yeast 2-hybrid screening method.43 IKK-2 is a cytoplasmic serine kinase (reviewed by Karin44). Thus, IKK-2 may be FANCA-PK, and we examined the possibility, using several approaches. Aspirin, which was reported to inhibit IKK-2,45 did not affect FANCA-PK either in vivo or in vitro (Table 1). No IKK-2 protein was detected in an anti-FANCA immune complex by Western blotting (not shown). Finally, we examined the effects of tumor necrosis factor (TNF)-α, a potent stimulator of IKK-244 and IKK-2(Lys44Ala), a dominant-negative form of IKK-236 on FANCA phosphorylation (Figure8). We cotransfected GM6914 cells with expression vectors containing FANCA, HA-IKK-2(Lys44Ala), and a nuclear factor (NF)-κB–dependent luciferase reporter gene. When HA-IKK-2(Lys44Ala) was expressed, TNF-α–induced activation of NF-κB was markedly suppressed, thus confirming a dominant-negative effect of IKK-2(Lys44Ala) in these cells. However, TNF-α stimulation or expression of IKK-2(Lys44Ala) had no effects on in vitro and in vivo phosphorylation of FANCA. Consistently, no HA-IKK-2(Lys44Ala) protein was detected in FANCA immune complexes by Western blotting (not shown). Taken together, these results indicate that FANCA-PK is not IKK-2.
Effects of TNF-α and a dominant-negative form of IKK-2 on FANCA phosphorylation.
GM6914 cells were transiently transfected with wt-FANCA, HA-IKK2(Lys44Ala) and NF-κB–luciferase reporter genes. At 48 hours after transfection, cells were used for further analyses. Whole cell extracts were used to confirm expression of HA-IKK2(Lys44Ala) by Western blotting with anti-HA monoclonal antibody. Cells were stimulated with TNF-α for 5 hours and cell lysates were prepared for assay of NF-κB activation. The values shown are averages ± SEM of triplicate samples for one representative experiment. To examine in vitro phosphorylation, lysates from cells stimulated with TNF-α (20 ng/mL) for 5 minutes were subjected to immunoprecipitation with anti-FANCA antibody. To examine in vivo phosphorylation, cells were metabolically labeled with 32P, stimulated with TNF-α (20 ng/mL) for 5 minutes and lysates were prepared. The lower panels show Western blots of FANCA.
Effects of TNF-α and a dominant-negative form of IKK-2 on FANCA phosphorylation.
GM6914 cells were transiently transfected with wt-FANCA, HA-IKK2(Lys44Ala) and NF-κB–luciferase reporter genes. At 48 hours after transfection, cells were used for further analyses. Whole cell extracts were used to confirm expression of HA-IKK2(Lys44Ala) by Western blotting with anti-HA monoclonal antibody. Cells were stimulated with TNF-α for 5 hours and cell lysates were prepared for assay of NF-κB activation. The values shown are averages ± SEM of triplicate samples for one representative experiment. To examine in vitro phosphorylation, lysates from cells stimulated with TNF-α (20 ng/mL) for 5 minutes were subjected to immunoprecipitation with anti-FANCA antibody. To examine in vivo phosphorylation, cells were metabolically labeled with 32P, stimulated with TNF-α (20 ng/mL) for 5 minutes and lysates were prepared. The lower panels show Western blots of FANCA.
Discussion
We earlier reported that FANCA is phosphorylated in EBV-immortalized lymphoblasts from normal controls.18 The present results show that FANCA is also phosphorylated in mitogen-stimulated lymphocytes from peripheral and cord blood, and transformed cell lines, including myeloid cells and fibroblastoid cells (Figure 1A,B), suggesting that FANCA is phosphorylated in various types of growing cells. We here present evidence indicating that the phosphorylation of FANCA is closely associated with its function, and that a PK phosphorylating FANCA forms a complex with the substrate, perhaps in the FA protein complex. FANCA-PK is a cytoplasmic serine kinase sensitive to wortmannin.
GM6914 and HSC72 are SV-40–immortalized fibroblasts and EBV-immortalized lymphoblasts, respectively, both derived from the same patient with FA-A. These cells express no endogenous FANCA protein and are hypersensitive to MMC.9,24 Retroviral gene transfer introduces wild-type and mutant FANCA gene cDNAs into these cells with high efficiency and enables stable expression of proteins at high levels.24,25 The wt-FANCA expressed in these cells was shown to correct MMC sensitivity, bind to FANCC and FANCG, and localize in the nuclei, like endogenous FANCA.24 25 Thus, this system should be useful for studying posttranslational modification and structure-function relationships of FANCA.
We hypothesized that FANCA phosphorylation is associated with its function because it is abrogated in multiple FA patient–derived cells of different complementation groups.18 Furthermore, a defective phosphorylation of FANCA(His1110Pro) protein was noted in patient-derived lymphoblasts.22 However, endogenous FANCA protein levels were moderately (sometimes, severely) decreased in the patient-derived cells, which could account for the defective phosphorylation. To overcome this problem, we exogenously expressed several patient-derived mutant FANCA proteins in GM6914 cells and analyzed functions and phosphorylation. FANCA(His1110Pro) and FANCA(Arg1117Gly) were previously shown to fail to correct MMC sensitivity of the cells.24,25 In addition, we tested another 2 mutant proteins, FANCA (Arg435Cys) and FANCA(1239-43del). These 2 also failed to correct MMC sensitivity of GM6914 cells (Figure 1C). Furthermore, all of the 4 mutants failed to induce monoubiquitination of FANCD2 (Figure 1D), indicating that they are incapable of activating the FA pathway.21 All of the mutant proteins, expressed in these cells at comparable levels to the wild-type protein, were defective for in vivo phosphorylation (Figure1E). Thus, the present results further support the correlation between functions and phosphorylation of FANCA.
The abolition of FANCA phosphorylation by various mutations in distinct regions, Arg435Cys, His1110Pro, Arg1117Gly and 1239-43del, suggests that the domains including these mutations are important for FANCA phosphorylation. Emerging evidence indicates that many PKs form a complex with their substrates through direct interactions between a part of the kinase and a part of the substrate, known as docking sites (reviewed by Holland and Cooper46 and Sharrocks and coworkers47). Intensive studies on mitogen-activated protein (MAP) kinases revealed that multiple docking sites of substrates are important for efficient and specific actions of the kinases.48 In some cases, a third protein known as an adapter protein or a scaffold protein plays a crucial role in PK-substrate interactions.49 Patient-derived mutations of FANCA may destroy such direct or indirect interactions with FANCA-PK and inhibit the efficient phosphorylation of FANCA.
FANCA(del NLS), which fails to correct MMC sensitivity and fails to bind to FANCC or FANCG in GM6914 cells,24 is phosphorylated similarly to wt-FANCA (Figure 6C,D). From this observation, we can draw at least 2 conclusions, in addition to cytoplasmic localization of FANCA phosphorylation. First, FANCA phosphorylation is not sufficient for its function. Second, FANCA-PK binds and phosphorylates FANCA without the need of FANCC or FANCG. Consistent with these conclusions, when FANCA was overexpressed in FA-C or FA-G cells, FANCA was phosphorylated but these cells were not complemented (data not shown). By contrast, FANCA(His1110Pro) and FANCA (Arg1117Gly) bind to FANCG but are defective for phosphorylation and complementation. Taken together, these results indicate that FANCG binding and FANCA phosphorylation are independent, and that either of the 2 events alone is inadequate for the function of FANCA.25,26 However, because a free form of FANCA is unstable,23 it is more likely that FANCA-PK phosphorylates the substrate in an FA protein complex, under physiologic conditions. Consistent with this notion, FANCA-PK appears to bind and phosphorylate FANCA, at least in part, in the FANCA/FANCG complex (Figure 3).
The molecular basis underlying the association between FANCA phosphorylation and its function remains to be elucidated. FANCA phosphorylation may be necessary for assembly of functional FA protein complex or vice versa; obviously, these 2 possibilities are not exclusive. To further study this subject, it is crucial to determine phosphorylation sites of FANCA. For this purpose, it is helpful to have an in vitro phosphorylation system. Several lines of evidence indicate that a physiologic kinase binds and phosphorylates FANCA in vitro: (1) a 2-step immunopurification study suggests that FANCA forms a specific complex with a PK (Figure 2A); (2) a phosphopeptide mapping analysis suggests that common sites are phosphorylated in vivo and in vitro (Figure 4); (3) FANCA is phosphorylated only on serine residues both in vivo and in vitro (Figure 5); (4) both in vitro kinase activity and occurrence of in vivo phosphorylation are localized to the cytoplasm (Figure 6); and (5) both in vivo and in vitro FANCA phosphorylation are sensitive to wortmannin (Figure 7).
FANCA-PK appears to be a wortmannin-sensitive cytoplasmic serine kinase (Figure 7). Wortmannin is a potent inhibitor of PI kinases42 and related PKs: ATM, DNA-dependent PK, and FKBP12-rapamycin–associated protein (FRAP), but not ATR, are sensitive to this compound.50,51 Substrates of PI kinases are phosphoinositides, although PI3K exceptionally autophosphorylates their subunits p85 and p110.52,53 However, FANCA was phosphorylated in ATM-deficient lymphoblasts (not shown). We detected no PI3K subunit p85, which tightly binds to p110, or DNA-PK in the anti-FANCA immune complexes by Western blot (not shown). FANCA-PK unlike FRAP54 was not sensitive to rapamycin (Table 1). Thus, FANCA-PK seems unlikely to be a known member of PI kinase–related PKs.
Recent findings indicate that DNA damage activates FANCD2, a downstream nuclear component of the FA pathway, depending on FA protein complex assembly and nuclear accumulation of the complex.21 Any inherited defect in the FA proteins results in an FA phenotype. However, little is known regarding physiologic regulations of the pathway. The present study raises the possibility that a cytoplasmic serine kinase regulates the upstream area of the FA pathway. Identification of the PK may provide new insights into functions of the FA pathway.
We thank Dr T. Nakahata for support, Dr H. Joenje for providing the FA-G lymphoblasts, and M. Ohara for language assistance. We are also grateful to Drs M. Karin, M. Hayakawa, M. Delhase, M. Takagi and H. Tokumitsu for the generous supply of reagents.
Supported by a Grant-in-Aid for Scientific Research from the Ministry of Education, Science, Technology, Sports and Culture of Japan and grants from the Ministry of Health, Labor and Welfare of Japan. The Division of Genetic Diagnosis is supported in part by Otsuka Pharmaceutical.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Takayuki Yamashita, Institute of Medical Science, University of Tokyo, Division of Genetic Diagnosis, 4-6-1 Shirokanedai Minato-ku Tokyo 108-8639, Japan; e-mail: y-taka@ims.u-tokyo.ac.jp.


![Fig. 2. In vitro phosphorylation of FANCA. / (A) In vitro phosphorylation of Flag-wt-FANCA in GM6914 cells. Cell lysates from GM6914 and GM6914/Flag-wt-FANCA were immunoprecipitated (IP) with anti-Flag antibody (M2) or anti-FANCA antibody. For a 2-step immunoprecipitation, Flag-wt-FANCA was absorbed to anti-Flag antibody-conjugated Sepharose, eluted with Flag peptide (100 μg/mL), then immunoprecipitated with anti-FANCA antibody. Immunoprecipitates were incubated in a kinase buffer containing [γ-32P] ATP (10 μCi [0.37 MBq]) for 20 minutes. (B) A time course of in vitro phosphorylation of Flag-wt-FANCA expressed in GM6914 cells. Anti-FANCA immunoprecipitates from GM6914/Flag-wt-FANCA were incubated in a kinase buffer containing [γ-32P] ATP (10 μCi [0.37 MBq]) for the indicated times. Western blots of FANCA were shown at the bottom.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/98/13/10.1182_blood.v98.13.3650/6/m_h82411875002.jpeg?Expires=1767751534&Signature=bz-i2tzHES5yLBQfSUBhWFx-wmW5QMRdcFiTxqMRbXOuFDc8cP4WrPTu~4ysMxTVHSQnIgeVpizSEJqJ3i6Oc7C7nuAsBWkYdqCglAsCz5FoagCRHvbL0FyZNz~XCmlrdi8hdgRx2lhxuhgc7wjqOury6uCxOGTNCngqBp5THZYZ8Z3PzloWTtGAecPjXWokQzV8pnOG2sY8J3hy-wCK7MsDKKgd83wh78E9MEgGz7yFIF3t-FsKDPlrqbFyaWGcZ8HBtL504P85DDnTQF8S6PsKvDV4B3O6a1PN23glSVVj9ohaMotsPcL~BoHi~JvmhHCAl995z8kbVkaDu2DSVw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal