Abstract
CCAAT displacement protein (cux/CDP) is an atypical homeodomain protein that represses expression of several developmentally regulated lymphoid and myeloid genes in vitro, including gp91-phox, immunoglobulin heavy chain, the T-cell receptor β and γ chains, and CD8. To determine how this activity affects cell development in vivo, a hypomorphic allele of cux/CDP was created by gene targeting. Homozygous mutant mice (cux/CDPΔHD/ΔHD) demonstrated a partial neonatal lethality phenotype. Surviving animals suffered from a wasting disease, which usually resulted in death between 2 and 3 weeks of age. Analysis of T lymphopoiesis demonstrated that cux/CDPΔHD/ΔHD mice had dramatically reduced thymic cellularity due to enhanced apoptosis, with a preferential loss of CD4+CD8+ thymocytes. Ectopic CD25 expression was also observed in maturing thymocytes. B lymphopoiesis was also perturbed, with a 2- to 3-fold reduction in total bone marrow B-lineage cells and a preferential loss of cells in transition from pro-B/pre-BI to pre-BII stages due to enhanced apoptosis. These lymphoid abnormalities were independent of effects related to antigen receptor rearrangement. In contrast to the lymphoid demise, cux/CDPΔHD/ΔHD mice demonstrated myeloid hyperplasia. Bone marrow reconstitution experiments identified that many of the hematopoietic defects were linked to microenvironmental effects, suggesting that underexpression of survival factors or overexpression of death-inducing factors accounted for the phenotypes observed. Tumor necrosis factor (TNF) levels were elevated in several tissues, especially thymus, suggesting that TNF may be a target gene for cux/CDP-mediated repression. These data suggest that cux/CDP regulates normal hematopoiesis, in part, by modulating the levels of survival and/or apoptosis factors expressed by the microenvironment.
Introduction
The development and function of the immune system are controlled, in part, by the regulation of tissue-specific gene expression. Orchestration of gene expression is modulated by a combination of both positively and negatively acting transcription factors. One such repressive factor is human CCAAT displacement protein (CDP), which belongs to a novel family of homeodomain proteins that appear to bind DNA through repeated domains termed “cut-repeats.”1,2 CDP was initially identified as a transcriptional repressor of the sea urchin sperm histoneH2B gene,3 and homologues include cux/CDP from mouse,4 Clox from canine,5 cut fromDrosophila,6 and CDP from rat.7Each of these 180- to 190-kd homologues contains 3 cut-repeats (CRs) in addition to an atypical homeodomain (HD) located near the carboxy-terminus. CDP requires cooperative interaction between either 2 CR domains or one CR domain and the homeodomain for DNA binding.8,9 CDP preferentially recognizes AT-rich DNA sequences9 that are often associated with nuclear matrix association regions (MARs; reviewed by Scheuermann and Garrard10). CDP forms homomeric complexes in coimmunoprecipitation experiments, presumably through a coiled-coil domain located near the amino-terminus.11 A C-terminal repression domain appears to associate with histone deacetylases.12 13
CDP appears to play vital roles in regulating genes involved in multilineage differentiation pathways. This has been exemplified inDrosophila, where cell fate determination in multiple lineages is altered by ectopic expression ofcut.14 The essential role of cut in controlling normal development is demonstrated by the observation that null mutations result in embryonic lethality.15,16 Within mammalian systems CDP and its homologues function as repressors of a wide variety of different genes within multiple systems (Wang and colleagues17 and references therein). The binding of CDP homologues to promoters, enhancers, or MARs of tissue-specific genes appears to be limited to developmental stages where target genes are not expressed. Target sites for cux/CDP binding and repression have been identified within myeloid- and lymphoid-specific genes and include the gp91-phox promoter,18 neutrophil collagenase promoter,19 lactoferrin promoter,20neutrophil gelatinase promoter,19 CD8a MAR,21T-cell receptor β (TCRβ) enhancer/MAR,22 and TCRδ enhancer (M. Krangel, written communication, July 1999) and the immunoglobulin heavy chain (IgH) intronic enhancer/MAR.17
cux/CDP appears to function through multiple mechanisms. In some cases, in vitro studies indicate that cux/CDP is able to compete for transcriptional activator binding, like BID/YY1, CP1, and IRF factors in the gp91-phox promoter23,24 and Bright in the IgH enhancer/MAR and promoter.17 Other experiments suggest cux/CDP might repress gene expression through higher-order changes in chromatin structure including histone deacetylation.13Many of the characterized cux/CDP binding sites also function as nuclear matrix attachment sites.10,22,25 cux/CDP binding can inhibit binding of MAR-containing DNA fragments to nuclear matrix preparations in vitro,26 suggesting that cux/CDP may influence gene expression by regulating nuclear localization.
To determine the roles of cux/CDP in controlling cell lineage determination and function in vivo, a cux/CDP allele with a C-terminal deletion encompassing the homeodomain and repression domain was generated using homologous gene targeting approaches in mice. Mutation of the cux/CDP protein affects several organ systems, including the hematolymphoid compartment, through cell-intrinsic and environmental effects. Our data suggest that cux/CDP functions to modulate the expression of lymphoid survival and/or death-inducing factors, which may include tumor necrosis factor (TNF).
Materials and methods
Generation of the ΔHD mice
Mouse cux/CDP genomic sequences were isolated from a 129/Sv strain λFix III library.27 A targeting construct was made to delete a 4-kilobase (kb) region encoding the homeodomain in exons 20 and 2127 (Figure 1A) by cloning a 696-bp HindIII (intron 19) and XhoI (exon 20-12 bp into the homeodomain) fragment upstream of theneo gene and a 6-kb BamHI fragment downstream. The neo gene was inserted in the opposite transcriptional orientation and provided an in-frame stop codon 30 bp downstream of the fusion site. An HSV-thymidine kinase gene was included further 5′ to allow for negative selection of random insertions.
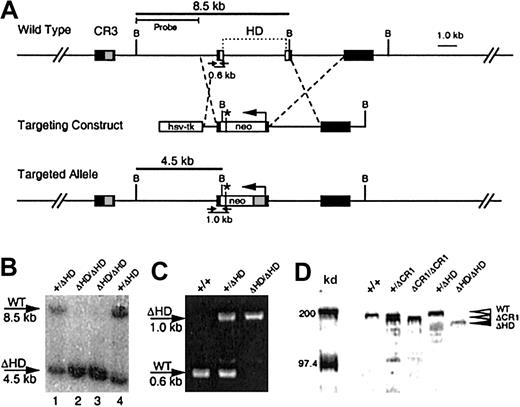
Generation of the cux/CDPΔHD hypomorphic allele.
(A) A targeting vector was designed to delete a 4-kb region encompassing the homeodomain within exons 20 and 21 and the intervening intron, with a PGK-neomycine resistance gene (neo), which provides a premature in-frame stop codon (*) for cux/CDP translation termination. Exons (black boxes) encoding cut repeat 3 (CR3, gray box) and the homeodomain (HD, open boxes) are shown above the wild-type gene. Primers for PCR detection of wild-type and targeted alleles with relative product sizes are shown as arrows below the schematics. The probe used for Southern detection and the fragments generated by digestion at the BamHI sites (letter “B”) are indicated. (B) Southern blot of BamHI-digested DNA from ES clones heterozygous (lanes 1 and 4) and homozygous (lanes 2 and 3) for the targeted allele. Southern blot was probed with a 4-kbBamHI/HindIII probe to detect the 8.5-kb wild-type and 4.5-kb targeted alleles. (C) Agarose gel demonstrating the products from multiplex PCR reactions to detect wild-type and knock-out alleles in wild type (+/+), heterozygous (+/ΔHD) and homozygous (ΔHD/ΔHD) mice. (D) Immunoblot of thymic nuclear extracts from wild- type mice (+/+), mice heterozygous for the ΔCR1 allele (+/ΔCR1), homozygous for the ΔCR1 allele (ΔCR1/ΔCR1), heterozygous for the ΔHD allele (+/ΔHD), and homozygous for the ΔHD allele (ΔHD/ΔHD), probed with anti-CDP antisera, demonstrates expression and size differences of the cux/CDPΔCR1 and cux/CDPΔHD proteins in comparison with wild-type cux/CDP. Molecular weight markers at 200 kd and 97.4 kd are shown.
Generation of the cux/CDPΔHD hypomorphic allele.
(A) A targeting vector was designed to delete a 4-kb region encompassing the homeodomain within exons 20 and 21 and the intervening intron, with a PGK-neomycine resistance gene (neo), which provides a premature in-frame stop codon (*) for cux/CDP translation termination. Exons (black boxes) encoding cut repeat 3 (CR3, gray box) and the homeodomain (HD, open boxes) are shown above the wild-type gene. Primers for PCR detection of wild-type and targeted alleles with relative product sizes are shown as arrows below the schematics. The probe used for Southern detection and the fragments generated by digestion at the BamHI sites (letter “B”) are indicated. (B) Southern blot of BamHI-digested DNA from ES clones heterozygous (lanes 1 and 4) and homozygous (lanes 2 and 3) for the targeted allele. Southern blot was probed with a 4-kbBamHI/HindIII probe to detect the 8.5-kb wild-type and 4.5-kb targeted alleles. (C) Agarose gel demonstrating the products from multiplex PCR reactions to detect wild-type and knock-out alleles in wild type (+/+), heterozygous (+/ΔHD) and homozygous (ΔHD/ΔHD) mice. (D) Immunoblot of thymic nuclear extracts from wild- type mice (+/+), mice heterozygous for the ΔCR1 allele (+/ΔCR1), homozygous for the ΔCR1 allele (ΔCR1/ΔCR1), heterozygous for the ΔHD allele (+/ΔHD), and homozygous for the ΔHD allele (ΔHD/ΔHD), probed with anti-CDP antisera, demonstrates expression and size differences of the cux/CDPΔCR1 and cux/CDPΔHD proteins in comparison with wild-type cux/CDP. Molecular weight markers at 200 kd and 97.4 kd are shown.
To generate cux/CDP targeted embryonic stem cell (ES) clones, the construct was linearized with NotI and ligated to hairpin oligos to protect the ends from exonuclease digestion. The construct was electroporated into the ES cell line CJ7 and G418 (200 μg/mL) and gancyclovir (2 μM) double-resistant clones were isolated. Targeting was confirmed by a Southern blot ofBamHI digested DNA, probed with a 3-kb HindIII probe, with a diagnostic band of 4.5 kb indicating the presence of a targeted allele. Two clones were identified, which had homologously recombined the targeting construct into the homeodomain and both had a normal diploid karyotype (2n = 40 chromosomes). One clone was selected for injection into C57BL/6J blastocysts, which were transferred to 2.5-day pseudopregnant females. Chimeras were evaluated for contribution of coat color by the ES cell agouti allele. Chimeras with high ES cell contribution were backcrossed to C57BL/6J females. The genotypes of offspring were determined by Southern blot and polymerase chain reaction (PCR).
The ES clones homozygous for the targeted allele were generated from heterozygous clones by increasing G418 concentrations to 2.8 mg/mL (Figure 1B, lanes 2 and 3) and one was selected for injection into RAG-2–deficient blastocysts to analyze the effect of the mutation on lymphoid differentiation.
PCR analysis of mice genotypes
Multiplex PCR was performed to genotype mice, usingcux/CDP intron 19 sense primer HD-F (CAGGGTTTTATTTGGGGGCTTTTT), which produced a 596-bp band with exon 20 antisense primer HD-R1 (AAGTTCCTCGATGGTTTTTGGTG) from the wild-type allele, or a 1.06-kb band with neo antisense primer HD-R2 (GCATCGCCTTCTATCGCCTTCTTG) from the targeted allele. PCR reactions were performed using 32 cycles of 94°C for 30 seconds, 62°C for 30 seconds, and 72°C for 1 minute.
Mice containing a germline rearranged V(D)J segment of Ig heavy chain specific for hapten 4-hydroxy-3-nitrophenyl28 were crossed to the cux/CDP+/ΔHD mice. The presence of the recombined 17.2.25 V(D)J segment was determined by PCR with primers GTCAATTCAGAGGTTCAGCTGCAG (sense) and CCAGTAGTCCATAGCATAGTAAGG (antisense).29 PCR reactions were performed using 35 cycles of 94°C for 30 seconds, 58°C for 30 seconds, and 72°C for 30 seconds to produce a specific band of 348 bp.
Immunoblotting
Thymuses were ground on nylon mesh in phosphate-buffered saline (PBS) to obtain a single-cell suspension. Cells were pelleted and nuclear extracts were prepared as described.25 Protein concentration was determined using Bio-Rad Protein Assay reagent (Bio-Rad, Hercules, CA) in comparison with a bovine serum albumin (BSA) standard curve. Thymic nuclear extracts (20 μg) were resolved by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE; 9% polyacrylamide), electrophoretically transferred to polyvinylidene difluoride (PVDF) membrane, blocked with 7.5% nonfat dry milk, washed, and then incubated with a 1:1000 dilution of guinea pig–anti-CDP antiserum6 for 2 hours at room temperature. The blot was washed, then incubated with 1:5000 dilution of a goat anti–guinea pig (AP-conjugated) antibody (Sigma Chemical, St Louis, MO) at room temperature for 1 hour. The blot was washed, then signal was developed with nitroblue tetrazolium/5-bromo-4-chloro-3-indolyl-phosphate (NBT/BCIP; Promega, Madison, WI) for 5 minutes at room temperature.
Histochemistry and TUNEL assays
Tissues for histochemistry were fixed in freshly prepared 4% paraformaldehyde in PBS overnight, embedded in paraffin wax, sectioned at 8 μm and counterstained with hematoxylin and eosin.
Thymuses for TUNEL assays were cryopreserved and cryosectioned at 8 μm. TUNEL assays were performed according to the manufacturer's instructions (Roche Diagnostics, Indianapolis, IN). Sections were mounted in Vectashield (Vector Laboratories, Burlingame, CA) containing 1 μg/mL DAPI (Sigma) before fluorescence microscopy.
Flow cytometry of hematopoietic organs
Thymus and spleen single-cell suspensions from 2- to 5-week-old mice were obtained by grinding and filtration through nylon mesh into FACS buffer (PBS, 2% fetal bovine serum [FBS], 0.01% sodium azide, 2 mM EDTA). Marrows from femurs were flushed and cells suspended by pipetting. Cells were pelleted and resuspended in FACS buffer; then 1 to 2 × 106 cells were stained with the following antibodies: anti-B220 (RA3-6B2), anti–c-Kit (2B8), anti-CD25 (PC615.3 or 7D4), anti-IgM (polyclonal), anti–Mac-1 (M1/70), anti-CD43 (S7), anti-CD61 (2C9.G2), anti-CD4 (CT-CD4), anti-CD8 (53-6.7), anti-CD44 (IM7), and anti-CD3ε (145-2C11). Cells not staining with anti–Gr-1 (RB6-8C5), anti–Mac-1 (M1/70), anti-CD19 (1D3), anti-CD4 (H129.19), anti-CD8 (53-6.7), and anti-CD3ε (145-2C11) were defined as lineage negative in thymus.
In reconstitution experiments, antibodies anti-CD45.1 (A20) and anti-CD45.2 (104) were used to distinguish between recipient Ly-5.1 (CD45.1) and donor Ly-5.2 (CD45.2) alloantigens, respectively. The presence of the Vβ8.1 TCRβ transgene30 in offspring resulting from compound crosses with cux/CDP+/ΔHD mice was determined by flow cytometry with an anti-Vβ8 antibody (F23.1).
Annexin V stain of apoptotic B cells was performed using an annexin V fluorescein isothiocyanate (FITC) kit (Oncogene, San Diego, CA) according to the manufacturer's instructions. Cells were stained with anti-B220-APC (RA3-6B2) antibody at the same time as annexin V in the supplied binding buffer.
Most antibodies were directly conjugated with fluorochromes. In some instances biotin conjugates were used, which required a secondary layer of streptavidin-PerCP (Pharmingen, San Diego, CA). All antibodies were purchased from Pharmingen except anti-IgM (polyclonal), anti-CD25 (PC615.3), and anti-CD4 (CT-CD4), which were purchased from Caltag (Burlingame, CA). Flow cytometry was performed using the FACS Calibur (Becton Dickinson, San Jose, CA) and analysis restricted to viable cells defined by forward and side scatter.
Colony-forming assays
For myeloid/erythroid colony-forming assays, 2 × 105 bone marrow cells were seeded in methylcellulose containing erythropoietin (Epo, 2 U/mL) plus stem cell factor (SCF, 250 ng/mL) for erythroid burst-forming units (BFU-Es), or granulocyte-macrophage colony-stimulating factor (GM-CSF, 15 U/mL) plus interleukin-3 (IL-3) (15 U/mL) for granulocyte-macrophage colony-forming units (CFU-GMs). Cells were plated in triplicate and cultures were incubated in a fully humidified incubator with 5% CO2 before scoring by morphology after 14 days.
Pre-BII cell colony-forming assays were performed by culturing 2 × 104 bone marrow cells in 0.9% base methylcellulose (Stem Cell Technologies, Vancouver, BC, Canada) containing 30% FBS (Stem Cell Technologies), 1:20 dilution of conditioned media containing IL-7 (kindly provided by Steven Bauer), 2 mM l-glutamine (Gibco BRL, Rockville, MD), and 0.1 mM 2-mercaptoethanol (Gibco BRL) in Iscoves modified Dulbecco medium (IMDM; Gibco BRL). Cultures were plated in duplicate and were incubated for 7 days (as above) prior to scoring of colonies by morphology.
Frequency determination of pro-B/pre-BI colony-forming cells was performed by limiting dilution analysis. Stromal line ST231was cultured at 3 × 104 cells/mL in 96-well dishes for 2 days in IMDM (Gibco BRL) with 10% FBS, 100 U penicillin, and 100 μg streptomycin antibiotics per milliliter and 50 μM 2-mercaptoethanol. Semiconfluent stroma plates were irradiated at 3000 rads and bone marrow cells added at limiting dilution (range, 150-0.62 cells/well) in media containing a 1:20 dilution of IL-7–conditioned media (equivalent to 100-200 U/mL). Cocultures were scored for the growth of lymphocyte pro-B/pre-BI colonies containing more than 10 cells after 7 days. The frequency of pro-B/pre-BI colony precursors was determined at the value of 37% negative wells when the fraction of negative wells was plotted against cells/well, according to the Poisson distribution.32
Bone marrow reconstitution
Femurs and tibias were flushed with Hanks buffered saline solution (HBSS; Gibco BRL) containing 5% FBS. Viable cell concentration was adjusted to 5 × 106 cells/mL in HBSS with 0.5% FBS. Recipient 8- to 15-week-old C57BL/6-SJL mice, congenic for Ly-5.1 alloantigen, were irradiated with 950 rads (2 doses of 475 rads separated by 3 hours) from a 137Cs source and 1 × 106 bone marrow cells were injected into the lateral tail vein. Mice were housed in specific pathogen-free (SPF) facilities and were provided with neomycin sulfate (1.1 g/L) in drinking water. Contributions of donor cells to the reconstituted hematopoietic system were determined by flow cytometry. In most cases, reconstitution with donor cells was over 80%.
TNF enzyme-linked immunosorbent assays
Cells from bone marrow, spleen, and thymus were cultured at 5 × 106 cells/mL. Levels of secreted TNF in supernatants were measured using a mouse-specific TNF enzyme-linked immunosorbent assay (ELISA) kit according to the manufacturer's instructions (Biosource, Camarillo, CA). Bone marrow culture media was IMDM, 10% FBS, 100 U penicillin, and 100 μg streptomycin per milliliter, and 50 μM 2-mercaptoethanol. Thymocytes and splenocytes were cultured in Dulbecco modified Eagle medium (DMEM)–high glucose, 10% FBS, 55 μM 2-mercaptoethanol, 100 U penicillin and 100 μg streptomycin per milliliter, and 0.1 mM nonessential amino acids supplemented with concanavalin A (ConA; Sigma) at 6.25 mg/L. All media supplies were from Gibco BRL. Protein lysates were produced by homogenizing approximately 0.1 g tissue in 500 μL lysis buffer (25 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1 μg/mL aprotinin, 10 μg/mL leupeptin, 50 mM NaF, 1 mM sodium orthovanadate, and 2% NP40) placed on ice for 30 minutes with vortexing every 10 minutes. Nuclei were pelleted at 14 000 rpm at 4°C for 10 minutes and supernatants were frozen in aliquots. Protein concentrations were determined as previously described. Equal amounts of protein per set ofcux/CDPΔHD/ΔHD and control tissues (range, 4.5-370 μg) were evaluated for TNF expression by ELISA.
Results
Generation of the cux/CDP ΔHD hypomorphic allele
Mutation of the cux/CDP gene was performed using a targeting vector that disrupted the C-terminal homeodomain (Figure 1A). The targeted allele was predicted to express a truncated cux/CDP protein containing only 4 amino acids of the homeodomain and a deletion of the C-terminal repression domain. Two clones were identified to be heterozygous for the mutation by Southern blot analysis (Figure 1B, lanes 1 and 4). One clone was selected to generate chimeric mice that were subsequently backcrossed to the C57BL/6 strain. Heterozygous mice were healthy and fertile with no apparent phenotypic abnormalities when compared to wild-type littermates. Genotypes of mice resulting from heterozygous intercrosses were determined by genomic PCR (Figure 1C). Homozygous (cux/CDPΔHD/ΔHD) mutant mice were present at 5.4% rather than the expected mendelian ratio of 25% (1219 mice analyzed). Genotype analysis of fetuses at 16 to 17 days found a normal mendelian ratio (28%) of homozygous mutants, demonstrating that death occurs at or shortly after birth.
Because the cux/CDP homeodomain was targeted for truncation, we investigated whether a truncated protein was synthesized. An immunoblot of thymic nuclear extracts from wild-type (+/+), heterozygous (+/ΔHD), and homozygous (ΔHD/ΔHD) mice were probed with an anti-CDP antiserum (Figure 1D). For size comparison, thymic extracts from heterozygous (+/ΔCR1) and homozygous (ΔCR1/ΔCR1) mice with a deleted CR1 domain (B6,129-Cutl1tm1Ejn) were included.27 A band smaller in size than wild-type CDP (180-190 kd) and ΔCR1 protein (160-170 kd) was observed in lanes containing thymic extracts from cux/CDP+/ΔHDand cux/CDPΔHD/ΔHD mice, which corresponded in size to the predicted cux/CDPΔHD protein (150-160 kd) (Figure 1D). The intensity of the cux/CDPΔHD protein band was approximately 5-fold lower than the wild-type protein band. This suggests that the targeted mutation reduces the level of transcription or translation of the mutated allele, or results in the generation of unstable truncated messenger RNA (mRNA) or protein. Given that the truncated cux/CDPΔHD protein retains the CR DNA binding domains, it is possible the remaining protein has partial function (ie, a hypomorph).
cux/CDPΔHD/ΔHDmice have pleiotropic developmental abnormalities
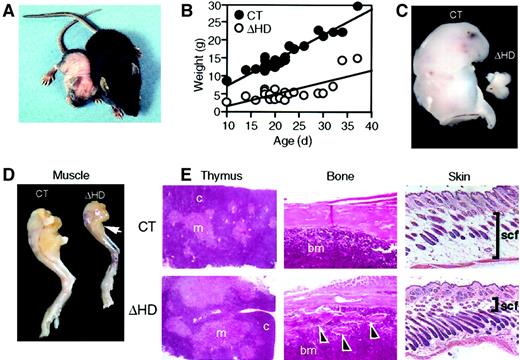
Surviving cux/CDPΔHD/ΔHD mice are distinguishable from their heterozygous and wild-type (control) littermates as early as 3 to 4 days postpartum due to their reduced size and skin pallor. These mice have difficulty thriving, have a reduced stature, gain little weight, have wavy whiskers, and lose fur between 2 and 3 weeks of age (Figure2A-B). A sexual dimorphism in mutant mice viability was observed with 89.4% of the mutants surviving neonatal lethality being phenotypically male. Few mice lived beyond 4 weeks of age. Anatomic analysis of cux/CDPΔHD/ΔHDmice revealed that the size and cellularity of the thymus was dramatically reduced in comparison with control littermates (Figure 2C and Table 1) with a preferential loss of thymic cortex (Figure 2E). Furthermore, these mice suffered from cachexia due to muscle wasting (Figure 2D) and loss of body fat, including subcutaneous fat (Figure 2E). Bones appeared thin and flaky (Figure 2E) and the hair follicles within skin appeared greater in number and somewhat disorganized (Figure 2E). No obvious explanation for the neonatal lethality was apparent by histologic examination of vital organs.
cux/CDPΔHD/ΔHD mice have pleiotropic abnormalities.
(A) Comparison of 14-day-old male littermates heterozygous (right) and homozygous (left) for the cux/CDPΔHD allele. (B) Comparative graph of the relationship between age and weights of wild-type/heterozygous (CT) and homozygous (ΔHD) littermates aged between 10 and 37 days. Comparison of thymus (C) and (D) hind leg sizes fromcux/CDPΔHD/ΔHD and control littermates. Arrow indicates reduced size of calf muscle. (E) Histologic comparisons of cux/CDPΔHD/ΔHD(ΔHD) andcux/CDP+/ΔHD (CT) thymus, femur (arrowheads show bone fragmentation), and skin (bars show thickness of subcutaneous fat); c indicates cortex; m, medulla; bm, bone marrow; scf, subcutaneous fat. Magnification 100 ×.
cux/CDPΔHD/ΔHD mice have pleiotropic abnormalities.
(A) Comparison of 14-day-old male littermates heterozygous (right) and homozygous (left) for the cux/CDPΔHD allele. (B) Comparative graph of the relationship between age and weights of wild-type/heterozygous (CT) and homozygous (ΔHD) littermates aged between 10 and 37 days. Comparison of thymus (C) and (D) hind leg sizes fromcux/CDPΔHD/ΔHD and control littermates. Arrow indicates reduced size of calf muscle. (E) Histologic comparisons of cux/CDPΔHD/ΔHD(ΔHD) andcux/CDP+/ΔHD (CT) thymus, femur (arrowheads show bone fragmentation), and skin (bars show thickness of subcutaneous fat); c indicates cortex; m, medulla; bm, bone marrow; scf, subcutaneous fat. Magnification 100 ×.
Flow cytometric evaluation of lymphoid and myeloid populations in primary cux/CDPΔHD/ΔHD mice, crossed to IgH “knock-in” and TCRβ transgenics
| Populations* . | CT . | ΔHD . | IgH knock-in CT1-153 . | IgH knock-in ΔHD1-153 . | TCR-β TG CT1-153 . | TCR-β TG ΔHD1-153 . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| % cells† . | No. cells × 105‡ . | % cells† . | No. cells × 105‡ . | % cells† . | No. cells × 105‡ . | % cells† . | No. cells × 105‡ . | % cells† . | No. cells × 105‡ . | % cells† . | No. cells × 105‡ . | |
| Bone marrow | N/A | (14.2 ± 3.7) | N/A | (16.7 ± 3.1) | N/A | (21.1 ± 2.4) | N/A | (18.7 ± 0.7) | ND | ND | ND | ND |
| B220+ | 37.8 ± 6.5 | (5.0 ± 1.2) | 15.1 ± 7.21-155 | (2.6 ± 1.3)1-155 | 37.5 ± 3.1 | (8.0 ± 1.5) | 13.7 ± 5.31-154 | (2.5 ± 0.9)1-154 | ND | ND | ND | ND |
| B220+c-Kit+ | 5.7 ± 1.1 | (0.8 ± 0.2) | 3.4 ± 0.91-155 | (0.6 ± 0.1) | 2.4 ± 0.4 | (0.5 ± 0.1) | 2.4 ± 0.8 | (0.4 ± 0.1) | ND | ND | ND | ND |
| B220+CD25+ | 12.8 ± 3.0 | (1.9 ± 0.6) | 3.7 ± 2.01-155 | (0.6 ± 0.3)1-155 | 15.7 ± 3.4 | (3.4 ± 1.0) | 3.9 ± 3.01-154 | (0.7 ± 0.5)1-154 | ND | ND | ND | ND |
| B220+IgM+ | 12.7 ± 2.6 | (1.6 ± 0.4) | 7.8 ± 3.11-155 | (1.2 ± 0.5) | 9.1 ± 2.0 | (1.9 ± 0.4) | 3.8 ± 1.51-154 | (0.7 ± 0.3)1-154 | ND | ND | ND | ND |
| Mac-1+B220− | 28.3 ± 4.6 | (4.2 ± 1.2) | 55.8 ± 7.21-155 | (9.2 ± 2.3)1-155 | 38.8 ± 1.0 | (8.2 ± 1.1) | 58.6 ± 8.61-154 | (11.0 ± 1.9) | ND | ND | ND | ND |
| Spleen | N/A | (109 ± 31) | N/A | (75 ± 63) | N/A | (70.0 ± 24.4) | N/A | (82.1 ± 38.2) | N/A | ND | N/A | ND |
| B220+ | 59.2 ± 7.3 | (64.2 ± 6.2) | 47.6 ± 8.2 | (46.4 ± 20.7) | 35.2 ± 5.8 | (23.2 ± 3.9) | 20.6 ± 3.71-154 | (16.6 ± 7.8) | ND | ND | ND | ND |
| Mac-1+B220− | 5.3 ± 0.7 | (6.0 ± 1.7) | 18.6 ± 6.41-154 | (20.4 ± 14.8) | 10.4 ± 3.1 | (6.6 ± 0.9) | 20.5 ± 10.1 | (13.1 ± 4.8) | ND | ND | ND | ND |
| CD4+ | 6.9 ± 0.9 | (6.8 ± 1.6) | 12.5 ± 4.31-154 | (9.0 ± 4.4) | ND | ND | ND | ND | ND | ND | ND | ND |
| CD8+ | 2.7 ± 0.6 | (2.7 ± 0.5) | 4.1 ± 1.4 | (3.2 ± 1.8) | ND | ND | ND | ND | ND | ND | ND | ND |
| Thymus | N/A | (202 ± 97) | N/A | (43 ± 44)1-155 | ND | ND | ND | ND | N/A | (140 ± 30.8) | N/A | (10 ± 16)1-155 |
| lin−CD4−CD8− | 2.0 ± 0.6 | (3.9 ± 1.7) | 3.0 ± 2.8 | (0.8 ± 0.4)1-154 | ND | ND | ND | ND | 5.8 ± 2.2 | (8.2 ± 3.7) | 9.0 ± 6.2 | (0.5 ± 0.5)1-155 |
| CD4+CD8+ | 83.8 ± 2.8 | (171 ± 84) | 61.8 ± 28.71-154 | (37 ± 38)1-155 | ND | ND | ND | ND | 82.8 ± 5.2 | (113 ± 17.8) | 54.2 ± 33.8 | (6.1 ± 12)1-155 |
| CD4+CD8− | 9.9 ± 1.7 | (19.6 ± 9.3) | 24.4 ± 181-155 | (13.4 ± 22) | ND | ND | ND | ND | 8.7 ± 3.5 | (13.6 ± 6.9) | 29.8 ± 23.8 | (2.8 ± 3.4)1-154 |
| CD4−CD8+ | 3.94 ± 1.2 | (7.7 ± 3.7) | 8.3 ± 8.9 | (1.6 ± 1.0)1-155 | ND | ND | ND | ND | 5.5 ± 3.4 | (8.4 ± 6.6) | 11.1 ± 9.3 | (0.9 ± 0.9)1-154 |
| CD3intCD25int | 2.4 ± 0.9 | (4.8 ± 3.3) | 16.2 ± 5.51-155 | (9.5 ± 11.3) | ND | ND | ND | ND | ND | ND | ND | ND |
| Populations* . | CT . | ΔHD . | IgH knock-in CT1-153 . | IgH knock-in ΔHD1-153 . | TCR-β TG CT1-153 . | TCR-β TG ΔHD1-153 . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| % cells† . | No. cells × 105‡ . | % cells† . | No. cells × 105‡ . | % cells† . | No. cells × 105‡ . | % cells† . | No. cells × 105‡ . | % cells† . | No. cells × 105‡ . | % cells† . | No. cells × 105‡ . | |
| Bone marrow | N/A | (14.2 ± 3.7) | N/A | (16.7 ± 3.1) | N/A | (21.1 ± 2.4) | N/A | (18.7 ± 0.7) | ND | ND | ND | ND |
| B220+ | 37.8 ± 6.5 | (5.0 ± 1.2) | 15.1 ± 7.21-155 | (2.6 ± 1.3)1-155 | 37.5 ± 3.1 | (8.0 ± 1.5) | 13.7 ± 5.31-154 | (2.5 ± 0.9)1-154 | ND | ND | ND | ND |
| B220+c-Kit+ | 5.7 ± 1.1 | (0.8 ± 0.2) | 3.4 ± 0.91-155 | (0.6 ± 0.1) | 2.4 ± 0.4 | (0.5 ± 0.1) | 2.4 ± 0.8 | (0.4 ± 0.1) | ND | ND | ND | ND |
| B220+CD25+ | 12.8 ± 3.0 | (1.9 ± 0.6) | 3.7 ± 2.01-155 | (0.6 ± 0.3)1-155 | 15.7 ± 3.4 | (3.4 ± 1.0) | 3.9 ± 3.01-154 | (0.7 ± 0.5)1-154 | ND | ND | ND | ND |
| B220+IgM+ | 12.7 ± 2.6 | (1.6 ± 0.4) | 7.8 ± 3.11-155 | (1.2 ± 0.5) | 9.1 ± 2.0 | (1.9 ± 0.4) | 3.8 ± 1.51-154 | (0.7 ± 0.3)1-154 | ND | ND | ND | ND |
| Mac-1+B220− | 28.3 ± 4.6 | (4.2 ± 1.2) | 55.8 ± 7.21-155 | (9.2 ± 2.3)1-155 | 38.8 ± 1.0 | (8.2 ± 1.1) | 58.6 ± 8.61-154 | (11.0 ± 1.9) | ND | ND | ND | ND |
| Spleen | N/A | (109 ± 31) | N/A | (75 ± 63) | N/A | (70.0 ± 24.4) | N/A | (82.1 ± 38.2) | N/A | ND | N/A | ND |
| B220+ | 59.2 ± 7.3 | (64.2 ± 6.2) | 47.6 ± 8.2 | (46.4 ± 20.7) | 35.2 ± 5.8 | (23.2 ± 3.9) | 20.6 ± 3.71-154 | (16.6 ± 7.8) | ND | ND | ND | ND |
| Mac-1+B220− | 5.3 ± 0.7 | (6.0 ± 1.7) | 18.6 ± 6.41-154 | (20.4 ± 14.8) | 10.4 ± 3.1 | (6.6 ± 0.9) | 20.5 ± 10.1 | (13.1 ± 4.8) | ND | ND | ND | ND |
| CD4+ | 6.9 ± 0.9 | (6.8 ± 1.6) | 12.5 ± 4.31-154 | (9.0 ± 4.4) | ND | ND | ND | ND | ND | ND | ND | ND |
| CD8+ | 2.7 ± 0.6 | (2.7 ± 0.5) | 4.1 ± 1.4 | (3.2 ± 1.8) | ND | ND | ND | ND | ND | ND | ND | ND |
| Thymus | N/A | (202 ± 97) | N/A | (43 ± 44)1-155 | ND | ND | ND | ND | N/A | (140 ± 30.8) | N/A | (10 ± 16)1-155 |
| lin−CD4−CD8− | 2.0 ± 0.6 | (3.9 ± 1.7) | 3.0 ± 2.8 | (0.8 ± 0.4)1-154 | ND | ND | ND | ND | 5.8 ± 2.2 | (8.2 ± 3.7) | 9.0 ± 6.2 | (0.5 ± 0.5)1-155 |
| CD4+CD8+ | 83.8 ± 2.8 | (171 ± 84) | 61.8 ± 28.71-154 | (37 ± 38)1-155 | ND | ND | ND | ND | 82.8 ± 5.2 | (113 ± 17.8) | 54.2 ± 33.8 | (6.1 ± 12)1-155 |
| CD4+CD8− | 9.9 ± 1.7 | (19.6 ± 9.3) | 24.4 ± 181-155 | (13.4 ± 22) | ND | ND | ND | ND | 8.7 ± 3.5 | (13.6 ± 6.9) | 29.8 ± 23.8 | (2.8 ± 3.4)1-154 |
| CD4−CD8+ | 3.94 ± 1.2 | (7.7 ± 3.7) | 8.3 ± 8.9 | (1.6 ± 1.0)1-155 | ND | ND | ND | ND | 5.5 ± 3.4 | (8.4 ± 6.6) | 11.1 ± 9.3 | (0.9 ± 0.9)1-154 |
| CD3intCD25int | 2.4 ± 0.9 | (4.8 ± 3.3) | 16.2 ± 5.51-155 | (9.5 ± 11.3) | ND | ND | ND | ND | ND | ND | ND | ND |
N/A indicates not applicable; ND, no data.
B and T lymphoid and myeloid cell populations in cux/CDPΔHD/ΔHD (ΔHD) and control wild-type/heterozygous littermate (CT) hematopoietic tissues were analyzed by flow cytometry using lineage- and stage-specific antibodies.
Data presented are the percentages of cells staining positive for specific surface antigens within the population analyzed (viable gate for bone marrow and spleen; viable lymphoid gate for thymus).
Population cell numbers (× 105), shown in parentheses, are normalized for body weight (spleen and thymus) and additionally per femur (bone marrow).
Population percentages and cell numbers are also shown for cux/CDPΔHD/ΔHD and control mice crossed to the IgH “knock-in” (Ig) and TCRβ (TCR) transgenic lines.
P < .005.
P < .05.
cux/CDPΔHD/ΔHDmice have defects in lymphoid and myeloid development
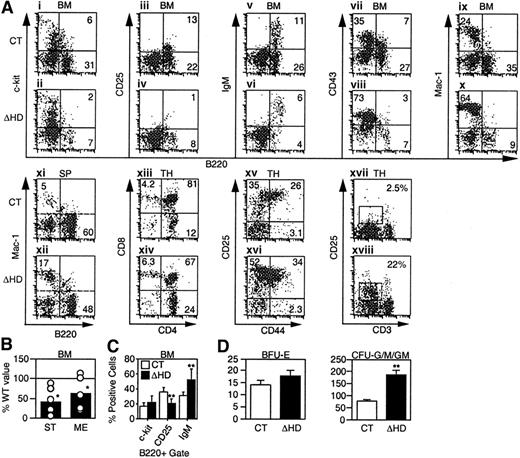
We investigated the effects of the cux/CDPΔHD mutation on lymphopoiesis and myelopoiesis by flow cytometry in mice that survived the neonatal period. Analysis of mutant bone marrow with pan-B lineage-specific marker B220 identified a 2- to 3-fold reduction in the percentage and absolute numbers of B cells when compared to littermate controls (Figure 3A and Table 1). Analysis of c-Kit (Figure 3Ai-ii,), CD25 (Figure 3Aiii-iv), and IgM (Figure 3Av-vi) within the B220 population to identify pro-B/pre-BI, pre-BII and immature/mature B-lineage cells indicated that the percentages of all B-lineage subpopulations were reduced in mutant bone marrow (Table 1). This overall reduction was also observed in pro-B/pre-BI and pre-BII colony-forming assays (Figure 3B). Although all B-lineage subsets were reduced in number to some degree, the CD25+ pre-BII population was preferentially affected. When a B220+ gate was used, the CD25+ population was reduced 2-fold in comparison with other B-lineage populations (Figure3C). Decreases in these B-lineage populations could arise due to suppressed differentiation or proliferation, or enhanced apoptosis.
cux/CDP ΔHD/ΔHD mice have defects in lymphoid and myeloid development.
Flow cytometry for lineage- and stage-specific cell surface markers was performed on cells isolated from wild-type/heterozygous (CT) and homozygous (ΔHD) bone marrow (BM), spleen (SP), and thymus (TH) from mice 2 to 5 weeks of age. (A) Representative FACS plots for each set of markers are shown for cells within viable gates for bone marrow and spleen and a viable lymphocyte gate for thymus. Analysis of CD25 and CD44 staining in thymus also included a lineage-negative gate. (B) Frequencies of pre-B colony-forming units in control and homozygous bone marrow were determined using stroma and IL-7 (ST) for pro-B/pre-BI cells and methylcellulose and IL-7 (ME) for pre-BII cells. Data are expressed as the percentage ofcux/CDPΔHD/ΔHD colonies obtained relative to control samples. (C) Comparison of the percentages of B-lineage bone marrow cells within a B220+ viable gate staining positive for differentiation-specific antigens c-Kit (pro-B/pre-BI), CD25 (pre-BII), and IgM (immature/mature). (D) Comparison of the numbers of erythroid (BFU-E) and myeloid colony-forming units (CFU-G/M/GM), which include granulocyte, macrophage, and mixed colonies. Studentt test P less than .05 is indicated with one asterisk and less than .005 with 2 asterisks.
cux/CDP ΔHD/ΔHD mice have defects in lymphoid and myeloid development.
Flow cytometry for lineage- and stage-specific cell surface markers was performed on cells isolated from wild-type/heterozygous (CT) and homozygous (ΔHD) bone marrow (BM), spleen (SP), and thymus (TH) from mice 2 to 5 weeks of age. (A) Representative FACS plots for each set of markers are shown for cells within viable gates for bone marrow and spleen and a viable lymphocyte gate for thymus. Analysis of CD25 and CD44 staining in thymus also included a lineage-negative gate. (B) Frequencies of pre-B colony-forming units in control and homozygous bone marrow were determined using stroma and IL-7 (ST) for pro-B/pre-BI cells and methylcellulose and IL-7 (ME) for pre-BII cells. Data are expressed as the percentage ofcux/CDPΔHD/ΔHD colonies obtained relative to control samples. (C) Comparison of the percentages of B-lineage bone marrow cells within a B220+ viable gate staining positive for differentiation-specific antigens c-Kit (pro-B/pre-BI), CD25 (pre-BII), and IgM (immature/mature). (D) Comparison of the numbers of erythroid (BFU-E) and myeloid colony-forming units (CFU-G/M/GM), which include granulocyte, macrophage, and mixed colonies. Studentt test P less than .05 is indicated with one asterisk and less than .005 with 2 asterisks.
In the thymus, an average 5-fold reduction (1.2- to 64-fold range, n = 17) in the number of thymocytes per gram body weight was observed (Table 1). Although each of the CD4/CD8 populations were reduced in number, the CD4+CD8+ population was preferentially affected (Figure 3Axiii-xiv, and Table 1). Within the lin−CD4−CD8− compartment, an accumulation of the CD25+ subset was observed in half of the mice (Figure 3Axv-xvi). Interestingly, although the thymus cellularity was dramatically reduced, the number of T cells in the spleen was not affected (Table 1).
In normal mice, the early expression of CD25 disappears in the CD4−CD8− population and is not observed in the more mature CD4+CD8+ or single positive populations where the CD3/TCR begins to be expressed. In contrast,cux/CDPΔHD/ΔHD mice retained CD25 expression in these more mature cells in conjunction with intermediate CD3 expression (Figure 3Axvii-xviii, and Table 1). These data suggest that CDP may regulate the expression of CD25 in T lymphocytes.
Reductions in lymphoid populations at the pre-BII and CD4+CD8+ stages of development could arise ifcux/CDPΔHD/ΔHD mice were defective in IgH or TCRβ antigen receptor rearrangement, due to the process of heavy chain selection. To determine whether the provision of recombined antigen receptors could rescuecux/CDPΔHD/ΔHD B- and T-cell development,cux/CDP+/ΔHD mice were crossed to an IgH “knock-in” line28 or a TCRβ transgenic line.30 Flow cytometric evaluation of B and T lymphopoiesis identified similar reductions in B220+populations in bone marrow and CD4/CD8 populations in the thymus, regardless of whether the mice carried an IgH or TCRβ transgene (Table 1). These data argue against the hypothesis that defective antigen receptor rearrangement is responsible for the changes in lymphocyte subsets.
In contrast to the decrease in lymphocytes incux/CDPΔHD/HD mice, an increase in the percentage and numbers of myeloid cells was observed in bone marrow, spleen (Figure 3A,D and Table 1), and peripheral blood (data not shown). Increases in cells positive for myeloid markers CD43 (Figure 3Avii-viii), Mac-1 (Figure 3Aix-x), CD61 (not shown), and Gr-1 (not shown) in bone marrow, and Mac-1 (Figure 3Axi-xii) and Gr-1 (not shown) in spleen were observed (summarized in Table 1). Myeloid progenitors capable of colony formation in vitro were also expanded 2- to 3-fold within cux/CDPΔHD/ΔHD bone marrow (Figure3D). Flow cytometry confirmed that c-Kit+Mac-1+ cells were expanded 2- to 3-fold in mutant bone marrow (data not shown). In contrast, erythroid progenitors were not increased (Figure 3D). Taken together, these data suggest that cux/CDP plays opposing roles in regulating lymphoid and myeloid development.
Cell-intrinsic and environmental effects cause hematopoietic defects in cux/CDPΔHD/ΔHDmice
The changes observed in lymphoid and myeloid populations incux/CDPΔHD/ΔHD mice could be due to cell-intrinsic or microenvironmental influences. To distinguish between these possibilities, bone marrow cells fromcux/CDPΔHD/ΔHD mice and littermate controls were transplanted into lethally irradiated C57BL/6-SJL recipients. Reconstitution was assayed at 4 and 8 weeks and 10 to 11 months after transplantation. Donor cells were distinguished from recipient cells using antibodies for the Ly-5 alloantigens.
At all time points, the total numbers and percentages of B220+ B-cells in bone marrow were reduced in mice reconstituted with cux/CDPΔHD/ΔHD bone marrow compared to control bone marrow (Figure4A-D, Table2, and data not shown). However, analysis of the subpopulations within a B220+ gate failed to reveal preferential effects on the pre-BII population within the reconstituted mice as was seen in primary mutant animals (data not shown). These data suggest that both cell intrinsic and environmental influences affect B cells in cux/CDP mutant mice.
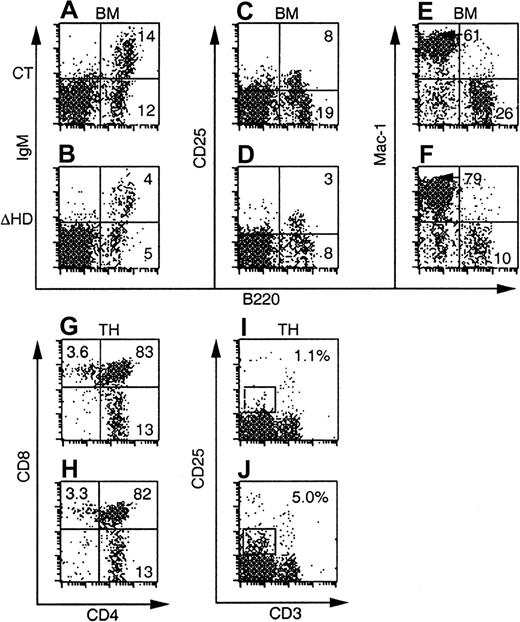
Many of the hematopoietic defects incux/CDPΔHD/ΔHD mice are not cell intrinsic.
FACS plots of bone marrow and thymus from C57BL/6-SJL mice reconstituted with control orcux/CDPΔHD/ΔHD mice and analyzed 8 weeks after transplantation are shown. Ly-5.2+donor-derived cells were analyzed for the expression of lineage- and stage-specific antigens within a viable gate for bone marrow and a viable lymphocyte gate for thymus. Plots for CD25/B220 were obtained from mice reconstituted for 10 to 11 months.
Many of the hematopoietic defects incux/CDPΔHD/ΔHD mice are not cell intrinsic.
FACS plots of bone marrow and thymus from C57BL/6-SJL mice reconstituted with control orcux/CDPΔHD/ΔHD mice and analyzed 8 weeks after transplantation are shown. Ly-5.2+donor-derived cells were analyzed for the expression of lineage- and stage-specific antigens within a viable gate for bone marrow and a viable lymphocyte gate for thymus. Plots for CD25/B220 were obtained from mice reconstituted for 10 to 11 months.
Flow cytometric evaluation of lymphoid and myeloid populations in mice repopulated with cux/CDPΔHD/ΔHDbone marrow
| Populations* . | CT . | ΔHD . | ||
|---|---|---|---|---|
| % cells† . | No. cells × 107‡ . | % cells† . | No. cells × 107‡ . | |
| Bone marrow | N/A | (3.29 ± 0.42) | N/A | (3.11 ± 0.39) |
| B220+ | 37.3 ± 3.2 | (1.16 ± 0.21) | 22.4 ± 1.72-153 | (0.67 ± 0.09)2-153 |
| B220+c-Kit+ | 4.9 ± 1.2 | (0.16 ± 0.06) | 3.2 ± 0.82-155 | (0.09 ± 0.02)2-153 |
| B220+CD25+ | 4.8 ± 2.9 | (0.20 ± 0.15) | 2.0 ± 1.0 | (0.07 ± 0.04) |
| B220+IgM+ | 16.3 ± 3.0 | (0.52 ± 0.16) | 8.1 ± 0.52-153 | (0.24 ± 0.04)2-153 |
| Mac-1+B220− | 52.8 ± 2.3 | (1.64 ± 0.25) | 68.3 ± 5.02-153 | (2.05 ± 0.37) |
| Spleen | N/A | (10.5 ± 3.8) | N/A | (13.3 ± 4.7) |
| B220+ | 70.0 ± 2.9 | (7.4 ± 3.0) | 69.4 ± 4.6 | (9.2 ± 3.4) |
| Mac-1+B220− | 4.7 ± 1.1 | (0.48 ± 0.22) | 5.9 ± 2.1 | (0.74 ± 0.36) |
| CD4+ | 16.1 ± 3.9 | (1.4 ± 0.4) | 16.7 ± 4.1 | (1.84 ± 0.8) |
| CD8+ | 6.5 ± 1.1 | (0.55 ± 0.21) | 5.7 ± 1.9 | (0.77 ± 0.33) |
| Thymus | N/A | (11.3 ± 2.3) | N/A | (9.2 ± 2.9) |
| lin−CD4−CD8− | 0.8 ± 0.1 | (0.1 ± 0.0) | 0.7 ± 0.3 | (0.1 ± 0.0) |
| CD4+CD8+ | 83 ± 0.9 | (9.3 ± 1.9) | 86 ± 1.82-155 | (7.9 ± 2.5) |
| CD4+CD8− | 12 ± 1.1 | (1.3 ± 0.3) | 9.9 ± 1.52-155 | (0.9 ± 0.2)2-155 |
| CD4−CD8+ | 3.8 ± 0.6 | (0.4 ± 0.1) | 3.3 ± 0.7 | (0.3 ± 0.1) |
| CD3intCD25int | 1.0 ± 0.2 | (0.1 ± 0.0) | 3.9 ± 0.32-153 | (0.4 ± 0.1)2-153 |
| Populations* . | CT . | ΔHD . | ||
|---|---|---|---|---|
| % cells† . | No. cells × 107‡ . | % cells† . | No. cells × 107‡ . | |
| Bone marrow | N/A | (3.29 ± 0.42) | N/A | (3.11 ± 0.39) |
| B220+ | 37.3 ± 3.2 | (1.16 ± 0.21) | 22.4 ± 1.72-153 | (0.67 ± 0.09)2-153 |
| B220+c-Kit+ | 4.9 ± 1.2 | (0.16 ± 0.06) | 3.2 ± 0.82-155 | (0.09 ± 0.02)2-153 |
| B220+CD25+ | 4.8 ± 2.9 | (0.20 ± 0.15) | 2.0 ± 1.0 | (0.07 ± 0.04) |
| B220+IgM+ | 16.3 ± 3.0 | (0.52 ± 0.16) | 8.1 ± 0.52-153 | (0.24 ± 0.04)2-153 |
| Mac-1+B220− | 52.8 ± 2.3 | (1.64 ± 0.25) | 68.3 ± 5.02-153 | (2.05 ± 0.37) |
| Spleen | N/A | (10.5 ± 3.8) | N/A | (13.3 ± 4.7) |
| B220+ | 70.0 ± 2.9 | (7.4 ± 3.0) | 69.4 ± 4.6 | (9.2 ± 3.4) |
| Mac-1+B220− | 4.7 ± 1.1 | (0.48 ± 0.22) | 5.9 ± 2.1 | (0.74 ± 0.36) |
| CD4+ | 16.1 ± 3.9 | (1.4 ± 0.4) | 16.7 ± 4.1 | (1.84 ± 0.8) |
| CD8+ | 6.5 ± 1.1 | (0.55 ± 0.21) | 5.7 ± 1.9 | (0.77 ± 0.33) |
| Thymus | N/A | (11.3 ± 2.3) | N/A | (9.2 ± 2.9) |
| lin−CD4−CD8− | 0.8 ± 0.1 | (0.1 ± 0.0) | 0.7 ± 0.3 | (0.1 ± 0.0) |
| CD4+CD8+ | 83 ± 0.9 | (9.3 ± 1.9) | 86 ± 1.82-155 | (7.9 ± 2.5) |
| CD4+CD8− | 12 ± 1.1 | (1.3 ± 0.3) | 9.9 ± 1.52-155 | (0.9 ± 0.2)2-155 |
| CD4−CD8+ | 3.8 ± 0.6 | (0.4 ± 0.1) | 3.3 ± 0.7 | (0.3 ± 0.1) |
| CD3intCD25int | 1.0 ± 0.2 | (0.1 ± 0.0) | 3.9 ± 0.32-153 | (0.4 ± 0.1)2-153 |
N/A indicates not applicable.
Hematopoietic tissues from six 8-week-posttransplantation recipients (10-11 months for B220+CD25+populations) reconstituted with cux/CDPΔHD/ΔHD (ΔHD) and control (CT) bone marrow were analyzed by flow cytometry using lineage- and stage-specific antibodies. Donor cells were identified using antibodies to donor-specific alloantigen Ly-5.2 and subsequent population analyses were performed on an Ly-5.2–positive gate.
Data presented are the percentages of cells staining positive for specific surface antigens within the total Ly-5.2–positive population analyzed.
Population cell numbers (× 107) are shown in parentheses and bone marrow data are expressed on a per femur basis.
P < .005.
P < .05.
Thymuses from mice reconstituted withcux/CDPΔHD/ΔHD bone marrow at all time points analyzed showed no reduction in thymus cellularity (Table 2 and data not shown). This strongly contrasted with the dramatic thymic phenotype in primary cux/CDPΔHD/ΔHD mice. Analysis of various stages of thymocyte development by flow cytometry demonstrated no differences in the CD4 and CD8 subpopulations in mice reconstituted with either control orcux/CDPΔHD/ΔHD bone marrow (Figure 4G-H, and Table 2). However, the increased percentages and absolute numbers of CD25+ thymocytes, intermediate for CD3 staining seen in primary mice were observed in thecux/CDPΔHD/ΔHD reconstituted mice (Figure 4I-J, and Table 2). These data support the notion that the diminution of primary cux/CDPΔHD/ΔHD thymuses is due to environmental effects, whereas the overrepresentation of CD25+CD3intermediate thymocytes is due to cell-intrinsic influences. These data again implicate cux/CDP in CD25 repression in thymocytes undergoing selection.
Bone marrow myeloid cells positive for Mac-1+ (Figure 4 and Table 2), CD43+ (not shown) or CD61+ (not shown) but negative for B220, were again increased in percentage in mice reconstituted with cux/CDPΔHD/ΔHD bone marrow. This increase was less dramatic than that in primarycux/CDPΔHD/ΔHD mice and parallels the reduced phenotype of the lymphoid compartment in reconstituted mice. Taken together, these transplantation experiments suggest that in addition to cell-intrinsic effects of the ΔHD mutation, strong environmental influences cause reductions in the lymphoid compartment and stimulation of myelopoiesis.
Levels of thymic and bone marrow B-cell apoptosis are elevated incux/CDPΔHD/ΔHDmice
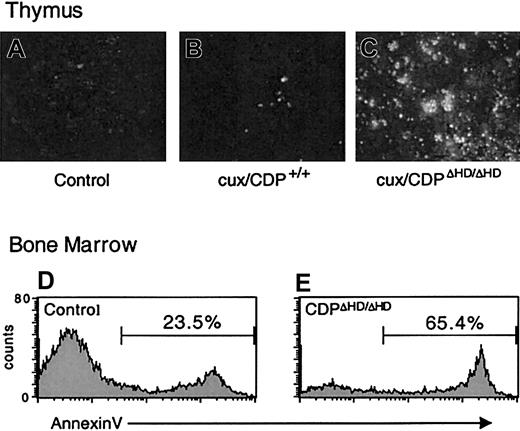
Because cell-intrinsic developmental defects did not appear to be responsible for the dramatic thymus size reduction or bone marrow B-cell loss in primary cux/CDPΔHD/ΔHD mice, we sought to determine whether these effects were due to enhanced cell death. TUNEL assays on cux/CDPΔHD/ΔHD thymus sections indicated that the level of apoptosis was dramatically increased in the thymus (Figure 5A-C). Furthermore, analysis of B220+ B cells in bone marrow by annexin V staining demonstrated that B-cell apoptosis was elevated 2- to 3-fold in primary cux/CDPΔHD/ΔHD mice (Figure 5D-E). No difference was seen between the percentages of annexin V+ cells in the B220− control andcux/CDPΔHD/ΔHD bone marrow populations indicating that the enhanced apoptosis was confined to the B-cell compartment (data not shown). Therefore, enhanced cell death is responsible for thymic atrophy and loss of bone marrow B cells in primary cux/CDPΔHD/ΔHD mice.
Levels of apoptosis are elevated incux/CDPΔHD/ΔHD thymuses and bone marrow B cells.
TUNEL apoptosis assay on sections from wild-type andcux/CDPΔHD/ΔHD thymuses demonstrates enhanced apoptosis in cux/CDPΔHD/ΔHDthymuses. The cux/CDPΔHD/ΔHD thymuses were incubated without (A) or with (C) TdT, and (B) wild-type thymus with TdT. Histograms of B-lineage cells in a B220+ gate from control (D) and cux/CDPΔHD/ΔHD (E) bone marrow stained with annexin V are shown. The percentages of annexin V+ cells within the B220+ gate from a second pair of mice were control, 20.9% andcux/CDPΔHD/ΔHD, 39.4%.
Levels of apoptosis are elevated incux/CDPΔHD/ΔHD thymuses and bone marrow B cells.
TUNEL apoptosis assay on sections from wild-type andcux/CDPΔHD/ΔHD thymuses demonstrates enhanced apoptosis in cux/CDPΔHD/ΔHDthymuses. The cux/CDPΔHD/ΔHD thymuses were incubated without (A) or with (C) TdT, and (B) wild-type thymus with TdT. Histograms of B-lineage cells in a B220+ gate from control (D) and cux/CDPΔHD/ΔHD (E) bone marrow stained with annexin V are shown. The percentages of annexin V+ cells within the B220+ gate from a second pair of mice were control, 20.9% andcux/CDPΔHD/ΔHD, 39.4%.
TNF levels are elevated incux/CDPΔHD/ΔHDmice
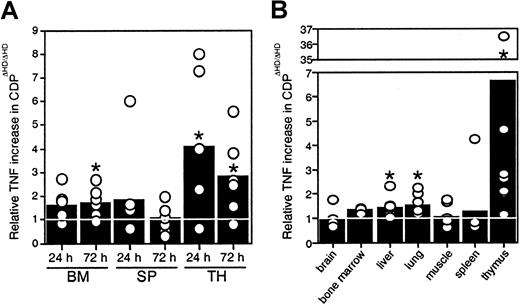
Because no thymic atrophy was seen in mice reconstituted withcux/CDPΔHD/ΔHD bone marrow, our data indicated that microenvironmental effects were responsible for the thymic phenotype, suggesting that cux/CDP might be repressing expression of a death-inducing factor or stimulating the expression of a survival factor in normal mice. Many hematopoietic and nonhematopoietic anomalies in thecux/CDPΔHD/ΔHD mice are similar to those observed under specific conditions of TNF overexpression, particularly the alopecia, cachexia, lymphopenia, and myeloid hyperplasia.33 34 Therefore, we hypothesized that overexpression of TNF may be contributing to these abnormalities. To determine whether hematopoietic organs were overexpressing TNF, bone marrow cells, splenocytes, and thymocytes fromcux/CDPΔHD/ΔHD and littermate controls were cultured and levels of TNF secretion quantified by ELISA. Significantly higher levels of TNF were observed in culture supernatants fromcux/CDPΔHD/ΔHD bone marrow (1.5- to 2-fold at 72 hours) and thymus (3- to 4-fold) (Figure6A). TNF levels were also elevated in protein extracts from hematopoietic and nonhematopoietic tissues. Although variable, TNF protein levels were significantly higher incux/CDPΔHD/ΔHD liver, lung, and thymus relative to control tissues (Figure 6B). TNF levels in brain, muscle, spleen, bone marrow, small intestine, heart, kidney, skin, and stomach were not significantly different from control tissues (Figure 6B and data not shown). Thymuses fromcux/CDPΔHD/ΔHD mice expressed the highest levels of TNF, approximately 3- to 7-fold higher than controls (Figure6B). These data suggest that TNF could be a target gene for cux/CDP-mediated repression and that TNF overexpression in hematopoietic and nonhematopoietic tissues could be partly responsible for the phenotypes observed incux/CDPΔHD/ΔHD animals.
Levels of TNF expression incux/CDPΔHD/ΔHD tissues are elevated.
(A) Cells from 6 pairs of control andcux/CDPΔHD/ΔHD bone marrow, and ConA- stimulated spleen and thymus were cultured for 24 and 72 hours, after which supernatants were harvested and analyzed for levels of TNF by ELISA. Levels of secreted TNF fromcux/CDPΔHD/ΔHD cells relative to cells from control littermates are depicted. (B) Levels of TNF in tissue extracts from 8 pairs of mice (n = 3 for bone marrow) were analyzed for TNF protein by ELISA. Mean levels are indicated with bars. Asterisks indicate directional Wilcoxon signed-rank tests of statistical significance P less than .05.
Levels of TNF expression incux/CDPΔHD/ΔHD tissues are elevated.
(A) Cells from 6 pairs of control andcux/CDPΔHD/ΔHD bone marrow, and ConA- stimulated spleen and thymus were cultured for 24 and 72 hours, after which supernatants were harvested and analyzed for levels of TNF by ELISA. Levels of secreted TNF fromcux/CDPΔHD/ΔHD cells relative to cells from control littermates are depicted. (B) Levels of TNF in tissue extracts from 8 pairs of mice (n = 3 for bone marrow) were analyzed for TNF protein by ELISA. Mean levels are indicated with bars. Asterisks indicate directional Wilcoxon signed-rank tests of statistical significance P less than .05.
Discussion
In this study we demonstrate that mice expressing low levels of a truncated cux/CDP protein have a complex phenotype, including partial neonatal lethality, runting, wavy fur and whiskers with latent alopecia, and muscle wasting with loss of body fat akin to cachexia. This complex phenotype is consistent with a role for cux/CDP in the transcriptional regulation of multiple target genes.
Our findings also establish a pivotal role for cux/CDP in B and T lymphopoiesis and myelopoiesis. B and T lymphoid demise could be explained by several abnormalities—a block in differentiation, a block in proliferative expansion, or enhanced cell death. A block in differentiation is unlikely due to the fact that mature B and T cells are present in normal proportions and numbers in peripheral lymphoid organs and there is no accumulation of undifferentiated cells, as seen in mice deficient in RAG-235 and Pax5.36
Clonal expansion of developing lymphocytes occurs after productive rearrangement of the IgH chain in B-lineage cells and TCRβ chain in thymocytes. In vivo bromodeoxyuridine (BrdU) incorporation assays demonstrated that although there was a general reduction in the incorporation of BrdU, the lymphocytes proliferating after IgH and TCRβ chain rearrangement were not preferentially reduced in primarycux/CDPΔHD/ΔHD mice (data not shown). Moreover, proliferation anomalies were not observed by in vivo BrdU incorporation assays in mice reconstituted withcux/CDPΔHD/ΔHD bone marrow (data not shown). Therefore, the cux/CDPΔHD mutation does not affect the cell-intrinsic abilities of developing lymphocytes to proliferate after antigen receptor rearrangement, but may affect environmentally derived proliferative signals. However, given the enhanced apoptosis observed in the lymphoid populations, it is difficult to rule out the possibility that the reduction in BrdU incorporation is not due to a preferential death of cells progressing through the cell cycle.
Our data strongly suggest that cell death accounts for lymphopenia incux/CDPΔHD/ΔHD mice, particularly in the thymic T-lineage cells and bone marrow B cells. However, mice reconstituted with cux/CDPΔHD/ΔHD bone marrow had normal thymus size and cellularity. This suggests that thymic apoptosis is induced by the overexpression of lymphocyte death-inducing factors or the absence/underexpression of survival factors derived from the tissue stroma. These possibilities are not mutually exclusive. Alterations in the expression of death or survival factors by some hematopoietic cells are also possible because B lymphopoiesis was affected in bone marrow of transplant recipients, though not as severely as in primary animals. Our finding that B and T lymphopoiesis are severely affected in RAG-2−/− mice complemented with cux/CDPΔHD/ΔHD ES cells (data not shown) is consistent with the notion that thecux/CDPΔHD/ΔHD hematopoietic microenvironment is abnormal. Because these mice are chimeric for both hematopoietic cells and the supportive cellular matrix, we suggest that the cux/CDPΔHD/ΔHD-derived stroma either induces cell death or is deficient in providing survival signals. These factors may also act systemically and cause neonatal lethality in primary cux/CDPΔHD/ΔHD mice.
Due to the pleiotropic abnormalities observed incux/CDPΔHD/ΔHD mice, and given that TNF can have either death-inducing or growth-stimulating effects depending on the cell type,37 38 we hypothesized that TNF may be inducing the lymphocyte death observed. This hypothesis was supported by our findings that TNF was elevated in the thymus of knock-out mice, which correlated with enhanced apoptosis. This suggests that the TNF gene may be a target for cux/CDP-mediated repression. However, at this time, we cannot exclude that its overexpression is a secondary effect. Investigations are currently underway to determine whether cux/CDPΔHD/ΔHDmice on a TNF null background are rescued from many of the pleiotropic abnormalities. Preliminary data suggest that most of the nonhematopoietic phenotypes are not rescued by loss of TNF but that some of the lymphoid abnormalities are. Thus, cux/CDP may influence the development and function of various organ systems through the coordinated regulation of several factors that regulate cell growth and survival, including TNF.
In contrast to the lymphopenia seen incux/CDPΔHD/ΔHD mice, myeloid hyperplasia was observed in bone marrow, spleen, and peripheral blood, in both percentages and absolute numbers, indicating that this is not due to the decrease in lymphocytes. Mice repopulated withcux/CDPΔHD/ΔHD bone marrow also had elevated numbers of myeloid cells, though not as dramatic. It is interesting to note that human CDP is located on chromosome 7q22, a region that is often deleted in 7q-related acute myeloid leukemia and myeloid dysplasia.39-43 In addition, loss of CDP is found in breast tumors44 and uterine leiomyomas.45,46These studies suggest that CDP is a tumor suppressor and that its loss is a significant event in the generation or progression (or both) of myeloid disorders. It will be interesting to determine if thecux/CDPΔHD/ΔHD mutation can accelerate the genesis of myeloid hyperplasia when combined with a PML/RARα transgene.47
Due to the complex phenotype of thecux/CDPΔHD/ΔHD mice, which is caused by both cell-intrinsic and microenvironmental effects on various populations, it has been difficult to examine the expression levels of putative cux/CDP target genes. cux/CDP binds to the IgH Eμ/MARs,17 TCRβ enhancer/MAR,22 and CD8a silencer/MAR.21 However, flow cytometric analysis of IgM and IgD (not shown) on B cells, and CD8 and TCRβ on T cells in primary cux/CDPΔHD/ΔHD mice and transplant recipients, demonstrated no changes in surface expression of these receptors. Both IgH and TCRβ enhancers also function in VDJ recombination.48-51 However, provision of recombined antigen receptors incux/CDPΔHD/ΔHD mice failed to rescue B- or T-cell development, suggesting that antigen receptor recombination is not affected by the cux/CDPΔHD mutation.
Although the deletion of the homeodomain and C-terminal repression domain has dramatic effects on protein function, the remaining cut repeats in the cux/CDPΔHD protein could still bind and compete for transcriptional activator binding in some enhancers/MARs.12 However, recent data have demonstrated that the cux/CDPΔHD protein is localized in the cytoplasm (A. van Wijnen, written communication, May 2001). Therefore, the cux/CDPΔHD protein would be unable to access the nucleus to bind DNA in vivo. Thus the cux/CDPΔHD/ΔHD mice are likely to be null for cux/CDP-mediated transcriptional repression.
We have shown that cux/CDP may play a role in repressing thymic TNF expression and CD25 expression in thymocytes undergoing selection. Indeed, CDP overexpression studies in a myeloid cell line has identified that CDP can repress lipopolysaccharide-induced TNF expression (unpublished observation, 2001). Consistent with the notion that CD25 may be a target gene for cux/CDP-mediated repression, the DNA binding activity of cux/CDP is up-regulated in CD4+CD8+ thymocytes,22,52 a point at which CD25 expression is down-modulated. However, it is unclear if cux/CDP has a direct effect on CD25 transcription because CD25 expression can be induced by cytokines, including IL-1 and TNF, on thymocyte precursors.53 It is possible that these or other factors may directly or indirectly induce ectopic CD25 expression incux/CDPΔHD/ΔHD mice. Future analyses will be required to define the role of cux/CDP in mediating repression of the putative novel target genes, such as CD25 and TNF, identified here.
We thank Fred Alt and Barry Sleckman for assistance with the RAG-2−/− blastocyst complementation studies, Matthias Wabl for providing the IgH “knock-in” mice, Mark Greene for supplying the TCR-Vβ8.1 transgenic mice, Steven Bauer for providing the IL-7, and Andre van Wijnen for communication of results prior to publication.
Supported by Public Health Service grants GM50329 (R.H.S.), HL49196 (E.N.J.), and CA31534 and GM31689 (P.W.T.). E.J.N. is a fellow of the Lucille P. Markey Foundation.
A.M.S., J.A.L., and A.G. contributed equally to the work in this article.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Richard H. Scheuermann, Department of Pathology and Laboratory of Molecular Pathology, 5323 Harry Hines Blvd, Dallas, TX 75390-9072; e-mail: scheuerm@utsw.swmed.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal