Abstract
The transcription factor PU.1 (also known as Spi-1) plays a critical role in the development of the myeloid lineages, and myeloid cells derived from PU.1−/− animals are blocked at the earliest stage of myeloid differentiation. Expression of the PU.1 gene is tightly regulated during normal hematopoietic development, and dysregulation of PU.1 expression can lead to erythroleukemia. However, relatively little is known about how the PU.1 gene is regulated in vivo. Here it is shown that myeloid cell type–specific expression of PU.1 in stable cell lines and transgenic animals is conferred by a 91-kilobase (kb) murine genomic DNA fragment that consists of the entire PU.1 gene (20 kb) plus approximately 35 kb of upstream and downstream sequences, respectively. To further map the important transcriptional regulatory elements, deoxyribonuclease I hypersensitive site mapping studies revealed at least 3 clusters in the PU.1 gene. A 3.5-kb fragment containing one of these deoxyribonuclease I hypersensitive sites, located −14 kb 5′ of the transcriptional start site, conferred myeloid cell type–specific expression in stably transfected cell lines, suggesting that within this region is an element important for myeloid specific expression of PU.1. Further analysis of this myeloid-specific regulatory element will provide insight into the regulation of this key transcriptional regulator and may be useful as a tool for targeting expression to the myeloid lineage.
Introduction
PU.1 is an Ets family transcription factor that plays an essential role in the development of both myeloid (granulocytes and monocytes/macrophages) and lymphoid lineages.1-3 PU.1 regulates almost every myeloid gene promoter studied to date, such as CD11b.4 PU.1 null mice have an absolute block in the development of monocytes/macrophages and B lymphoid cells. Granulocytes and T cells are also affected in that there is a delay in their development as well as abnormalities in neutrophil function.5,6 Subsequent analysis of PU.1−/− cells demonstrated that they are blocked at the earliest stage of myeloid maturation and lack expression of the granulocyte-macrophage colony-stimulating factor (GM-CSF), M-CSF, and G-CSF receptor genes, whose promoters were previously shown to be regulated directly by PU.1.7-12 Restoration of expression of PU.1, but not of the receptor genes, was able to rescue myeloid differentiation, demonstrating the importance of transcription factor function in establishing differentiation of these cells.12 13
Expression of PU.1 is restricted to hematopoietic lineages. PU.1 is expressed in multipotential progenitors, including erythroblasts.14 PU.1 expression is tightly regulated during subsequent hematopoietic differentiation: PU.1 is up-regulated with myeloid and down-regulated with erythroid commitment.15 In mature hematopoietic cells, PU.1 is expressed in myeloid and B lymphoid cells but not erythrocytes or T cells.1,14,16 17
As a critical determinant of hematopoietic lineage development, it is essential that PU.1 is properly regulated. PU.1 was originally isolated as the gene product of the Spi-1 locus as a protooncogene identified as a common spleen focus-forming virus integration site in murine erythroleukemia cells.18,19 The spleen focus-forming virus insertion leads to activation of PU.1 expression and blocks terminal differentiation of erythroblasts.18,19 Furthermore, overexpression of PU.1 in a transgenic model results in erythroleukemia.20 During normal hematopoietic development, PU.1 expression increases as stem cells commit to the myeloid lineage,15 and PU.1 expression is turned off with erythroid differentiation as GATA-1 expression increases.15 The importance of proper regulation of PU.1 is demonstrated by several recent studies showing that PU.1 suppresses GATA-1 activity to block erythroid differentiation.21-24
Previous promoter studies using transient transfections demonstrated myeloid cell type–specific regulation confined to a 334–base pair (bp) promoter sequence.25,26 Within this region, functional binding sites for octamer binding factors, Sp1, and PU.1 were identified.25,26 We have previously shown that the PU.1 promoter is autoregulated by PU.1 itself and that PU.1 in conjunction with Sp1 is important for myeloid expression, while regulation by octamer binding proteins in conjunction with the B cell–specific coactivator Bob-1 is likely to direct expression in B cells.25,26 A recent study demonstrated that C/EBPα can induce PU.1 gene expression in as short as 4 hours, suggesting a direct role for regulation of the PU.1 gene.27 Other studies have suggested that PU.1 expression might be regulated by DNA methylation and chromatin structure, as assessed by deoxyribonuclease (DNase) I hypersensitive studies.28 However, no studies to date have performed functional analyses regarding regulation of PU.1 gene expression in the context of chromatin or in transgenic mice.
Therefore, we undertook additional experiments to ask how PU.1 is regulated in vivo. Here we show that cell type– and tissue-specific chromatin-independent expression of the murine PU.1 gene can be conferred by a 91-kb genomic DNA fragment in both stable cell lines and transgenic mice. Within this 91-kb PU.1 locus, 3 potential transcriptional regulatory elements were identified using DNase I hypersensitive site (HS) studies. One of the 3 elements was shown to greatly enhance basal promoter activity and is likely to contain an important previously unidentified regulatory element.
Materials and methods
Cell culture
Human myeloid U937 cells (No. CRL1593, America Type Culture Collection, Rockville, MD) were grown in RPMI 1640 medium containing 10% fetal bovine serum (FBS) and 2 mM L-glutamine. The murine myeloid progenitor line 416B29 was maintained in Dulbecco modified Eagle medium with 15% FBS. The murine T-lymphoblast cell line BW514730 was cultured in Dulbecco modified Eagle medium with 10% heat-inactivated FBS. Also, 2 μg/mL puromycin was used for selection of stable cell lines.
Isolation of the murine PU.1 P1 clone and plasmid constructions
The 91-kb murine PU.1 clone (PU.1-P1-37) was obtained by screening a P1 library from Genome Systems (St Louis, MO). P1 clone mPU.1-Thy1.1-P1 consisted of a 544-bp murine Thy1.1 complementary DNA (cDNA) reporter31 inserted into the 91-kb murine PU.1 gene (PU.1-P1-37) between exons 4 and 5 through homologous recombination in bacteria.32 pXP2/−0.334 kb/Luc was generated by subcloning a 500 bp BamHI fragment spanning −334 to +147 of the murine PU.1 promoter region as described previously.25 pXP2−14 kb/−0.334 kb/Luc was generated by ligating a 3.5-kb HindIII fragment containing the −14 kb DNase I HS (see “Results”) into the HindIII site in front of the 334-bp mPU.1 promoter of pXP2/−0.334 kb/Luc.
Preparation of linearized DNA from P1 constructs for generation of transgenic mice and stable cell lines
The mPU.1-Thy1.1-P1 plasmid DNA was purified using a Qiagen Midi Kit followed by phenol extraction and ethanol precipitation. The plasmid was linearized with MluI and purified on a Sepharose CL4b column (Pharmacia, Valencia, CA) as previously described.32 The same linearized mPU.1-Thy1.1-P1 fragment, which contained the P1 vector, was used for making either transgenic mice or stable cell lines.
Transfections
Transient transfections were performed as described previously.33 U937, 416B, and BW5147 cells were electroporated at 300, 270 and 250 V, respectively, and 960 microfarads. Cells were harvested approximately 5 hours after electroporation. Transfection efficiency was normalized with the activity of cotransfected Renilla luciferaseactivity.34 Individual transfections were repeated 3 times. For stable cell lines, plasmid DNA (except for the mPU.1-Thy1.1-P1 clone) was purified with the Qiagen Maxi DNA purification kit. Stable cell lines were generated by electroporation of 2 × 107 416B or BW5147 cells, respectively, and 1 × 107 U937 cells as described for the transient transfections. Plasmids used for stable lines included 5 μgNdeI-linearized pXP2/−0.334 kb/Luc,AatII-linearized pXP2/−14 kb/−0.334 kb/Luc,SalI-linearized pPU.1/−1.3 kb/Thy1.1, orXhoI-linearized pPU.1/−1.3 kb/Intron I/Thy1.1 cotransfected with 0.2 μg NaeI-linearized pGK-puro.35 Cells were plated on 96-well plates and selected for transfected stable clones with 2 μg/mL puromycin 48 hours after electroporation.
Luciferase assays
Luciferase assays were performed according to the manufacturer's (Promega) protocol. Cell lysates were quantitated with the Bradford protein assay (Bio-Rad Laboratories, Hercules, CA).
Southern blot analysis
Genomic DNA was separated on agarose gels, blotted onto BioTrans+ membranes, hybridized, and washed as described.36 Stable chromosomal integration of the murine Thy1.1 or luciferase genes was detected by either a random-primed labeled 544-bp BamHI fragment of the murine Thy1.1 cDNA or a 1.7-kb XbaI fragment of the luciferase gene from pXP2.37
Northern blot analysis
Total RNA was isolated from stably transfected cell lines by guanidium extraction38 and from transgenic animals following tissue homogenization in 4 M guanidine isothiocyanate solution and separated on cushions of cesium trifluoroacetate (Amersham Pharmacia Biotech, Piscataway, NJ). Murine Thy1.1 RNA was detected with a labeled 544-bp BamHI fragment of the murine Thy1.1 cDNA,31 murine PU.1 messenger RNA (mRNA) with a 431-bp PU.1 5′ cDNA fragment (which is specific for murine PU.1),17 and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA with a 1.3- kb PstI fragment of rat GAPDH cDNA.39 Blots were subsequently hybridized for detection of Thy1, murine PU.1, and GAPDH mRNAs, respectively, and the blot exposed for several days in between hybridization to ensure that the previous probe had indeed been removed prior to subsequent hybridization.
DNase I hypersensitive site studies
DNase I hypersensitivity was assayed as described.36 Probes were as follows: probe 1 is a 1.4-kbEcoRI-PstI fragment located −13 kb 5′ of the transcriptional start site; probe 2 is a 2-kbBamHI-EcoRI fragment including 1.9 kb of intron 1 and exon 2; probe 3 is a 1.5-kb BamHI-EcoRI fragment from the 3′ end of the gene (located 1 kb downstream from exon 5); probe 4 is a 1.1-kb EcoRI-PstI fragment located 12 kb 5′ of the transcriptional start site; probe 5 is a 657-bpBamHI-DraIII fragment of intron 1; and probe 6 is 1.5 kb EcoRI-PstI fragment located 15 kb 5′ of the transcriptional start site.
Transgenic mice
MluI-linearized mPU.1-Thy1.1-P1 purified as described above was resuspended in 1 mM Tris, pH 8.0, 0.01 mM ethylenediaminetetraacetic acid, injected into fertilized oocytes of FVB mice, and implanted in uteri of pseudopregnant FVB mice according to standard procedures.31
Isolation of lineage-specific cell populations from transgenic animals
B220+ and CD4+/CD8+(meaning positive for either or both T-cell markers) cells were isolated from spleen and thymus, respectively, of PU.1 transgenic mice. Mac-1+ cells are isolated from peritoneal macrophages collected 48 hours after thioglycollate injection. To obtain Gr-1+ cells, 5 mg G-CSF was injected subcutaneously every 12 hours for 4 days prior to injection of thioglycollate. Granulocytes were collected from the peritoneal wash 4 hours later. This procedure yields high numbers of more than 90% granulocytic cells.17 40 Each cell population was incubated with rat anti-B220, anti-CD4 and anti-CD8, anti–Mac-1, or anti–Gr-1 antibody, respectively (Pharmingen). Subsequently, cells were incubated with goat antirat immunoglobulin G Microbeads and purified over a RS+ Separation Column and VarioMACS magnetic system (Miltenyi Biotec).
RT-PCR analysis of transgene expression in purified Mac-1+ myeloid and TER119+ erythroid cells
Mac-1+ cells and TER119+ cells were isolated from bone marrow cells of PU.1 transgenic mice. Bone marrow cells were stained with anti–Mac-1 antibody and anti-TER119 antibody (BD Pharmingen, San Diego, CA), and each 10 000 Mac-1+cells and TER119+ cells were sorted using a highly modified triple laser (488 nm argon laser, 599 nm dye laser, and ultraviolet laser) Cytomation system (MoFlo, Fort Collins, CO). The purity of sorted cells was more than 99%. Total RNA was extracted with Tri-reagent (Molecular Research Center, Cincinnati, OH) with 10 μg yeast transfer RNA as a carrier. The cDNA was synthesized with Superscript II RNase H-Reverse Transcriptase (Gibco, Gaithersburg, MD). Reverse transcriptase–polymerase chain reaction (RT-PCR) was performed with RNA equivalent to 200 cells each and Pfx DNA polymerase (Gibco). Primers for transgene-derived mRNA were PU.1 5′ sense primer 5′-AAACCTTGTCCCCAGCCCACCAG-3′ and Thy1.1 3′ antisense primer 5′-GCAGTCCAGGCGAAGGTTTTG-3′. Primers for murine GATA-1 were exon 2 5′-GCATCAGCACTGGCCTACTACAG-3′ and exon 3 5′-CCGTAAGCACTGCCGGTGACAGG-3′.41 Primers for murine GAPDH were 5′-GGTGCTGAGTATGTCGTGGAGTCTA-3′ and 5′-CAAAGTTGTCATGGATGACC-3′.
Western blot analysis of transgenic PU.1 proteins
Cell lysates were extracted with RIPA buffer. Proteins were detected with a PU.1 antibody recognizing an N-terminal PU.1 peptide,4 a PU.1 antibody that recognizes a C-terminal PU.1 peptide (Santa Cruz), and an anti-Thy 1.1 antibody (Santa Cruz).
Results
PU.1 promoter fragments that confer high-level activity in transient transfections do not express in transgenic mice
Previously published transient transfection studies had established that a 500-bp BamHI fragment containing 334 bp of the murine PU.1 promoter and 166 bp of 5′ untranslated region (“0.5-kb promoter”), as well as a 2.1-kb SacI fragment containing this promoter and additional 5′ sequences (“2.1-kb promoter”), conferred high-level promoter activity in myeloid and B-cell lines but not in other cell types.25,26 We asked whether either of these 2 constructs could direct expression in transgenic mice. We analyzed 4 founder lines with the 2.1 kb and 5 lines with the 0.5-kb promoter using a β-galactosidase reporter,31 and in none of the lines did we observe significant expression (detectable by β-gal staining) in B cells, T cells, or peritoneal macrophages (data not shown). Because we were concerned that the β-gal reporter system we were using might not work efficiently in granulocytes, we remade the transgenic constructs with the murine Thy-1 reporter gene that we had used successfully to measure CD11b promoter activity in transgenic mice both by fluorescence-activated cell sorter (FACS) and Northern blot analysis.31 In 12 independent transgenic founder lines—6 with the 2.1-kb and 6 with the 500-bp PU.1 promoter—we observed no detectable Thy1.1 surface expression by FACS of peripheral blood, spleen (source of B cells), peritoneal macrophages, or thymus (source of T cells) or mRNA by Northern blot of peritoneal macrophages, spleen, or thymus (data not shown). In summary, these experiments indicated that elements in addition to the promoter are important for expression in transgenic mice.
A murine PU.1 91-kb genomic fragment confers high-level reporter gene expression in myeloid cell lines
To direct cell type–specific gene expression in a chromatin-independent manner, additional regulatory elements must be needed. Therefore, we isolated a 91-kb murine genomic P1 clone containing the murine PU.1 gene (Figure1). Using a combination of field inversion and traditional agarose gel electrophoresis, we established a restriction map of the clone (Figure 1). The 20-kb murine PU.1 gene is located almost in the center of the P1 clone, with approximately 35 kb of 5′ and 3′ flanking sequence. A few hundred base pairs from each end of the 91-kb PU.1 genomic fragment were sequenced. The 5′ end sequence demonstrated identity with the 3′ end of murine tat binding protein-1 (GenBank accession No. D49686), while the 3′ end sequence of the PU.1 fragment did not demonstrate similarity to any known sequences (data not shown).
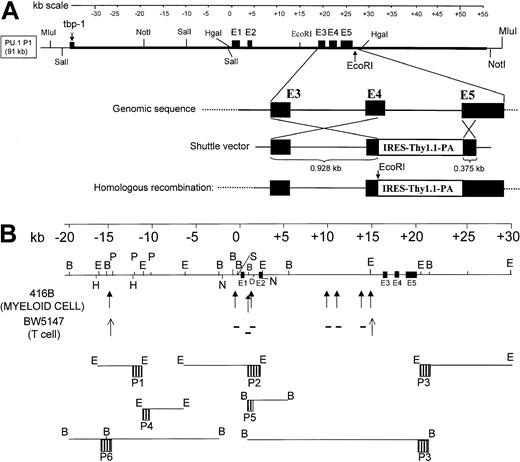
Isolation and characterization of a murine PU.1 91-kb P1 genomic clone.
(A) Map of the genomic clone and strategy for insertion of the Thy1.1 reporter by homologous recombination. The top cartoon is a kilobase scale. The middle panel is a map of the murine PU.1 91-kb genomic insert (thick black line), with sequences from the P1 vector shown by the thinner black line. Shown are the relative locations of the 5 coding exons, labeled E1 to E5; restriction sites of enzymes used to map the clone; and the location of the murine equivalent of the 3′ end of the human tbp-1 gene at the extreme 5′ end of the insert. The bottom panel shows the strategy used for insertion of the Thy1.1 reporter by homologous recombination. “Genomic sequence” represents an expanded view of the murine PU.1 genomic region between exons 3 to 5. The panel below depicts the shuttle vector in which a 928-bp homologous fragment, including parts of exon 3, intron 3, and exon 4, were placed upstream of an IRES followed by the 544-bp murine Thy1.1 cDNA, a polyadenylation signal, and a 375-bp homologous portion of exon 5. “Homologous recombination” represents the final structure of the modified P1 vector, referred to as mPU.1-Thy1.1-P1 in the text. Following homologous recombination into the same region as was done for PU.1 knockout animals, we predict a genomic structure of the P1 as shown in the bottom figure in which a single transcript would encode both the PU.1 gene and Thy1.1 reporter. The normal PU.1 reading frame terminates at the junction of the fragment of exon 4 and the IRES, and this construct would not be expected to encode a functional PU.1 protein. Also shown are restriction enzyme sites, including a PCR-generated EcoRI site in front of the IRES, Thy1.1 cDNA, and exon 5. This extra EcoRI site allows detection of the transgene from the endogenous PU.1 gene using a 544-bp BamHI Thy1.1 cDNA probe on Southern blots to establish transgene copy number.31 (B) Map of DNase I HSs and genomic probes. The top panel is a kilobase scale. The middle panel is a map of the murine PU.1 genomic sequence in this region, showing restriction enzymes used for DNase I mapping. Also shown are the 5 coding exons (E1-E5). DNase I HSs identified in this paper are indicated below the genomic map in either bold arrows (for sites in 416B myeloid cells) or in thin arrows (for sites in BW5147 T cells); sites identified in the myeloid cells but not T cells are indicated with a minus (−) symbol. The relative positions of each of the 6 probes (P1 to P6) and the restriction enzymes used with each probe are indicated at the bottom of the figure.
Isolation and characterization of a murine PU.1 91-kb P1 genomic clone.
(A) Map of the genomic clone and strategy for insertion of the Thy1.1 reporter by homologous recombination. The top cartoon is a kilobase scale. The middle panel is a map of the murine PU.1 91-kb genomic insert (thick black line), with sequences from the P1 vector shown by the thinner black line. Shown are the relative locations of the 5 coding exons, labeled E1 to E5; restriction sites of enzymes used to map the clone; and the location of the murine equivalent of the 3′ end of the human tbp-1 gene at the extreme 5′ end of the insert. The bottom panel shows the strategy used for insertion of the Thy1.1 reporter by homologous recombination. “Genomic sequence” represents an expanded view of the murine PU.1 genomic region between exons 3 to 5. The panel below depicts the shuttle vector in which a 928-bp homologous fragment, including parts of exon 3, intron 3, and exon 4, were placed upstream of an IRES followed by the 544-bp murine Thy1.1 cDNA, a polyadenylation signal, and a 375-bp homologous portion of exon 5. “Homologous recombination” represents the final structure of the modified P1 vector, referred to as mPU.1-Thy1.1-P1 in the text. Following homologous recombination into the same region as was done for PU.1 knockout animals, we predict a genomic structure of the P1 as shown in the bottom figure in which a single transcript would encode both the PU.1 gene and Thy1.1 reporter. The normal PU.1 reading frame terminates at the junction of the fragment of exon 4 and the IRES, and this construct would not be expected to encode a functional PU.1 protein. Also shown are restriction enzyme sites, including a PCR-generated EcoRI site in front of the IRES, Thy1.1 cDNA, and exon 5. This extra EcoRI site allows detection of the transgene from the endogenous PU.1 gene using a 544-bp BamHI Thy1.1 cDNA probe on Southern blots to establish transgene copy number.31 (B) Map of DNase I HSs and genomic probes. The top panel is a kilobase scale. The middle panel is a map of the murine PU.1 genomic sequence in this region, showing restriction enzymes used for DNase I mapping. Also shown are the 5 coding exons (E1-E5). DNase I HSs identified in this paper are indicated below the genomic map in either bold arrows (for sites in 416B myeloid cells) or in thin arrows (for sites in BW5147 T cells); sites identified in the myeloid cells but not T cells are indicated with a minus (−) symbol. The relative positions of each of the 6 probes (P1 to P6) and the restriction enzymes used with each probe are indicated at the bottom of the figure.
We next used homologous recombination in bacteria32to insert a Thy1.1 cDNA as a reporter into the PU.1 locus. We selected Thy1.1 because we have experience using it as a sensitive and effective reporter of protein and mRNA expression in macrophages and neutrophils of transgenic mice.31 We successfully inserted the internal ribosome entry site (IRES)–Thy1.1 construct into the PU.1 P1 clone, replacing part of exons 4 and 5. The targeted P1 clone (mPU.1-Thy1.1-P1) would be expected to express Thy1.1 mRNA from the PU.1 transcriptional unit with a predicted 3-kb mRNA species. We would also predict that the clone would efficiently translate Thy1.1 protein from the IRES but that the PU.1 coding region would be interrupted and a functional PU.1 protein not expressed.
We generated 9 independent stable lines of human U937 myeloid cells with the targeted mPU.1-Thy1.1-P1 clone. No deletions or rearrangements were noted following separation of restriction fragments by field inversion gel electrophoresis (data not shown). Thy1.1 mRNA was expressed in every clone harboring the transgene in both U937 (Figure2) and 416B (data not shown) stable cell lines. No Thy1.1 mRNA was detected in parental U937 cells (Figure 2A, lane 11) or clone 14 (Figure 2A, lane 5), which does not contain the transgene (data not shown). The T-cell line BW5147, which served as a positive control for Thy1 expression, did indeed express Thy1.1 mRNA but with a more slowly migrating species (1.85 kb vs the major band of 1.4 kb observed in the U937 transfectants). We observed 3 major mRNA species, which hybridized to the murine Thy1.1 probe in the transfectants of 3 kb, 2 kb, and 1.4 kb. The same pattern was consistently observed in every stable cell line as well as in transgenic mice in vivo (see below); therefore, the multiple RNA species observed are independent of the integration site and more likely an inherent property of the transgene. As noted above, we predicted a predominant 3-kb mRNA species but, in addition, observed a 2-kb species resulting from a cryptic splicing event (see below) as well as a predominant 1.4-kb mRNA, which presumably arises from yet another cryptic splice donor or acceptor we have not yet mapped.
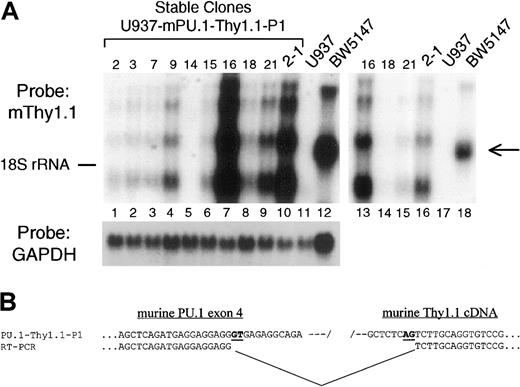
The 91-kb murine PU.1 genomic clone directs high-level reporter gene expression in stably transfected cell lines.
(A) Northern blot analysis of Thy1 mRNA in stable cell-line transfectants harboring the murine 91-kb PU.1 P1 clone. The 91-kb P1 DNA was linearized and stable transfectants established in myeloid U937 cells. The blot was sequentially probed with murine Thy1 cDNA (top) and GAPDH (bottom). Numbers at the top of the blot indicate clone number (lanes 1-10) as well as the negative control U937 cells (lane 11) and positive control BW5147 T-cell line (lane 12). Lanes 13 to 18 represent a limiting exposure of the left-hand blot (lanes 7-12). The position of migration of 18S ribosomal RNA is shown on the left, and the arrow on the right indicates the position of endogenous Thy1.1 mRNA in BW5147 T cells. (B) Sequence of the cryptic splice resulting in loss of Thy1.1 protein expression. Flanking primers were used to derive cDNA from the region spanning PU.1 exon 4, the IRES, and Thy1.1 cDNA (Figure1A). Shown in the upper line is the sequence of the PU.1-Thy1.1-P1 plasmid, and below it is the actual sequence derived from mRNA from stably transfected U937 cells. Alignment of the 2 sequences demonstrated a gap in the cDNA in which the IRES and 5′ end of the Thy1.1 cDNA (including the start ATG) are missing. Also shown in bold and underlined are nucleotides presumptively serving as the signals for the cryptic splice donor (GT) and acceptor (AG).
The 91-kb murine PU.1 genomic clone directs high-level reporter gene expression in stably transfected cell lines.
(A) Northern blot analysis of Thy1 mRNA in stable cell-line transfectants harboring the murine 91-kb PU.1 P1 clone. The 91-kb P1 DNA was linearized and stable transfectants established in myeloid U937 cells. The blot was sequentially probed with murine Thy1 cDNA (top) and GAPDH (bottom). Numbers at the top of the blot indicate clone number (lanes 1-10) as well as the negative control U937 cells (lane 11) and positive control BW5147 T-cell line (lane 12). Lanes 13 to 18 represent a limiting exposure of the left-hand blot (lanes 7-12). The position of migration of 18S ribosomal RNA is shown on the left, and the arrow on the right indicates the position of endogenous Thy1.1 mRNA in BW5147 T cells. (B) Sequence of the cryptic splice resulting in loss of Thy1.1 protein expression. Flanking primers were used to derive cDNA from the region spanning PU.1 exon 4, the IRES, and Thy1.1 cDNA (Figure1A). Shown in the upper line is the sequence of the PU.1-Thy1.1-P1 plasmid, and below it is the actual sequence derived from mRNA from stably transfected U937 cells. Alignment of the 2 sequences demonstrated a gap in the cDNA in which the IRES and 5′ end of the Thy1.1 cDNA (including the start ATG) are missing. Also shown in bold and underlined are nucleotides presumptively serving as the signals for the cryptic splice donor (GT) and acceptor (AG).
We performed Southern blot analysis to determine the number of copies of the transgene in each stable line (data not shown). In general, expression of Thy1.1 mRNA correlated very well with copy number. Therefore, it appears that the transgene is expressed in a copy number–dependent manner, although more clones with intermediate transgene copy numbers would be required to state this with certainty.
Initially, we analyzed reporter Thy1.1 protein expression by FACS analysis, but no detectable Thy1.1 surface expression was observed in any of the expressing clones (data not shown). We therefore performed further RT-PCR and sequencing analysis using total RNA isolated from the highest Thy1.1 mRNA expressing clone (No. 16). Sequencing of the RT-PCR product derived from this clone demonstrated that the IRES sequence intended to provide with ribosomal binding sites for efficient translation initiation had been spliced out using a cryptic splice donor found in murine PU.1 exon 4 (nucleotide 555, in the middle of exon 4) and splice acceptor in the middle of the murine Thy1.1 cDNA (Figure 2B).
DNase I hypersensitivity assays detect 3 sites within the murine PU.1 locus
We next sought to localize the important regulatory element(s) within the 91-kb murine PU.1 fragment that conferred expression in cell lines. Transcriptionally active regions in chromatin have an open configuration that allows transcription factors to bind to and are therefore more susceptible to DNase I digestion. To define the regulatory elements in the 91-kb PU.1 genomic DNA fragment, a 50-kb region spanning from −20 kb upstream to 10 kb downstream of the PU.1 gene was systematically searched for DNase I HSs (Figure3). Three DNase I HS regions were observed (Figure 3) as shown by Southern analysis of DNase I–treated genomic DNA. The first hypersensitive region (Figures 1B and 3A) is located 14- kb upstream of the transcriptional start site. This −14-kb HS appears not to be myeloid specific, because it was observed in both myeloid and the T-cell line BW5147, which does not express PU.1 (Figure3A). The second hypersensitive region mapped to the proximal promoter and intron 1 consisted of at least 3 distinct sites observed in 416B murine myeloid cells but not in BW5147 T cells. The +0.149 kb HS was localized near the proximal promoter, while the other 2 localized to intron 1 (Figure 3B). The third hypersensitive region was located in intron 2 with +9 kb, +12 kb, and +15 kb HSs observed only in 416B myeloid cells, while the +17 kb site was observed in both 416B myeloid and BW5147 T cells (Figure 3C). We did not observe specific DNase I HSs with probes 4, 5, and 6.
DNase I HS mapping detects 3 major regions in the PU.1 gene.
(A) DNase I HSs detected with probe 1 (P1 in Figure 1B). The arrow indicates a DNase I HS mapping approximately 14-kb upstream of the transcription start site detected in both myeloid 416B cells (left) and T-cell BW5147 cells (right). “0′” indicates no treatment with Dnase I; the triangle above the lanes indicates increasing time of digestion of genomic DNA with DNase I. DNA was digested with EcoRI. (B) DNase I HSs detected with probe 2 (P2 in Figure 1B). The arrows indicate 3 DNase I HSs at 149 bp, 1.4 kb, and 1.8 kb downstream of the transcription start site detected only in myeloid 416B cells (left) but not in T-cell BW5147 cells (right). DNA was digested withEcoRI. (C) DNase I HSs detected with probe 3 (P3 in Figure1B). The arrows indicate DNase I HSs at +9 kb, +12 kb, +15 kb, and +17 kb downstream of the transcription start site. The first 3 are observed only in myeloid 416B cells (left) but not in T-cell BW5147 cells (right); the +17-kb site is observed in both cell types. DNA was digested with BamHI.
DNase I HS mapping detects 3 major regions in the PU.1 gene.
(A) DNase I HSs detected with probe 1 (P1 in Figure 1B). The arrow indicates a DNase I HS mapping approximately 14-kb upstream of the transcription start site detected in both myeloid 416B cells (left) and T-cell BW5147 cells (right). “0′” indicates no treatment with Dnase I; the triangle above the lanes indicates increasing time of digestion of genomic DNA with DNase I. DNA was digested with EcoRI. (B) DNase I HSs detected with probe 2 (P2 in Figure 1B). The arrows indicate 3 DNase I HSs at 149 bp, 1.4 kb, and 1.8 kb downstream of the transcription start site detected only in myeloid 416B cells (left) but not in T-cell BW5147 cells (right). DNA was digested withEcoRI. (C) DNase I HSs detected with probe 3 (P3 in Figure1B). The arrows indicate DNase I HSs at +9 kb, +12 kb, +15 kb, and +17 kb downstream of the transcription start site. The first 3 are observed only in myeloid 416B cells (left) but not in T-cell BW5147 cells (right); the +17-kb site is observed in both cell types. DNA was digested with BamHI.
The −14-kb DNase I HS does not act as an enhancer but confers high-level myeloid-specific expression of the minimal PU.1 promoter in stably transfected myeloid cell lines
To determine if the −14-kb DNase I HS had a positive regulatory effect on the minimal promoter activity, a 3.5-kb HindIII fragment containing this site was subcloned in both orientations in front of the −334 PU.1 promoter directing luciferase reporter gene expression along with appropriate control plasmids (Figure4A). To ask whether this 3.5-kb fragment could function as a classical enhancer, these constructs were transiently transfected into either myeloid (U937 and 416B) or T-cell (BW5147) lines. As shown in Figure 4A, the −334-bp PU.1 promoter by itself demonstrated relatively high levels of luciferase activity in U937 cells, which express relatively high levels of PU.1.17 Lesser activity was observed in 416B cells, which represent a multipotential line and express lower levels of endogenous PU.1 mRNA (H.S.R. and D.G.T., unpublished results, 2001), and lesser still in BW5147 T cells, which do not express detectable levels of PU.1 (Figure 4A). When the 3.5-kb BamHI fragment containing the −14-kb HS was placed in either the genomic or reverse genomic orientation, no significant augmentation of either activity or specificity was observed compared with the −334-bp minimal promoter in transient transfections. Therefore, in the assay used, the −14-kb HS does not act as a classical enhancer element.
A region containing the DNase I HS located −14 kb upstream of the murine PU.1 gene directs cell type–specific expression in stably transfected cells.
(A) The DNase I HS region does not confer specificity (or activity) in transient transfections. The 3.5-kb HindIII fragment located at −14 kb (Figure 1B), containing a DNase I HS (Figure 3A), was placed in front of a 500-bp fragment containing 334 bp of 5′ upstream promoter sequence and 152 bp of 5′ untranslated sequences in front of a luciferase reporter gene in both normal and reverse orientation, as shown in the top diagram. These constructs were transiently transfected into myeloid and T-cell lines and luciferase activity measured (bottom panels) and compared with the activity of the promoter fragment or luciferase vector only. Also shown is the SD of at least 3 independent experiments. (B) The murine PU.1 upstream DNase I HS region confers activity in stable cell lines in myeloid cell lines but not a T-cell line. The PU.1 334-bp promoter constructs, with (top) or without (bottom) the 3.5-kb HindIII fragment containing the −14-kb DNase I HS, were stably transfected into myeloid U937 and 416B as well as T-cell BW5147 lines, and multiple independent clones assayed for luciferase activity normalized to the plasmid copy number as determined by Southern blot analysis. Shown is the SD of at least 10 independent clones for each construct in each cell line.
A region containing the DNase I HS located −14 kb upstream of the murine PU.1 gene directs cell type–specific expression in stably transfected cells.
(A) The DNase I HS region does not confer specificity (or activity) in transient transfections. The 3.5-kb HindIII fragment located at −14 kb (Figure 1B), containing a DNase I HS (Figure 3A), was placed in front of a 500-bp fragment containing 334 bp of 5′ upstream promoter sequence and 152 bp of 5′ untranslated sequences in front of a luciferase reporter gene in both normal and reverse orientation, as shown in the top diagram. These constructs were transiently transfected into myeloid and T-cell lines and luciferase activity measured (bottom panels) and compared with the activity of the promoter fragment or luciferase vector only. Also shown is the SD of at least 3 independent experiments. (B) The murine PU.1 upstream DNase I HS region confers activity in stable cell lines in myeloid cell lines but not a T-cell line. The PU.1 334-bp promoter constructs, with (top) or without (bottom) the 3.5-kb HindIII fragment containing the −14-kb DNase I HS, were stably transfected into myeloid U937 and 416B as well as T-cell BW5147 lines, and multiple independent clones assayed for luciferase activity normalized to the plasmid copy number as determined by Southern blot analysis. Shown is the SD of at least 10 independent clones for each construct in each cell line.
However, we obtained a significantly different result when the same −334-bp minimal PU.1 promoter construct, with and without the −14-kb HS fragment, was analyzed in stably transfected lines. In myeloid lines, the average luciferase activity with the −14-kb HS–containing fragment was approximately 194-fold higher (U937) and 46-fold higher (416B) than that obtained with the −334-bp minimal promoter alone (Figure 4B). Moreover, the difference was less than 2-fold with or without the −14-kb HS in stably transfected BW5147 T cells (Figure4B), in which both constructs were expressed at extremely low levels, more than 100-fold less compared with the myeloid lines. Therefore, the 3.5-kb BamHI fragment located at −14 kb with respect to the PU.1 transcriptional start site confers high-level activity to the PU.1 promoter in a chromosomal context. The number of stable clones obtained was not significantly different between the constructs (Table 1), suggesting a chromatin “opening” (or locus control region [LCR]) effect rather than increased integration into favorable chromatin.
Analysis of expression of clones from stable cell lines
| Cell type . | Construct . | Clones expressing the reporter/total clones with transgene . |
|---|---|---|
| U937 human myeloid (PU.1+) | Dnase I HS + PU.1 promoter | 14/14 |
| U937 human myeloid (PU.1+) | PU.1 promoter only | 10/14 |
| 416B murine myeloid (PU.1+) | Dnase I HS + PU.1 promoter | 18/18 |
| 416B murine myeloid (PU.1+) | PU.1 promoter only | 14/16 |
| BW5147 murine T cell (PU.1−) | Dnase I HS + PU.1 promoter | 18/18 |
| BW5147 murine T cell (PU.1−) | PU.1 promoter only | 15/18 |
| Cell type . | Construct . | Clones expressing the reporter/total clones with transgene . |
|---|---|---|
| U937 human myeloid (PU.1+) | Dnase I HS + PU.1 promoter | 14/14 |
| U937 human myeloid (PU.1+) | PU.1 promoter only | 10/14 |
| 416B murine myeloid (PU.1+) | Dnase I HS + PU.1 promoter | 18/18 |
| 416B murine myeloid (PU.1+) | PU.1 promoter only | 14/16 |
| BW5147 murine T cell (PU.1−) | Dnase I HS + PU.1 promoter | 18/18 |
| BW5147 murine T cell (PU.1−) | PU.1 promoter only | 15/18 |
Clones were considered to be expressing the reporter if luciferase activity was 3-fold higher than background. The luciferase activity of the average of expressing clones is shown in Figure4B.
The murine PU.1/91–kb genomic fragment confers tissue-specific reporter gene expression in transgenic mice
The results from analysis of the expression of the PU.1 transgene in cell lines suggested that this clone, in contrast to the smaller 2.1-kb promoter construct, contained sufficient regulatory sequences to direct expression in vivo. To test this, we established transgenic mouse lines containing this P1 clone. To date we have been able to establish lines from 2 independent founders that harbored approximately 6 copies of the transgene. In these 2 lines, high-level reporter gene mRNA expression was detected using a Thy1.1 probe in RNA derived from peritoneal macrophages, bone marrow, and spleen from transgenic animals (Figure 5A), with the same multiple RNA species as observed in stable cell lines (Figure 2A). No Thy1.1 mRNA was detectable in any nontransgenic animal tissue except for in the thymus, in which both the transgenic and nontransgenic littermates gave the same pattern, which is likely a result of endogenous Thy1.1 expression. After very long exposures, weak Thy1.1 hybridization signals could be detected in other tissues of transgenic animals, most likely arising from the presence of macrophages or granulocytes in these tissues (data not shown). Using a murine PU.1 probe, we observed a pattern of expression similar to that of the Thy1.1 probe in peritoneal macrophages, bone marrow, and spleen in transgenic animals but only a single strong mRNA species consistent with the endogenous PU.1 RNA17 in nontransgenic tissues and very weak hybridization in the thymus. These results indicate that the transgene expresses an mRNA species hybridizing to both the Thy1.1 and the PU.1 probe in tissues in which endogenous PU.1 is strongly expressed (peritoneal macrophages, bone marrow, spleen) but not in which PU.1 is weakly expressed (heart, kidney, lung, thymus). Both founder lines (including the one depicted in Figure 5A) have approximately 6 copies of the PU.1 transgene (data not shown). The mRNA expression levels are 3-fold and 3.5-fold of that of endogenous PU.1 by Northern blot analysis; therefore, we estimate that the amount of mRNA per transgene is very similar to that of the endogenous murine PU.1 gene. However, the number of founder lines is too small to draw precise conclusions as to whether expression correlates with transgene copy number. We predicted that no significant amount of PU.1-Thy1.1 hybrid protein would be produced because both an efficient ribosome binding site and start ATG codon were missing from the mRNA produced by the transgene (Figure 2B). By Western blot analysis using amino-terminal– and carboxyl-terminal–specific anti-PU.1 and anti-Thy1 antibodies, we were unable to detect such a PU.1-Thy1 fusion protein in transgenic mice expressing this transgene (data not shown). Therefore, it is very unlikely that a fusion peptide encoded by the transgene affects its expression.
The 91-kb murine PU.1 genomic clone directs high-level reporter gene expression in myeloid and B cells of transgenic mice.
(A) Northern blot analysis demonstrates that the transgene is specifically expressed in the same tissues as the endogenous murine PU.1 gene. Total RNA was isolated from the different tissues as indicated at the top from either a nontransgenic littermate (Tg −) or a murine PU.1 91- kb P1 transgenic mouse (Tg +). The blot was successively hybridized with a probe for murine Thy1 (top), murine PU.1 (middle), and GAPDH (bottom). The Thy1 probe hybridized to transgene mRNA strongly in peritoneal macrophages (MΦ), bone marrow (BM), and spleen and weakly in liver and lung. The hybridization observed in both nontransgenic and transgenic thymus is derived from the endogenous murine Thy1 gene, which is expressed strongly in thymus, weakly in spleen and heart, and undetectable in the other tissues examined and indicated by an arrow on the right of the panel. Similar results were obtained with a second transgenic founder line. (B) Northern blot analysis of fractionated cells demonstrate myeloid- and B cell–specific expression of the PU.1 transgene. Cells from the P1 transgenic line shown in panel A were fractionated into B220+ (B cells); Gr-1+ (granulocytes); Mac-1+ (granulocytes and macrophages); and CD4+and/or CD8+ (T-cell) populations as described in “Materials and methods.” Murine Thy1.1, PU.1, and GAPDH RNAs were detected by Northern blot analysis as described for panel A. The endogenous murine Thy1 mRNA species was detected in the CD4+/CD8+ lane slightly below the position of the second fastest migrating exogenous transgene band, as indicated by the arrow to the right of the panel. The purity of cells for each lineage was more than 95% for Gr-1+ cells and more than 90% for Mac-1+, B220+, and CD4+/CD8+ cells. (C) RT-PCR analysis demonstrates expression of transgenic RNA in Mac-1+ myeloid cells but not TER119+ erythroid cells. Highly purified (> 99%) Mac-1+ (lane 1) and TER119+ cells (lane 2) were isolated by FACS. Complementary DNA from 200 cells was used in each RT-PCR reaction. Transgene-derived mRNA was detected by a combination of 5′ PU.1 sense and 3′ Thy1.1 antisense primers. GATA-1 and GAPDH RT-PCR were performed as controls for RNA integrity.
The 91-kb murine PU.1 genomic clone directs high-level reporter gene expression in myeloid and B cells of transgenic mice.
(A) Northern blot analysis demonstrates that the transgene is specifically expressed in the same tissues as the endogenous murine PU.1 gene. Total RNA was isolated from the different tissues as indicated at the top from either a nontransgenic littermate (Tg −) or a murine PU.1 91- kb P1 transgenic mouse (Tg +). The blot was successively hybridized with a probe for murine Thy1 (top), murine PU.1 (middle), and GAPDH (bottom). The Thy1 probe hybridized to transgene mRNA strongly in peritoneal macrophages (MΦ), bone marrow (BM), and spleen and weakly in liver and lung. The hybridization observed in both nontransgenic and transgenic thymus is derived from the endogenous murine Thy1 gene, which is expressed strongly in thymus, weakly in spleen and heart, and undetectable in the other tissues examined and indicated by an arrow on the right of the panel. Similar results were obtained with a second transgenic founder line. (B) Northern blot analysis of fractionated cells demonstrate myeloid- and B cell–specific expression of the PU.1 transgene. Cells from the P1 transgenic line shown in panel A were fractionated into B220+ (B cells); Gr-1+ (granulocytes); Mac-1+ (granulocytes and macrophages); and CD4+and/or CD8+ (T-cell) populations as described in “Materials and methods.” Murine Thy1.1, PU.1, and GAPDH RNAs were detected by Northern blot analysis as described for panel A. The endogenous murine Thy1 mRNA species was detected in the CD4+/CD8+ lane slightly below the position of the second fastest migrating exogenous transgene band, as indicated by the arrow to the right of the panel. The purity of cells for each lineage was more than 95% for Gr-1+ cells and more than 90% for Mac-1+, B220+, and CD4+/CD8+ cells. (C) RT-PCR analysis demonstrates expression of transgenic RNA in Mac-1+ myeloid cells but not TER119+ erythroid cells. Highly purified (> 99%) Mac-1+ (lane 1) and TER119+ cells (lane 2) were isolated by FACS. Complementary DNA from 200 cells was used in each RT-PCR reaction. Transgene-derived mRNA was detected by a combination of 5′ PU.1 sense and 3′ Thy1.1 antisense primers. GATA-1 and GAPDH RT-PCR were performed as controls for RNA integrity.
To determine which bone marrow cells expressed PU.1 mRNA, we fractionated bone marrow cells into granulocytes, monocytes, B cells, and T cells. As expected, the Thy1.1 probe hybridized to a single strong RNA species in CD4+ and CD8+ T cells, consistent with hybridization to the endogenous Thy1.1 mRNA (Figure5B). In addition, the Thy1.1 probe detected 3 RNA species in granulocytes, monocytes, and B cells that do not normally express it, in a pattern similar to that observed in cell lines (Figure 2A) and mice (Figure 5A) harboring the P1 transgene. The expression pattern of Thy1.1 mRNA in these cells (granulocytes, monocytes, and B cells but not in T cells) is similar to that detected with the PU.1 probe (Figure5B). By RT-PCR (Figure 5C), we could easily detect transgene RNA expression in Mac-1+ myeloid cells, but we were unable to detect any evidence for transgene RNA in TER119+ erythroid cells. In summary, these results, combined with those obtained in stable cell lines, suggest that all of the regulatory elements necessary for high-level, cell type–specific PU.1 expression are contained within this 91-kb fragment.
Discussion
Our laboratory and others demonstrated that as little as 334 bp of murine PU.1 promoter sequence confers cell type–specific gene expression in transient transfection assays.3,25,26However, this promoter, as well as a longer fragment extending up to 2.1 kb, could not direct detectable reporter gene expression in multiple lines of transgenic mice, suggesting that additional transcriptional regulatory elements are needed to function in the context of chromatin. Because studies from other laboratories have demonstrated that elements that direct high-level expression in transgenic animals are often located tens of kilobases from the promoter,36,42-46 we expanded our search for these elements. We isolated a 91-kb PU.1 genomic DNA fragment that contains PU.1 gene locus and approximately 35 kb of DNA both 5′ and 3′ of the coding exons. We also inserted a murine Thy1.1 cDNA reporter gene31 between exons 4 and 5 using a RecA-based homologous recombination method.32 This 91-kb PU.1-Thy1.1 construct confers high-level, cell line–specific expression in stable transfection assays. Furthermore, the same construct directs tissue-specific reporter gene expression in transgenic mice in a manner similar to that observed by the endogenous PU.1 gene. These results indicate that distal elements located more than 2 kb from the transcription start site are required for proper expression of the PU.1 gene in vivo. Similar results were recently obtained for another gene expressed in very early hematopoietic cells, CD34.36 In the case of CD34, a minigene that included 5 kb of the CD34 promoter and a 3′ enhancer, both of which were active in transient transfection assays,47 failed to express CD34 in transgenic mice. However, a 160-kb PAC clone expressed CD34 very efficiently in vivo,36 and recent studies indicate that the distal elements required for CD34 expression are likely to be located approximately 18 kb 5′ of the transcription start site (Y. O. and D.G.T., unpublished results).
Because we initially planned to use FACS analysis to detect Thy1.1 protein expression, an IRES sequence that provides the binding site for translation initiation was placed in front of the Thy1.1 reporter cDNA. Unfortunately, we found that the IRES sequence was spliced out as a result of a cryptic splice donor site in the murine PU.1 exon 4 and a cryptic splice acceptor at the junction of Thy1.1 exon 1 and intron 1 (Figure 2B). As a result, we failed to detect Thy1.1 protein expression in the presence of abundant Thy1.1 mRNA. However, Thy1.1 reporter mRNA was detected in every clone containing the transgene in both U937 and 416B stable cell lines (Figure 2 and data not shown). Different-sized Thy1.1 mRNAs were detected in these stable clones, with a major transcript of 1.4 kb that is about 0.7 kb smaller than the expected size (Figure 2). This 0.7-kb decrease in the apparent size of the transcript matches the size of the IRES sequence plus those sequences from both PU.1 exon 4 and Thy1.1 exon 1. The nature of these transcripts was further verified by the finding of no detectable Thy1.1 mRNA expression in either parental U937 or 416B cell lines or clones without the transgene (Figure 2), while a 1.8-kb Thy1.1 mRNA was observed in the BW5147 T-cell line.
In addition to high-level and chromatin-independent reporter gene expression as that observed from the stable cell lines, this 91-kb PU.1 genomic DNA also confers cell type– and tissue-specific gene expression in transgenic mice. Expression of the Thy1.1 reporter gene was detected at high levels in peritoneal macrophages, bone marrow, and spleen, with very low levels in heart and liver (perhaps from contaminating blood or macrophages). No reporter gene expression was observed in kidney or lung. Importantly, the expression of the exogenous transgene was almost identical to that of the endogenous PU.1 gene (Figure 5). These results suggest that all of the elements necessary for proper control of PU.1 expression are contained within this 91-kb PU.1 genomic region.
One of the main features for these types of regulatory elements, such as LCRs, is chromatin opening ability.42,43,45 48-51 When genes in a chromosome are actively transcribed, they have an open configuration that allows transcription regulators to bind to and are therefore sensitive to DNase I treatment. To map specific transcriptional regulatory regions in this 91-kb PU.1 locus, we searched through an approximately 50-kb region spanning from 20 kb upstream to 10 kb downstream of the murine PU.1 gene. We identified 3 clusters of DNase I HSs, located −14 kb 5′ of the transcriptional start site, in intron 1, and in intron 2, respectively. The function of the first cluster of DNase I HSs at −14 kb was analyzed in both stably transfected 414B and U937 myeloid cells and BW5147 T-cell lines. This HS-containing region enhanced basal promoter activities approximately 100 to 200 times higher. Although the −14-kb DNase I HS was observed both in myeloid (416B and U937) and T (BW5147) cell lines, it appears to function differently in these 2 cell types. In myeloid cells, it enhances promoter function, while in T cells it does not. We find this result surprising, and in fact we can extend these results to demonstrate that a construct similar to one shown in Figure 4B, containing the −14-kb DNaseI HS and a 2-kb PU.1 promoter, can direct expression of a Thy1.1 reporter gene in transgenic mice in a specific manner similar to the 91-kb P1 (Y.O., unpublished data, 2001). It is possible that different transcription factors bind to the −14-kb DNaseI HS—perhaps activators in myeloid cells and repressors in T cells. Additional studies investigating this upstream region will be required to obtain further insight into why this element works in a specific manner in either stable cell lines or transgenic animals. In addition, the studies presented here demonstrate that the 91-kb genomic PU.1 DNA fragment is sufficient to direct lineage-specific expression, but the failure to express Thy1.1 surface protein due to the cryptic splice (Figure 2B) has prevented us from determining whether the 91-kb fragment confers development-specific expression. We plan to address this question in the future using constructs that express a surface marker (such as Thy1.1).
Recent studies have shown that LCRs are both distance and orientation dependent52 and that the DNase I HSs work together to confer chromatin opening as well as tissue-specific cell-type transcriptional activation. Further studies performed in vivo in transgenic mice will be required to determine the specific functions of the PU.1 DNase I HSs. If these DNase I HSs are shown to be the equivalent of myeloid-specific LCR-like elements, they could potentially provide powerful tools for directing heterologous gene expression for gene therapy applications in acute myeloid leukemia.
We thank Zheng Pan for suggestions with DNase I HS studies, Xiangdong Yang and Nathaniel Heintz for sharing information regarding targeted modification of the P1 by homologous recombination in bacteria, laboratory mates from the Tenen and Zhang laboratories for their support, and Mary Singleton and Alison Lugay for expert assistance with preparation of the manuscript.
Supported by grants CA41456 (D.G.T.) and AI30656 (R.A.M.).
Y. L. and Y. O. contributed equally to this work.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Daniel G. Tenen, Harvard Institutes of Medicine, Rm 954, 77 Ave Louis Pasteur, Boston, MA 02115; e-mail:dtenen@caregroup.harvard.edu.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal