Abstract
The development of blood cells proceeds from pluripotent stem cells through multipotent progenitors into mature elements belonging to at least 8 different lineages. The lineage choice process during which stem cells and progenitors commit to a particular lineage is regulated by a coordinated action of extracellular signals and transcription factors. Molecular mechanisms controlling commitment are largely unknown. Here, the transcription factor v-Myb and its leucine zipper region (LZR) are identified as regulators of the commitment of a common myeloid progenitor and progenitors restricted to the myeloid lineage. It is demonstrated that wild-type v-Myb with the intact LZR directs development of progenitors into the macrophage lineage. Mutations in this region compromise commitment toward myeloid cells and cause v-Myb to also support the development of erythroid cells, thrombocytes, and granulocytes, similar to the c-Myb protein. In agreement with that, the wild-type v-Myb induces high expression of myeloid factors C/EBPβ, PU.1, and Egr-1 in its target cells, whereas SCL, GATA-1, and c-Myb are more abundant in cells expressing the v-Myb LZR mutant. It is proposed that Myb LZR can function as a molecular switch, affecting expression of lineage-specifying transcription factors and directing the development of hematopoietic progenitors into either myeloid or erythroid lineages.
Introduction
Differentiation of hematopoietic stem cells and progenitors into various lineages is controlled by a complex array of extrinsic and intrinsic factors.1-4 Myeloid and erythroid blood cells develop from a common myeloid progenitor, which differentiates into either megakaryocytes and erythrocytes, or granulocytes and macrophages. Several experimental strategies including gene targeting, expression pattern analysis, antisense, and overexpression studies led to the identification of transcription factors that are required for formation, survival, and proliferation of multilineage progenitors and that direct the differentiation and maturation of individual lineages. Specifically, factors like SCL, Rbtn2, GATA-2, and c-Myb were found to be essential for multipotent cells.5 In more mature cells, the expression of SCL, GATA-1, c-Myb, Rbtn2, FOG, and EKLF is a prerequisite for proper development of the erythroid lineage. On the other hand, factors of the C/EBP family, PU.1, Egr-1, c-Myb, and AML-1 play an important role in the myeloid lineage.6-9
It has been suggested that in the common myeloid progenitor the concentrations of intracellular lineage-determining regulators are to be low and balanced.10,11 During the commitment process this balance is disturbed either in a stochastic or instructed manner, resulting in the prevalence of a particular set of factors and the subsequent development of the respective lineage. Several factors were identified that instruct progenitor cells to develop along a specific lineage. Using the model of primary chicken myb-ets–transformed multipotent progenitors (MEP),12 it was demonstrated that PU.1 and C/EBPβ forced differentiation of MEP progenitors into myeloid cells, and C/EBPα only into eosinophils.13,14 In mammalian cells, Egr-1 was found to commit myeloid cells into the macrophage lineage with the concomitant block to differentiation into granulocytes.15
The c-myb proto-oncogene is essential for early definitive myeloid and erythroid cells as documented by gene targeting experiments.16 It is required for the expansion of immature cells of the myeloid, erythroid, and lymphoid lineages and is down-regulated during their terminal differentiations.17
The v-myb gene transduced by avian myeloblastosis virus (AMV), as well as its truncated homologue, transduced as themyb-ets fusion by E26 leukemia virus, are oncogenes that specifically affect the developmental programs of avian hematopoietic cells. AMV v-Myb interferes exclusively with the development of the macrophage lineage of both primitive and definitive hematopoietic cells by blocking terminal differentiation of macrophage precursors and activating their self-renewal capacities in vitro. It causes fatal acute monoblastic leukemia in chicks. E26 v-Myb-Ets fusion protein transforms multipotent progenitors of primitive hematopoietic cells (blastoderm), and committed erythroid and myeloid cells of bone marrow. It induces erythroid leukemia in infected chicks.18-22 Thus, the E26 v-Myb-Ets protein affects a broader spectrum and more immature cells than the AMV v-Myb. In this respect the biologic activities of E26 v-Myb-Ets resemble those of c-Myb, which influence the development of many immature hematopoietic cells. The very specific biologic activities of AMV v-Myb are thought to result from the loss of some c-Myb functions due to N- and C-terminal deletions and point mutations. Therefore, it was speculated that the mechanisms by which AMV v-Myb deregulates development of hematopoietic cells might also differ significantly from the mechanisms of action of E26 v-Myb-Ets and c-Myb.23
In this paper we demonstrate that the long-observed restriction of AMV v-Myb transforming potential to macrophage precursors is mainly due to the activity of its leucine zipper region (LZR), which programs development of hematopoietic progenitors into the macrophage lineage. Mutations in the LZR rescue the Myb activity to affect uncommitted progenitors as well as cells of erythroid and granulocytic lineages. The LZR of Myb therefore appears to be a part of a mechanism that regulates a lineage choice in hematopoietic tissues.
Materials and methods
Cells and cell culture
Blastoderm cells were obtained from one-day-old Brown Leghorn C/E gs− leukosis-free chick embryos in Hamburger and Hamilton stages 5 through 8.24 Blastoderms were mechanically dissociated by gentle pipetting and filtered through a cell strainer to obtain single-cell suspensions. Cells (2 × 106) were infected by cocultivation with virus-producing chick embryo fibroblasts (CEFs) or QT6 cells.25 Two days later, nonadherent cells were transferred to a new dish and grown in the modified erythroid colony-forming unit (CFU-E) medium,26 referred to as standard growth medium, or seeded in a semisolid methylcellulose (0.8%) in standard growth medium (2 × 105 to 5 × 105 cells per 30-mm dish). Standard growth medium contained Dulbecco modified Eagle medium (DMEM, D6171; Sigma, St Louis, MO) supplemented with 11% fetal calf serum (FCS); 4% chicken serum (ChS); 580 mg/L l-glutamine; 25 mg/L hypoxanthine; 300 mg/L conalbumine; 0.13 mM β-mercaptoethanol; 0.75% detoxified bovine serum albumin (BSA); 100 units/mL penicillin/streptomycin (Gibco-BRL, Carlsbad, CA). In some experiments, liquid and methylcellulose cultures were supplemented with growth factors (bFGF, 5 ng/mL; transforming growth factor alpha (TGFα), 1 ng/mL [both Promega, Madison, WI]; stem cell factor [SCF], 50 ng/mL27). Cell numbers were determined at regular time intervals by counting cells with a Multisizer II cell counter (Coulter, Miami, FL) and cell morphology was evaluated by microscopy of cytospin preparations stained with histologic dyes. Colonies were isolated from methylcellulose assays 7 to 14 days after infection and expanded in liquid culture. The cell lines BM228 and HD429 were grown in DMEM (D5546; Sigma) supplemented with 8% FCS, 2% ChS; 580 mg/Ll-glutamine, and 100 units/mL of penicillin/streptomycin.
Differentiation assays
Infected cultures were grown for 14 days in the mixture of factors (bFGF, TGFα, SCF) and assayed for the expression of surface antigens. Factors were then withdrawn, and cells were treated with 30 nM TPA (12-O-Tetradecanoylphorbol 13-acetate; Sigma) for 7 days and analyzed again. Alternatively, cells infected with wild type (wt) and ΔP1 v-myb were grown for 14 days in the presence of growth factor mixture and then either kept without factors for 21 days or treated with 30 nM TPA for 7 days and cultivated for an additional 14 days in the absence of TPA and growth factors. Eight independent experiments were performed.
Cytochemical assays
To evaluate cell morphology and hemoglobin content, cytospin preparations were stained with neutral benzidine and counterstained with Diff-Quik (Baxter, Deerfield, IL).26,30May-Grünwald-Giemsa staining followed standard procedures. To detect heterophilic granules, cells were stained with Astra blue.31 The peroxidase activity in eosinophilic granules was determined as described.32 In each experiment, 300 to 500 cells were evaluated and the proportion of individual cell types was determined. Microphotographs were taken from representative fields.
Plasmids
The following AMV–v-myb mutants were used: ΔC contains a deletion of 27 C-terminal amino acid codons (362-388); ΔLZ1, ΔLZ2, ΔP1, ΔP2, and ΔP3 represent internal in-frame deletions of codons 325-362, 311-325, 301-311, 294-300, and 279-300, respectively. The numbering of codons is based on the v-mybcDNA sequence.33 All deletion mutants were generated by polymerase chain reaction (PCR) and verified by DNA sequencing. The L3,4A mutant was described previously.25 Mutant genes were inserted into the pNeoAMV vector34 and transfected into CEFs or QT6 cells along with MAV-1 helper virus DNA by calcium phosphate.25
Antibodies, flow cytometry, and immunofluorescence
The following antibodies were used: MC51/2, MC47/83, MC22/3,35 and JS436 (kindly provided by Dr H. Beug); 11C3 monoclonal antibody37 (generous gift of Dr M. Corbel); MEP26, MEP21, MEP17 antibodies38 (generously provided by Dr T. Graf); anti–GATA-1,39 anti-Myb monoclonal antibody 2.32,40 and polyclonal anti-C/EBPβ antibody41 (kind gift of Dr K.-H. Klempnauer).
Flow cytometry was performed as described25 using the EPICS Elite ESP Flow Cytometer (Coulter). For immunofluorescence analysis, cells were attached to coated slides (Bio-Rad, Hercules, CA) and processed according to the manufacturer's protocol with the following modifications. Briefly, the cells were recovered, resuspended in phosphate-buffered saline (PBS) and loaded onto adhesion slide reaction fields. The cells were fixed with 3% paraformaldehyde for 5 minutes, washed with PBS and permeabilized with 0.4% NP-40 in PBS for 2 minutes. After washing in PBS the reaction fields were blocked with 2% BSA in PBS for 1 hour. Then, the cells were reacted with primary monoclonal antibodies (1 hour), washed with PBS, 0.5% BSA, and stained with fluorescein isothiocyanate (FITC)–conjugated and/or rhodamin-conjugated secondary antibody (1 hour). All reactions were done at room temperature. Finally, the cells were mounted into Mowiol (Hoechst, Strasbourg, France) and images were taken with a Leica DMIRB microscope (Leica, Wetzlar, Germany) and processed with Leica Qwin and Adobe Photoshop software.
Northern blot analysis and RNase protection assay
Total RNA was prepared42 and 15 μg were separated on 1% agarose/formaldehyde gels, transferred to nylon membrane (GeneScreen, NEN, Boston, MA), and hybridized with specific probes. The following probes were used: 0.8-kb EcoRI fragment of chicken PU.1 cDNA43 (kind gift of Dr J. Ghysdael); 305–base pair (bp) BglII-NheI fragment of chicken c-myb cDNA44; chicken D-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) cDNA45; and full-length cDNA of chicken GATA-146 (kindly provided by Dr J. D. Engel). The chicken egr-1-, C/EBPβ-, and SCL-specific probes (589 bp, 610 bp, and 968 bp, respectively) were generated by reverse transcriptase (RT)–PCR (nucleotides: 1-589, 493-1102, and 208-1175, respectively; GenBank accession nos. AF026082, Z21646, and X63371). Probes were labeled by nick-translation and hybridized overnight in ULTRAhyb (Ambion, Austin, TX) at 42°C. Filters were washed twice in 2 × standard saline citrate (SSC)/0.1% sodium dodecyl sulfate (SDS) and then in 0.1 × SSC/ 0.1% SDS at 42°C, and exposed to BioMax MS film (Kodak, Rochester, NY) with intensifying screen.
For RNase protection assays, a 306-bp cDNA fragment of chicken GBX2 (nucleotide number 905-1211; GenBank accession no. AF022151) was subcloned into pGEM3Z. Similarly, a 182-bp cDNA fragment of chicken C/EBPβ (nucleotide number 919-1101; Genbank accession no.Z21646) was subcloned into pSP64. The constructs were linearized bySmaI and PvuII, and transcribed by SP6 polymerase (Ambion) in the presence of α-[32P]GTP to produce radioactive probes, which were used in RNase protection assays as described previously.47 48 The chicken actin cDNA fragment was used as a control.
Western blot analysis
Protein lysates were separated by 10% SDS-PAGE, blotted onto nitrocellulose membranes (BA85, Schleicher and Schuell, Dässel, Germany) and processed for western blot analysis as described.25 The 2.32 anti-Myb monoclonal antibody served as primary antibody.
Transient transactivation assay
CEFs (6 × 105 cells) were seeded onto 60-mm dishes and incubated in DMEM medium (D5546, Sigma) supplemented with 8% FCS and 2% ChS serum, 580 mg/L l-glutamine, and 100 units/mL penicillin/streptomycin (Gibco-BRL). The cells were transfected and processed as described earlier.25 The cells transfected with empty pNeo vector were used as a control. The chloramphenicol acetyl transferase (CAT) in transfected cells was determined by the CAT enzyme-linked immunosorbent assay (ELISA) (Boehringer Mannheim, Mannheim, Germany). The CAT value of wild-type v-Myb was assigned a value of 100%. Transcriptional activities of v-Myb mutants were calculated in percents of the wild-type v-Myb activity.
Results
Mutations in the LZR activate the transforming potential of Myb oncoprotein in erythroid cells
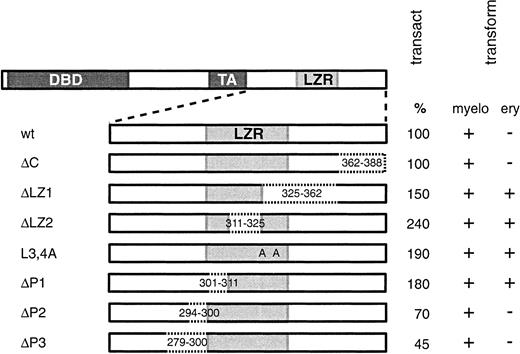
Previous work on myeloid cells demonstrated that v-Myb LZR is required for proliferation, growth factor independence, and leukemic properties of v-Myb–transformed monoblasts.25 49 In order to obtain more information about the biologic activity of v-Myb LZR, a series of deletion mutants were constructed that span the C-terminus of v-Myb protein (Figure 1). The point mutant v-MybL3,4A, where Leu3 and Leu4 of LZR are replaced by Ala25 was also used. Mutations affecting the LZR, namely ΔLZ1, ΔLZ2, ΔP1, and L3,4A are in the following text referred to as LZR mutations. Since C-terminal mutations could negatively influence the basic property of v-Myb, that is, its transcriptional activity, CAT assays in transiently transfected fibroblasts were performed. It was found that LZR mutations did not impair but rather slightly enhanced (1.5 fold-2.5 fold) the transcriptional activity of v-Myb (Figure 1). The activity of ΔP2 and ΔP3 mutants was reduced to 70% and 45%, respectively, of wild-type Myb.
LZR mutants of v-Myb exhibit elevated transcriptional activity and transform erythroid cells.
Schematic representation of full-length wild-type v-Myb protein with DNA-binding domain (DBD), transactivation domain (TA), and leucine zipper region (LZR). The specific deletion mutations in v-Myb C-terminus are delimited by dotted lines and characterized by the numbers of the first and last deleted amino acid. In the ΔC mutant, C-terminal amino acids were deleted; ΔLZ1, ΔLZ2, ΔP1, ΔP2, and ΔP3 mutants carry internal in-frame deletions as indicated. In the point mutant L3,4A, leucines (L) 325 and 332 were replaced by alanine (A). The transactivation activity of Myb proteins was determined by the standard CAT assay in transiently transfected fibroblasts. Results are representative of 2 independent experiments. Assays were performed in triplicate. The transformation potential of various Myb mutants for myeloid and erythroid cells (myelo and ery, respectively) is shown. Transform indicates transforming potential; transact, transactivation activity.
LZR mutants of v-Myb exhibit elevated transcriptional activity and transform erythroid cells.
Schematic representation of full-length wild-type v-Myb protein with DNA-binding domain (DBD), transactivation domain (TA), and leucine zipper region (LZR). The specific deletion mutations in v-Myb C-terminus are delimited by dotted lines and characterized by the numbers of the first and last deleted amino acid. In the ΔC mutant, C-terminal amino acids were deleted; ΔLZ1, ΔLZ2, ΔP1, ΔP2, and ΔP3 mutants carry internal in-frame deletions as indicated. In the point mutant L3,4A, leucines (L) 325 and 332 were replaced by alanine (A). The transactivation activity of Myb proteins was determined by the standard CAT assay in transiently transfected fibroblasts. Results are representative of 2 independent experiments. Assays were performed in triplicate. The transformation potential of various Myb mutants for myeloid and erythroid cells (myelo and ery, respectively) is shown. Transform indicates transforming potential; transact, transactivation activity.
Next, the transforming potential of the Myb mutants was analyzed. Retroviruses carrying mutant myb genes were used to infect blastoderm cells from 20- to 28-hour-old chicken embryos. The ΔC, ΔP2, and ΔP3 mutants with intact LZR as well as wild-type v-Myb caused an outgrowth only of cells resembling monoblasts (Figure 1). Interestingly, in cultures infected with the LZR mutants, numerous hemoglobinized erythroid cells were present in addition to myeloid cells. In 15 independent experiments the following percentage of erythroid cells was recorded: on days 10 to 20 after infection, erythroid cells represented approximately 20% of the entire cell population for ΔLZ1, ΔLZ2, and L3,4A mutants, and about 50% for ΔP1 Myb. These values were calculated from flow cytometry profiles obtained with JS4 and MC51/2 antibodies (shown for ΔP1 Myb in Figure2D), and from cytospins and smears stained with cytochemical dyes. Some erythroid cells differentiated spontaneously into mature erythrocytes during the 30-day culture period. In ΔP1 cultures numerous immature erythroid cells were present even after prolonged culture periods. As expected, in control cultures infected with the empty retroviral vector, no growing cells were ever obtained (data not shown).
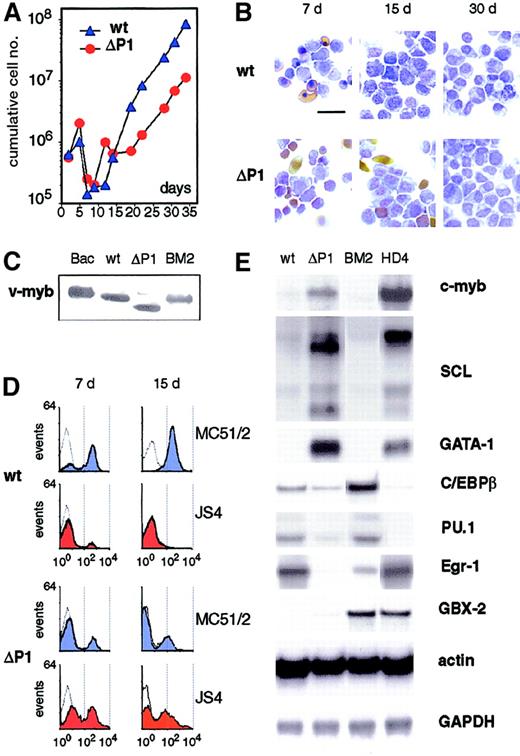
ΔP1 v-Myb cells express erythroid specific genes.
(A) Growth kinetics of chicken blastoderm cells infected with NeoAMVwt (wt) and NeoAMVΔP1 (ΔP1) viruses are shown. (B) Cytochemical staining of wild-type and ΔP1 v-Myb cells on days 7, 15, and 30 of culture. Benzidine staining reveals hemoglobinized erythroid cells (yellow-brown); cells were counterstained with Diff-Quik to show monoblasts (light blue). The bar indicates 15 μm.(C) v-Myb proteins in 106 wild-type and ΔP1 v-Myb cells (day 20) were analyzed by western blotting using the myb-specific monoclonal antibody. Equal amounts of proteins in loaded samples were verified by Ponceau staining of the western blot. v-Myb synthesized in the baculovirus expression system (Bac) and in 106 cells of the v-myb–transformed BM2 cell line (BM2) are shown as controls. (D) Flow cytometry of wild-type and ΔP1 v-Myb cells on days 7 and 15 using monoclonal antibodies MC51/2 (blue) for myeloid and JS4 (red) for erythroid surface antigens. Empty curves represent negative controls. Intensity of fluorescence is plotted on horizontal axes, amounts of measured cells on vertical axes. (E) mRNAs for c-Myb, GATA-1, PU.1, Egr-1, and multiple SCL mRNAs in wild-type and ΔP1 v-Myb cells (day 15) were analyzed by northern blotting; C/EBPβ and GBX2 mRNAs were analyzed by RNase protection. RNAs of myeloid BM2 and erythroid HD4 cell lines were analyzed for comparison. Actin and GAPDH probes were used as controls of equal loading in RNase protection and northern blot assays, respectively. Two independent experiments provided identical results.
ΔP1 v-Myb cells express erythroid specific genes.
(A) Growth kinetics of chicken blastoderm cells infected with NeoAMVwt (wt) and NeoAMVΔP1 (ΔP1) viruses are shown. (B) Cytochemical staining of wild-type and ΔP1 v-Myb cells on days 7, 15, and 30 of culture. Benzidine staining reveals hemoglobinized erythroid cells (yellow-brown); cells were counterstained with Diff-Quik to show monoblasts (light blue). The bar indicates 15 μm.(C) v-Myb proteins in 106 wild-type and ΔP1 v-Myb cells (day 20) were analyzed by western blotting using the myb-specific monoclonal antibody. Equal amounts of proteins in loaded samples were verified by Ponceau staining of the western blot. v-Myb synthesized in the baculovirus expression system (Bac) and in 106 cells of the v-myb–transformed BM2 cell line (BM2) are shown as controls. (D) Flow cytometry of wild-type and ΔP1 v-Myb cells on days 7 and 15 using monoclonal antibodies MC51/2 (blue) for myeloid and JS4 (red) for erythroid surface antigens. Empty curves represent negative controls. Intensity of fluorescence is plotted on horizontal axes, amounts of measured cells on vertical axes. (E) mRNAs for c-Myb, GATA-1, PU.1, Egr-1, and multiple SCL mRNAs in wild-type and ΔP1 v-Myb cells (day 15) were analyzed by northern blotting; C/EBPβ and GBX2 mRNAs were analyzed by RNase protection. RNAs of myeloid BM2 and erythroid HD4 cell lines were analyzed for comparison. Actin and GAPDH probes were used as controls of equal loading in RNase protection and northern blot assays, respectively. Two independent experiments provided identical results.
These results define LZR as a functional domain and demonstrate that its inactivation by mutation enables v-Myb to also affect erythroid cells in addition to monoblastlike cells. Importantly, mutations of LZR have no deleterious effect on transcriptional properties of v-Myb.
The ΔP1 v-Myb mutant best compares to wild-type v-Myb
In pilot experiments, cells infected with all Myb mutants were compared for their proliferation rate, survival, and colony-formation efficiency. The ΔC and ΔP2 mutants, containing an intact LZR, induced a monoblastic phenotype identical to that of wild-type v-Myb. The ΔP3 mutant displayed a rather weak transforming activity (data not shown). Among LZR mutants, ΔP1 induced in infected cells (a mixture of myeloid and erythroid cells) a proliferation rate, life span, and colony-formation efficiency that was comparable to wild-type v-Myb, whereas myeloerythroid cultures formed by ΔLZ1, ΔLZ2, and L3,4A mutants grew more slowly and the cells showed a somewhat reduced lifespan and colony-formation efficiency. Since the intracellular stability and subcellular distribution of ΔP1 and wild-type v-Myb proteins were also very similar,49 the ΔP1 mutant was selected for further studies and analyzed along with wild-type v-Myb.
The v-Myb LZR influences expression of lineage-specific genes
Blastoderm cells transformed by wild-type and ΔP1 v-Myb were first characterized for their growth potential, morphology, and surface marker expression, and second for expression of genes important for the development of the myeloid and erythroid lineages. Figure 2A,B shows that within the first 7 days after infection, wild-type v-Myb cultures were composed of monoblastlike cells and some erythroid cells, whereas from day 7 on only rapidly proliferating monoblasts were observed. In contrast, in ΔP1 cultures there was an initial outgrowth of primarily erythroid cells within the first 7 days after infection. Then, a high proportion of cells underwent spontaneous differentiation into erythrocytes, and resulting cultures contained erythroid cells of different maturation stages and monoblasts (Figure 2A,B). At later time points, cells belonging to additional hematopoietic lineages—thrombocytes, eosinophils, and heterophilic granulocytes (equivalent to neutrophils in mammals)—were also observed (see below). Protein analysis in cultured cells demonstrated essentially identical amounts of wild-type and ΔP1 v-Myb proteins, comparable with v-Myb expression in the myeloid cell line BM2 (Figure 2C). Thus, accumulation of different cells in wild-type and ΔP1 v-Myb cultures was not due to different expression levels of Myb proteins.
To further extend these observations, cells were analyzed for lineage-specific surface markers using monoclonal antibodies MC51/2 and JS4, which recognize myeloid and erythroid surface molecules, respectively. The ΔP1 culture contained a high proportion of cells stained for the erythroid marker JS4, whereas the wild-type v-Myb culture was predominantly myeloid (MC51/2 positive) and only a minority of cells were JS4 positive. At later stages (30 days after infection) wild-type v-Myb cultures contained only myeloid cells, whereas in ΔP1 cultures erythroid cells were also detectable, in line with the histochemical data (Figure 2B,D).
Next, wild-type and ΔP1 v-Myb cells were analyzed for expression of lineage-specific transcription factors by northern blotting and RNAse protection. Day 14 after infection was selected for analysis since at this time point ΔP1 cultures contained the highest proportion of undifferentiated erythroid cells. ΔP1 cells expressed high levels of c-Myb, SCL, and GATA-1 mRNAs, which are important for the development of the erythroid lineage (Figure 2E). In contrast, in wild-type v-Myb cells these mRNAs were essentially undetectable whereas mRNAs for the myelomonocytic factors C/EBPβ, PU.1, and Egr-1 were abundantly expressed. The expression of these molecules in myeloid BM2 and erythroid HD4 cell lines are shown for comparison.
Expression of the homeobox transcription factor GBX2 was also analyzed, as GBX2 was suggested to activate expression of chicken myelomonocytic growth factor (cMGF) and to generate an autocrine loop important for the development and proliferation of AMV v-Myb–transformed myeloid cells.50 In cultures of primary blastoderm cells, however, only very low levels of GBX2-specific transcripts were detected in ΔP1 v-Myb cells and there was no GBX2 mRNA in wild-type v-Myb cells. In myeloid BM2 and erythroid HD4 cell lines, however, high levels of GBX2 mRNA were detected (Figure 2E). Moreover, neither GBX2 nor cMGF mRNAs were present in primary bone marrow monoblasts transformed by wild-type v-Myb.49 Thus, in primary cells v-Myb appears not to activate the GBX2/cMGF pathway and the predominantly myeloid character of wild-type v-Myb cultures is not caused by cMGF. In contrast, as indicated here and in following experiments, wild-type v-Myb LZR positively influences the expression of myelomonocytic transcriptional regulators C/EBPβ, PU.1, and Egr-1. These are likely to dictate the strictly monoblastic character of wild-type v-Myb–transformed hematopoietic cells.
v-Myb cooperates with growth factors to block differentiation of multipotent progenitors
In early cultures of ΔP1 v-myb–infected and to a lesser extent also of wild-type v-myb–infected blastoderm hematopoietic cells, some cells expressed surface protein markers of multipotent progenitors recognized by MEP17, MEP21, and MEP26 monoclonal antibodies.12 38 We therefore reasoned that the different v-Myb cells described so far in this paper might represent the progeny of multipotent progenitors, which were not completely blocked in differentiation and hence spontaneously developed into myeloid or erythroid cells. To enhance the self-renewing capacity and/or delay spontaneous differentiation of putative Myb progenitors, different growth factors and combinations thereof were tested. TGFα, bFGF, and SCF, when applied simultaneously, were effective in increasing growth rates of both wild-type and ΔP1 v-Myb cells positive for MEP antigens. Maximal expression of MEP antigens was achieved on approximately days 14 to 21 of culture (Figure3A). At this point all cells also expressed the erythroid marker JS4. No myeloid cells were present, as documented by the absence of the myeloid marker MC51/2 (Figure4A). Thus, v-Myb and specific growth factors cause accumulation of early undifferentiated cells. These cells are progenitors since they can develop into different types of hematopoietic cells as will be documented in following experiments. Importantly, these progenitors accumulated in cultures only when growth factors were added at the time of infection with mybconstructs. Later addition of factors yielded only higher numbers of erythroid cells in ΔP1 cultures and had no effect on the strictly myeloid character of wild-type myb-infected cells. This observation ruled out the possibility that v-Myb monoblasts and erythroid cells were reprogrammed into progenitors by growth factors.
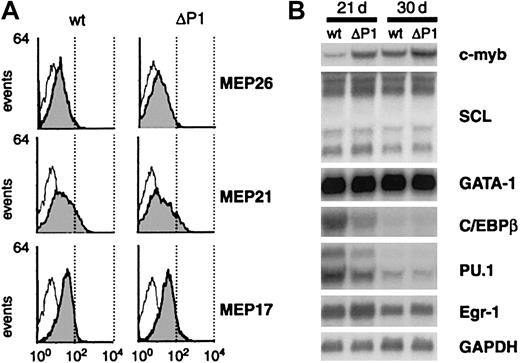
Wild-type v-Myb and ΔP1 v-Myb transform early progenitors in the presence of growth factors.
(A) Flow cytometry of wild-type and ΔP1 v-Myb cells grown in the presence of TGFα, bFGF, and SCF (day 21) using monoclonal antibodies that recognize multipotent cell surface antigens MEP 26, MEP 21, and MEP17. Negative controls are represented by empty curves. Plots axes are as in Figure 2D. (B) Northern blot analysis in wild-type and ΔP1 v-Myb cells of mRNAs for c-Myb, SCL, GATA-1, C/EBPβ, PU.1, Egr-1, and GAPDH on day 21 and day 30. Two identical blots were sequentially hybridized with SCL, PU.1, and Egr-1 probes, or with c-Myb, C/EBPβ, GATA-1, and GAPDH (loading control) probes.
Wild-type v-Myb and ΔP1 v-Myb transform early progenitors in the presence of growth factors.
(A) Flow cytometry of wild-type and ΔP1 v-Myb cells grown in the presence of TGFα, bFGF, and SCF (day 21) using monoclonal antibodies that recognize multipotent cell surface antigens MEP 26, MEP 21, and MEP17. Negative controls are represented by empty curves. Plots axes are as in Figure 2D. (B) Northern blot analysis in wild-type and ΔP1 v-Myb cells of mRNAs for c-Myb, SCL, GATA-1, C/EBPβ, PU.1, Egr-1, and GAPDH on day 21 and day 30. Two identical blots were sequentially hybridized with SCL, PU.1, and Egr-1 probes, or with c-Myb, C/EBPβ, GATA-1, and GAPDH (loading control) probes.
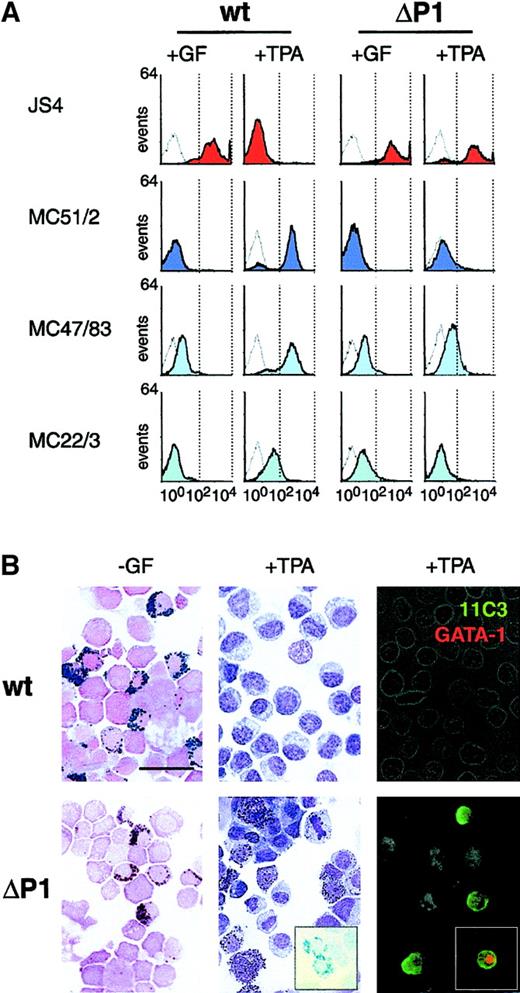
Wild-type v-Myb and ΔP1 v-Myb progenitors develop into different lineages.
(A) Flow cytometry of progenitor cells grown in the presence of TGFα, bFGF, and SCF (+GF) and induced to differentiate by removal of growth factors and TPA treatment (+TPA). Monoclonal antibodies specific for the erythroid JS4 antigen (red) or myeloid antigens MC51/2, MC47/83, and MC22/3 (shades of blue) were used. Negative controls are represented by empty curves. Plots axes are as in Figure 2D. (B) Cytochemical staining and immunofluorescence of progenitors induced to differentiate by growth factor withdrawal (-GF) and TPA treatment (+TPA). Left panels: myeloperoxidase staining for identification of eosinophilic granules. Middle panels: Giemsa staining to reveal cell morphology; Astra blue staining of heterophilic granules (inset in ΔP1 panel). Right panels: indirect immunofluorescence with 11C3 monoclonal antibody to reveal thrombocytes (green). Inset in ΔP1 panel, double-staining with 11C3 (green) and anti-GATA-1 (red) antibodies. The bar represents 15 μm.
Wild-type v-Myb and ΔP1 v-Myb progenitors develop into different lineages.
(A) Flow cytometry of progenitor cells grown in the presence of TGFα, bFGF, and SCF (+GF) and induced to differentiate by removal of growth factors and TPA treatment (+TPA). Monoclonal antibodies specific for the erythroid JS4 antigen (red) or myeloid antigens MC51/2, MC47/83, and MC22/3 (shades of blue) were used. Negative controls are represented by empty curves. Plots axes are as in Figure 2D. (B) Cytochemical staining and immunofluorescence of progenitors induced to differentiate by growth factor withdrawal (-GF) and TPA treatment (+TPA). Left panels: myeloperoxidase staining for identification of eosinophilic granules. Middle panels: Giemsa staining to reveal cell morphology; Astra blue staining of heterophilic granules (inset in ΔP1 panel). Right panels: indirect immunofluorescence with 11C3 monoclonal antibody to reveal thrombocytes (green). Inset in ΔP1 panel, double-staining with 11C3 (green) and anti-GATA-1 (red) antibodies. The bar represents 15 μm.
v-Myb LZR influences expression of lineage-specifying factors in progenitors
As stated above, the highest proportion of progenitors was obtained in 14- to 21-day-old cultures. Their prolonged cultivation in the presence of TGFα, bFGF, and SCF (for about 30 days) caused both wild-type and ΔP1 v-Myb progenitors to down-regulate MEP markers and to commit to the erythroid lineage, as evidenced by their ability to differentiate into hemoglobinized erythrocytes (P.B. et al, manuscript submitted, June 2001). To find out whether Myb LZR influences levels of C/EBPβ, PU.1, Egr-1, c-Myb, SCL, and GATA-1 mRNAs in progenitors and early erythroid cells, RNA from cells grown for 21 and 30 days in growth factor mixture was analyzed. Figure 3B shows that both wild-type and ΔP1 v-Myb cells abundantly expressed SCL and GATA-1 mRNAs at both time points. In contrast, c-Myb mRNA was lower in wild-type v-Myb cells than in ΔP1 cells on day 21, whereas its levels were essentially the same on day 30 when both cultures were erythroid. The mRNAs of myeloid factors C/EBPβ and PU.1 were more abundant in wild-type v-Myb cells than in ΔP1 cells on day 21 and barely detectable in both cultures on day 30. Virtually no difference was seen for Egr-1 mRNA levels at any time.
The data show that in v-Myb progenitors (day 21), wild-type LZR causes elevated levels of C/EBPβ and PU.1 mRNAs and a reduced level of c-Myb mRNA when compared with expression of these molecules in cells containing the ΔP1 LZR mutant. This specific expression pattern should predetermine the development of wild-type v-Myb progenitors into eosinophils and the macrophage lineage, as C/EBPβ and PU.1 were found to commit progenitors into these lineages.13 14 Indeed, the next experiments confirmed this assumption. The above data document the ability of v-Myb LZR to modulate expression of lineage-specifying factors.
Wild-type v-Myb progenitors differentiate predominantly into the macrophage lineage; ΔP1 v-Myb progenitors differentiate into multiple lineages
To reveal the differentiation potential of Myb progenitors, they were grown for 14 days in the presence of TGFα, bFGF, and SCF, and then their differentiation was induced by withdrawal of growth factors. As the phorbol ester TPA was found to be a potent inducer of differentiation of Myb progenitors,12 factor removal and TPA treatment were combined in some experiments. Resulting cells were first analyzed for the presence of lineage-specific surface proteins (Figure 4A). Immediately prior to differentiation induction both wild-type and ΔP1 v-Myb cells expressed the erythroid JS4 marker and no myeloid MC51/2 (Figure 4A, +GF). From additional myeloid markers the MC47/83 was weakly detectable in both cultures whereas the MC22/3 was present only on ΔP1 cells (Figure 4A, +GF). Several days after differentiation induction by growth factor removal and TPA treatment, wild-type v-Myb cells lost the erythroid marker JS4 and upregulated the myelomonocytic markers MC51/2, MC47/83, and MC22/3 (Figure 4A, +TPA). These cells clearly represented macrophage precursors as also judged by morphology (data not shown). In contrast, cells infected with ΔP1 v-myb still retained high JS4 expression and were negative for MC51/2 and MC22/3, and weakly positive for MC47/83 (Figure 4A, +TPA). Additionally, these cells kept their immature phenotype (not shown).
In similar experiments, differentiating cells were analyzed by cytochemical staining for the presence of granulocytes and by immunofluorescence for thrombocytes 20 days after differentiation induction (Figure 4B). In both wild-type and ΔP1 v-Myb cultures, some cells developed into eosinophils after removal of growth factors (Figure 4B; −GF panels). Following TPA treatment (Figure 4B; +TPA panels), only monoblastlike cells (wild-type, middle panel) and no thrombocytes (wild-type, right panel) were found in wild-type v-Myb cultures. In ΔP1 v-Myb cultures, however, numerous heterophilic granulocytes (ΔP1, middle panel) were revealed by Astra blue staining. Using the 11C3 antibody recognizing the GPIIb-IIIa integrin on chicken thrombocytes, numerous positive cells were also detected (ΔP1, right panel). Analyzed ΔP1 v-Myb cultures also contained monoblastlike cells and erythroid cells, which were, however, not specifically revealed by staining for granulocytes and thrombocytes. These findings indicate that ΔP1 progenitors have a broader differentiation potential than wild-type v-Myb cells and can differentiate into all lineages derived from the common myeloid progenitor. On the other hand, the wild-type v-Myb protein prevents progenitors from differentiating into erythroid cells, thrombocytes, and heterophilic granulocytes. The absence of heterophilic granulocytes in wild-type v-Myb cultures strongly indicates that wild-type v-Myb LZR forces the bipotent granulocyte/macrophage progenitor to differentiate only into the macrophage lineage. This could be due to the rather high expression of the zinc finger transcription factor Egr-1, which was found to be essential for both the development of monocyte/macrophage lineage and the block of granulocyte differentiation.15
The treatment of blastoderm cells transformed with ΔLZ1 and L3,4A v-Myb mutants with TGFα, bFGF, and SCF also resulted in a rapid accumulation of progenitors and early erythroid cells in cultures. However, their life span was limited, which did not allow us to perform differentiation experiments. Nevertheless, in short-term cultures grown in DMEM without growth factors, the ΔLZ1 v-Myb cells reproducibly contained 4% to 8% of heterophil and 2% to 3% of eosinophil granulocytes, and L3,4A v-Myb cells 3% to 5% of heterophil and 1% to 2% of eosinophil granulocytes. Under identical culture conditions ΔP1 v-Myb cultures contained 5% to 10% of heterophil and 1% to 2% of eosinophil granulocytes, whereas cultures transformed with wild-type v-Myb contained no granulocytes. Thus, the entire LZR appears to influence the potential of progenitors to differentiate into multiple cell types.
Myb LZR determines commitment of a common myeloid progenitor to the macrophage lineage
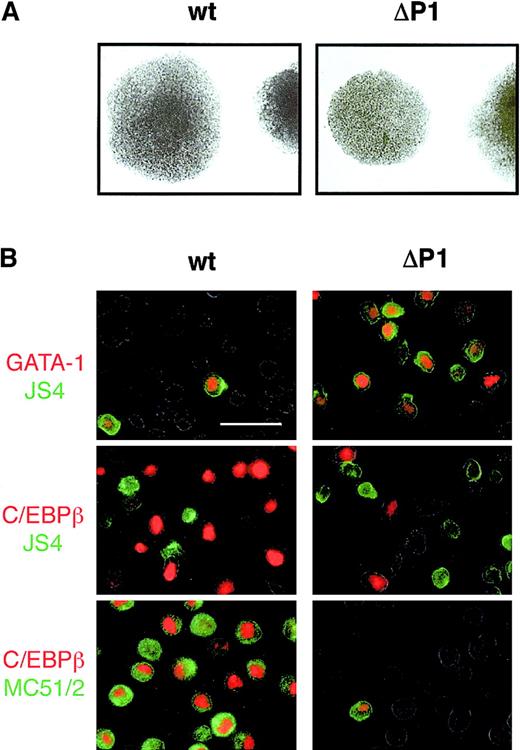
Our experiments suggest that LZR represents a critical element for development of progenitors into either erythroid or myeloid cells. To demonstrate this at the clonal level, blastoderm cells infected with wild-type or ΔP1 v-myb genes were seeded into semisolid media containing TGFα, bFGF, and SCF. Colonies were isolated 12 days later. In a representative experiment, 94 wild-type–myb and 155 ΔP1-myb colonies were obtained (Figure5A). More than 90% of both wild-type–myb and ΔP1-myb clones could be expanded in liquid culture. Ten representative clones of each type were finally studied in detail. At the time of isolation both wild-type and ΔP1 v-myb colonies contained cells displaying an immature phenotype, rather weak JS4 staining, and no MC51/2 staining.
v-Myb LZR regulates commitment of common myeloid progenitors.
(A) Colonies of wild-type and ΔP1 v-Myb cells grown in the presence of TGFα, bFGF, and SCF in semisolid medium. (B) Indirect immunofluorescence analysis of representative wild-type and ΔP1 v-Myb cell clones after expansion and differentiation induction by growth factors removal. Double-staining for nuclear antigens (GATA-1 or C/EBPβ, red) and surface antigens (JS4 or MC51/2, green). The bar represents 15 μm.
v-Myb LZR regulates commitment of common myeloid progenitors.
(A) Colonies of wild-type and ΔP1 v-Myb cells grown in the presence of TGFα, bFGF, and SCF in semisolid medium. (B) Indirect immunofluorescence analysis of representative wild-type and ΔP1 v-Myb cell clones after expansion and differentiation induction by growth factors removal. Double-staining for nuclear antigens (GATA-1 or C/EBPβ, red) and surface antigens (JS4 or MC51/2, green). The bar represents 15 μm.
Individual colonies were then grown for 10 to 14 days in the absence of factors to allow their differentiation and analyzed again for the presence of erythroid and myeloid markers. The analysis of one representative wild-type and one ΔP1 clone is shown in Figure 5B. Individual wild-type v-myb clones contained less than 10% of JS4+/GATA-1+ erythroid cells whereas about 90% of cells were myeloid- MC51/2+/C/EBPβ+. On the other hand, ΔP1 v-myb clones contained about 90% JS4+/GATA-1+ erythroid cells and only a few MC51/2+/C/EBPβ myeloid cells. Additionally, GATA-1+ cells were always JS4+ and MC51/2− and C/EBPβ−. On the other hand, C/EBPβ+ cells were always JS4−, GATA-1−, and MC51/2+.
These data demonstrate unequivocally that the colonies were composed of both myeloid and erythroid cells, which must have originated from a common myeloid progenitor. Additionally, wild-type v-Myb progenitors developed predominantly into MC51/2+/C/EBPβ+myeloid cells whereas the majority of ΔP1 v-myb–infected progenitors developed into JS4+/GATA-1+erythroid cells. Thus, the intact LZR of v-Myb has the potential to bias the differentiation of common myeloid progenitors toward myeloid cells.
Several well-growing clones were later analyzed for provirus integration sites by southern blot hybridization. The majority of clones contained a single integration site, indicating that they originated from a single cell (data not shown).
Discussion
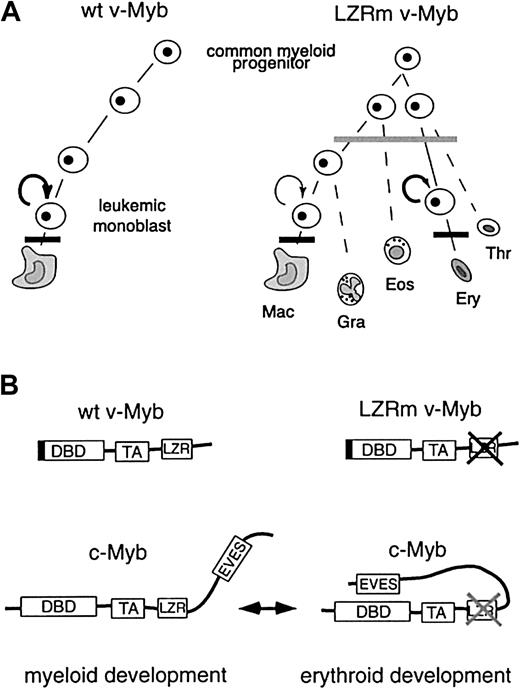
In this paper we describe the capacity of the Myb oncoprotein and its LZR to affect the lineage commitment of various hematopoietic progenitors. The lineage-determining activity of Myb LZR is particularly obvious when self-renewal of Myb progenitors is enhanced by specific growth factors (TGFα, bFGF, SCF). In the presence of these factors progenitors containing wild-type v-Myb or the LZR mutant ΔP1 v-Myb behave similarly and can be induced to develop into various lineages. While wild-type v-Myb progenitors differentiate predominantly into monoblastlike cells and to some extent also to eosinophils, cells with ΔP1 v-Myb differentiate into erythroid cells, monoblastlike cells, thrombocytes, and heterophilic granulocytes and eosinophils. As other LZR mutants, particularly the ΔLZ1 v-Myb, yielded similar results, the function in commitment was assigned to the entire LZR (Figure 6A). Therefore, we conclude that v-Myb LZR determines the commitment of progenitors predominantly into the macrophage lineage with a concomitant suppression of the development of erythroid cells, thrombocytes, and heterophilic granulocytes. On the other hand, v-Myb LZR mutants (ΔP1 v-Myb in particular) support development of erythroid cells and heterophilic granulocytes, similar to c-Myb. Thus, v-Myb still contains typical activities of c-Myb. These activities only appear to be overridden by wild-type LZR, presumably deregulated by C-terminal truncation of c-Myb. Therefore, we speculate that the LZR of c-Myb might also be involved in the lineage choice but in a regulated manner, for example, controlled by conformational changes in the c-Myb molecule (Figure 6B). It was established by previous work that the c-Myb C-terminus could form intramolecular interactions with the N-terminus.51 52We propose that this intramolecular interaction affects the accessibility of LZR for binding of other proteins. Depending on external signals, c-Myb LZR can either become accessible or not, and contribute in concert with other signaling pathways to enhanced formation of either myeloid or erythroid cells. We propose that the accessible state of c-Myb LZR is mimicked by wild-type v-Myb, whereas the inaccessible c-Myb LZR corresponds to v-Myb LZR mutants (Figure6B). The signals in the organism indicating the need of macrophages and granulocytes (eg, in infection and inflammation) might result in c-Myb conformation with the accessible (“active”) LZR that would induce expression of myeloid genes in progenitors and specify the development of granulocyte/macrophage lineages. Conversely, signals indicating a need for erythroid cells (eg, in hypoxia) might generate the c-Myb conformation with inaccessible (“inactive”) LZR. This conformation would cause accumulation of erythroid regulatory factors in progenitors and predominant formation of erythroid cells (Figure 6B). Posttranslational modifications and/or interaction with other regulatory proteins in response to external signals might qualify for causing such conformational changes. Thus, c-Myb could work as a molecular switch that determines the commitment of hematopoietic progenitors into myeloid or erythroid cells.
The model for Myb LZR activity in commitment of hematopoietic progenitors.
Scheme for the development of wild-type v-Myb and LZR mutant v-Myb (LZRm v-Myb) cells into various lineages. Macrophage (Mac), heterophilic granulocyte (Gra), eosinophil (Eos), erythrocyte (Ery), and thrombocyte (Thr). Horizontal bars denote strong (bold) or partial (gray) differentiation block induced by Myb proteins. Arrows indicate self-renewing potential of cells. (B) Model for regulation of lineage choice by c-Myb conformers based on biologic properties of v-Myb molecules (upper part) described in this paper. DNA binding domain (DBD), transactivation domain (TA), leucine zipper region (LZR), EVES domain.51
The model for Myb LZR activity in commitment of hematopoietic progenitors.
Scheme for the development of wild-type v-Myb and LZR mutant v-Myb (LZRm v-Myb) cells into various lineages. Macrophage (Mac), heterophilic granulocyte (Gra), eosinophil (Eos), erythrocyte (Ery), and thrombocyte (Thr). Horizontal bars denote strong (bold) or partial (gray) differentiation block induced by Myb proteins. Arrows indicate self-renewing potential of cells. (B) Model for regulation of lineage choice by c-Myb conformers based on biologic properties of v-Myb molecules (upper part) described in this paper. DNA binding domain (DBD), transactivation domain (TA), leucine zipper region (LZR), EVES domain.51
The hematopoietic progenitors transformed by v-Myb LZR mutants described in this paper, and exemplified by ΔP1 cells, resemble E26 v-myb-ets–transformed progenitors,12 since both progenitors can give rise to multiple hematopoietic lineages. Interestingly, in the E26 v-Myb-Ets protein the Myb LZR is absent due to the ets fusion, thus representing an LZR null mutation. In light of the results reported here this might be the reason why E26 v-Myb-Ets transforms multipotent and erythroid progenitors. In addition, the ets part of the E26 oncoprotein appears to be important, for self-renewal of early progenitors53 54 can be partially compensated by addition of growth factors to cells affected by the v-Myb LZR mutant.
The analysis of selected mRNAs and proteins with an impact on the development of myeloid and erythroid lineages revealed higher amounts of C/EBPβ, PU.1, and Egr-1 in wild-type Myb cells than in cells containing the ΔP1 Myb mutant. In marked contrast, ΔP1 cells have higher levels of c-Myb, SCL, and GATA-1. The rather high expression of Egr-1 in ΔP1 v-Myb progenitors appeared to be mainly caused by frequent addition of growth factors to progenitor cultures (J.K., unpublished data, May 2000).
Our data show that at least the elevated levels of C/EBPβ and PU.1 and reduction of c-Myb expression are determined by the LZR of Myb. We suggest that this specific expression pattern induced by Myb LZR is the molecular basis for development of wild-type v-Myb progenitors into the macrophage lineage since C/EBPβ and PU.1 were previously shown to commit E26 v-myb-ets progenitors to this lineage.13 14 The phenotype of ΔP1 v-Myb cells on the other hand seems to be determined in part by endogenous c-myb expressed in these cells. We have observed that overexpression of c-myb from the retroviral vector in blastoderm cells also results in accumulation of early erythroid cells. However, c-Myb cannot fully substitute for ΔP1 v-Myb in differentiation experiments, as c-Myb cells appear to need additional factors for survival whereas ΔP1 v-Myb cells are factor independent (P.B. et al, manuscript submitted, June 2001).
The data indicate that Myb can affect, through its LZR, expression of factors controlling development of myeloid and erythroid lineages. The exact mechanism of how this is achieved is not known at present. Recent work demonstrates that Myb can directly activate the C/EBPβ promoter in cooperation with the C/EBPβ factor itself,55suggesting that there exists an autoregulatory loop to generate high levels of C/EBPβ important for myeloid differentiation. Similarly, cooperation of Myb and MafB was also implicated in specific regulation of genes important for the development of myeloid cells.56,57 In addition, it was observed that Myb can both antagonize GATA-1 transcriptional activity and be inhibited by GATA-1.58 All these data document the Myb capacity to modulate expression and activity of other factors involved in regulation of hematopoiesis.
Additionally, a common feature of regulatory protein complexes in hematopoietic cells appears to be their association with the multifunctional regulator CBP, which recruits many hematopoietic transcription factors, including c-Myb.59 60 It is therefore tempting to speculate that Myb LZR (if present and accessible) favors the formation of a protein complex composed of CBP-Myb and myeloid factors (eg, C/EBPβ, PU.1). On the other hand, the inactivation or inaccessibility of Myb LZR (like in ΔP1 v-Myb or in the assumed “closed” c-Myb conformation) might result in CBP-Myb complexes enriched with erythroid factors (eg, GATA-1). As a result, myeloid or erythroid specific genes might become preferentially activated and determine the lineage choice. This hypothesis can be approached experimentally by analyzing the composition of Myb-CBP multiprotein complexes in early progenitor cells containing either wild-type or ΔP1 v-Myb.
This model also implies that Myb LZR itself binds specific protein factors. None of the above-mentioned factors bind Myb LZR, and therefore some others are likely to be involved. It can be expected that the complex biologic activity of Myb LZR is caused by rather complex interactions. At the present state of knowledge it is hard to indicate any protein whose interaction with Myb LZR might have been decisive for the lineage choice. The only well-characterized partner of Myb leucine zipper, the p160 protein,61 cannot be readily implicated in the regulation of hematopoietic commitment. Perhaps the analysis of recently identified transcription factors that bind to Myb leucine zipper (V.C., unpublished data, June 2000) might bring more information about mechanisms of action of Myb in hematopoietic progenitors.
We thank Drs J. D. Engel and J. Ghysdael for recombinant plasmids; and Drs H. Beug, C. Corbel, T. Graf, and K.-H. Klempnauer for providing antibodies. We are indebted to Drs Z. Kozmik and P. Urbanek for critical reading of the manuscript and I. Gallagher and S. Takacova for the help with manuscript preparation.
Supported by grants 301/98/K042 and 204/00/0554 from the Grant Agency of the Czech Republic (M.D.), A5052805 from the Grant Agency of the Academy of Sciences of the Czech Republic (M.D.), and 75195-540401 from the Howard Hughes Medical Institute (M.D. and M.Z.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Michal Dvořák, Institute of Molecular Genetics, Academy of Sciences of the Czech Republic, Flemingovo n 2, 166 37 Prague 6, Czech Republic; e-mail: mdvorak@img.cas.cz.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal