Abstract
Several studies point to multiple members of the Hox transcription factor family as playing key roles in normal hematopoietic development, and they link the imbalanced expression of these transcription factors, in particular of the Abd-like A cluster HOXgenes HOXA9 and HOXA10, to leukemogenesis. To test directly the hypothesis that HOXA10is involved in human hematopoietic development, the gene was retrovirally overexpressed in human highly purified CD34+/GFP+ hematopoietic progenitor cells derived from cord blood or fetal liver sources, and the impact of aberrant gene expression was analyzed on differentiation and proliferation in vitro and in vivo. HOXA10 misexpression profoundly impaired myeloid differentiation with a higher yield of blast cells in liquid culture and a greater than 100-fold increased generation of blast colonies after in vitro expansion or after replating of primary colonies first plated in methylcellulose directly after transduction (P < .01). Furthermore, aberrantHOXA10 expression almost completely blocked erythroid differentiation in methylcellulose (P < .02).HOXA10 deregulation also severely perturbed the differentiation of human progenitors in vivo, reducing B-cell development by 70% in repopulated NOD/SCID mice and enhancing myelopoiesis in the transduced compartment. The data provide evidence that the balanced expression of HOXA10 is pivotal for normal human hematopoietic development and that aberrant expression of the gene contributes to impaired differentiation and increased proliferation of human hematopoietic progenitor cells. These results also provide a framework to initiate more detailed analyses ofHOX regulatory domains and HOX cofactors in the human system in vitro and in vivo.
Introduction
HOX homeobox genes were first recognized as an evolutionarily conserved family of transcription factors critical to the control of early embryonic development. More recently, many of these genes have been implicated in the regulation of both normal and leukemic hematopoiesis.1,2 Examples of the latter include the frequent involvement of PBX1, a cofactor ofHOX function, or MLL, a putative upstream regulator ofHOX gene expression and of HOXA9 in human leukemia-associated fusion genes.3-7 More recently, we and others have shown that the leukemic blasts from many patients with acute myeloid leukemia (AML) show an aberrant pattern ofHOXA10 expression, further supporting the concept that dysregulated HOX gene expression may be a general feature of this malignancy.8,9 Notably, the expression ofHOXA9 has been identified as one of the most consistent diagnostic markers of AML in humans and as the only single gene expression marker of more than 6800 cDNAs tested by DNA micro-array analysis that correlated with clinical outcome.10Additional evidence of the leukemogenic potential of deregulatedHOX gene expression and, in particular, of certain members of the A cluster have come from analysis of mouse models of leukemia. In the BXH2 mouse, the development of AML is linked to the activation of HOXA9 or HOXA7 by retroviral insertion, often in concert with activation of the Meis cofactor.11Retrovirus-mediated overexpression of HOXA10 orHOXA9 in murine hematopoietic cells also leads to AML in mice transplanted with these cells after an initial “latency” period, suggesting the additional involvement of certain co-operating oncogenes.12,13 The relevance of normalHOX gene expression for hematopoietic development is demonstrated by the HOXA9−/− knock-out mouse, which exhibits several hematopoietic abnormalities including a 40% reduction in total leukocytes, a blunted response to the administration of granulocyte colony-stimulating factor (G-CSF), and a marked deficiency in transplantable stem cell activity.14
Both HOXA9 and HOXA10 belong to the group of so-called Abdominal B-like genes, which are homologous to the most 5′ gene Abdominal B (AbdB), initially identified and characterized in the Drosophila homeotic complex. The Abd-like genes share a number of distinguishing features, such as a conserved tryptophan motif mediating the binding of the Pbx protein, differences at various positions in the flexible N-terminal arm of the DNA-binding homeodomain, preferential recognition of a TTAT core in contrast to the Antennapedia (Antp) group of homeodomains, and an insensitivity to activation by retinoic acid.15-18HOXA10 and HOXA9 are expressed in the most primitive CD34+ cell compartment of normal adult human bone marrow cells, and their expression is virtually extinguished in the more prevalent differentiating CD34−compartment.19 We have, therefore, hypothesized that 5′-HOX A cluster genes may play an important role in early human hematopoietic progenitors but that the down-regulation of these genes may be necessary for their further differentiation. On the other hand, overexpression and prolonged expression of these genes may induce severe perturbations of the normal differentiation program.
To test this hypothesis directly in primitive human hematopoietic cells, we initiated the present study. We focused on the analysis of the effects of HOXA10 overexpression in the progeny of human cord blood or fetal liver cells transduced in vitro with a retroviral vector encoding the human HOXA10 cDNA linked by an internal ribosomal entry site (IRES) element to enhanced green fluorescent protein. Severe perturbations of the proliferation and differentiation of the transduced cells, detectable both in vitro and in vivo in repopulated NOD/SCID mice, were seen. These findings establish the feasibility of using this approach to dissect the consequences of altered expression of specific HOX genes on primitive human hematopoietic cell behavior and underline the importance of normal expression of the Abd-like HOXA10 gene for proper human hematopoietic development.
Materials and methods
Retroviral constructs
The full-length HOXA10 cDNA, representing the most abundant HOXA10 transcript expressed in adult human bone marrow cells,20 was cloned as an EcoRI fragment upstream of the IRES sequence from the encephalomyocarditis virus (ECM) virus linked to the gene encoding an enhanced green fluorescent protein (EGFP; Clontech, Palo Alto, CA) by standard procedures (A10-GFP virus). The IRES GFP cassette was originally provided by P. Leboulch (Boston, MA) in a vector developed from the murine stem cell virus (MSCV) 2.1 construct (provided by R. Hawley, Bethesda, MD). As a control, the MSCV vector carrying only the IRES GFP cassette (GFP virus) was used. High-titer, helper-free recombinant retrovirus was generated by first transfecting the amphotropic Phoenix packaging cell line and subsequently transducing PG13 packaging cells as previously described.21,22 High-titer producer clones were isolated for each virus (mean titers of 6.3 × 105/mL for the A10-GFP virus and 2.5 × 105/mL for the GFP-virus, as assessed by transduction and analysis of GFP expression in K562 cells). The presence of full-length provirus integrants in the PG13 producer cells was confirmed by Southern blot analysis using standard techniques.23 Expression by these cells of full-lengthHOXA10 transcripts was demonstrated by Northern blot analysis of total cellular RNA, as described previously.2332P-labeled probes used were the 0.67-kb homeodomain-free fragment of HOXA10, a 0.8-kb full-length GFPgene, and a 2.0-kb PstI fragment containing the β-actin gene.
Human cells
Cord blood was obtained from mothers undergoing cesarean delivery of normal, full-term infants. Fetal liver cells were obtained from 14- to 21-week-old aborted human fetuses. In all cases, approved institutional procedures for obtaining informed consent were followed. Single-cell suspensions from fetal liver samples were obtained by gentle trituration of dispase-treated fragments. Low-density (< 1.077g/cm3) cells were isolated by density centrifugation on Ficoll-Hypaque (Pharmacia Biotech AB, Uppsala, Sweden). Low-density cells were washed twice in Hanks balanced salt solution (Stemcell Technologies, Vancouver, BC, Canada) supplemented with 5% fetal calf serum (FCS; Stemcell Technologies). Before infection, a population enriched in CD34+ cells was obtained by the removal of cells expressing a number of lineage (Lin) markers using an immunomagnetic column, as described by the manufacturer (Stemsep; Stemcell Technologies).
Retroviral transduction of human hematopoietic progenitor cells
Lin− cord blood or fetal liver cells were transduced as previously described.21 Briefly, cells at 2 × 105/mL were prestimulated for 48 hours in Iscove minimum Dulbecco medium (IMDM) containing a serum substitute (BIT; Stemcell Technologies), 10−4 M mercaptoethanol (Sigma Chemical, St Louis, MO), and 40 μg/mL low-density lipoproteins (Sigma) supplemented with the following recombinant human cytokines: 100 ng/mL Flt-3 ligand (Immunex, Seattle, WA) 100 ng/mL steel factor (SF, prepared and purified in the Terry Fox Laboratory) 20 ng/mL interleukin-3 (IL-3) (Novartis, Basel, Switzerland), 20 ng/mL IL-6 (Cangene, Mississauga, ON), and 20 ng/mL granulocyte-colony stimulating factor (G-CSF; Stem Cell). After 48 hours, cells were harvested and resuspended in filtered virus-containing medium supplemented with the same cytokine combination and protamine sulfate (5 μg/mL) on Petri dishes that had been precoated with 5 μg/cm2 fibronectin (Sigma) and preloaded twice with virus-containing medium, each time for 30 minutes, as described.21 This procedure was repeated on the next 2 consecutive days, for a total of 3 infections. For subsequent in vitro studies, cells were transferred to fresh serum-free medium plus cytokines and incubated for an additional 48 hours before they were stained with Cy5-labeled anti-CD34 antibody (Becton Dickinson, San Jose, CA). Transduced GFP+/CD34+cells were then quantitated and isolated using a 3-laser Facstar Plus (Becton Dickinson). Immunodeficient mice repopulated with transduced cells were injected without preselection less than 24 hours after the third day of exposure to virus.
In vitro progenitor assays
Assays for in vitro colony-forming cells (CFCs) were carried out in methylcellulose cultures (GF H4434; Stemcell Technologies) supplemented with 50 ng/mL human SF, 20 ng/mL each of human IL-3, IL-6, GM-CSF (Novartis), and G-CSF, and 3 U/mL erythropoietin (Stemcell Technologies), as described previously.24 Secondary progenitor assays were performed by replating aliquots of cells obtained by harvesting all the cells in 14-day-old primary assays. Cell morphology was assessed by Wright-Giemsa–stained smears of individually plucked colonies. Long-term culture-initiating cell assays (LTC-ICs) were carried out as previously described using pre-established irradiated murine fibroblasts genetically engineered to produce human IL-3, G-CSF, and SF.24 For in vitro liquid expansion assays, cells were cultured in the same serum-free medium described above. The types of cells present in these cultures at various time points was determined by staining cytospin preparations with Wright-Giemsa and by plating cells in CFC assays.
Mice
NOD/LtSz-scid/scid (NOD/SCID) mice were bred and maintained in micro-isolators under sterile conditions in the animal facility of the BC Cancer Research Centre, as previously described.25Eight- to 10-week-old mice were irradiated with 350 cGy from a cesium Cs 137 source less than 24 hours before human cells were injected. Engraftment and lineage differentiation were analyzed 8 weeks after transplantation by the aspiration of cells from the femurs under light anesthesia. Mice were humanely killed 12 weeks after transplantation, and the bone marrow was obtained from femurs and tibias. Cell suspensions were prepared in cold Hanks balanced salt solution, supplemented with 5% pooled normal human serum (Stemcell Technologies) for fluorescence-activated cell sorter (FACS) and progenitor analyses. The absolute number of human engrafted cells per mouse was calculated based on the recovered cells obtained from femurs and tibias and represented 25% of the total marrow.
Human lymphomyeloid engraftment and lineage representation of cells obtained from the mice 8 and 12 weeks after transplantation were determined as previously described.21,26 Briefly, red cells were lysed with 7% ammonium chloride (Stemcell Technologies), washed, labeled with 1% propidium iodine (PI; Sigma), and incubated on ice for 10 minutes with human serum supplemented with an anti-mouse IgG Fc receptor antibody (2.4G2; Systemix, Palo Alto, CA) to block mouse Fc receptors and to minimize nonspecific binding. Separate aliquots were then incubated for 30 minutes on ice with a mouse isotype-matched control antibody (Becton Dickinson) or the following antibodies against human antigens: antihuman CD34-Cy5,27CD19-phycoerythrin (PE; Becton Dickinson), CD15-PE (Pharmingen, Ontario, Canada), and CD45-PE (Pharmingen). Viable (PI−) cells were analyzed using a Facscalibur with Cellquest software (Macintosh, Cupertino, CA). Twenty thousand events were analyzed to measure the proportion of positive cells present; positive cells were defined as those exhibiting a level of fluorescence exceeding 99.98% of that obtained with isotype-control antibodies labeled with the same fluorochromes. Mice were defined as positive for human lymphoid and myeloid engraftment when 5 or more cells/20 000 bone marrow cells were CD34−/CD19+ and CD15+, respectively. To assess in vivo regenerated human CFCs, cells stained with antihuman CD34-Cy5 were isolated by FACS and additionally fractionated according to their expression of GFP. Subsequent staining of pooled colonies with antihuman CD45-PE/CD71-PE antibodies confirmed that they were human.
Statistical analysis
Data were statistically tested using the t test for dependent or independent samples (Statistica 5.1; Statsoft, Tulsa, OK). Differences with P < .05 were considered statistically significant.
Results
Efficient retroviral transduction of HOXA10 in human hematopoietic progenitor cells
To facilitate the selection and tracking of retrovirally transduced cells by FACS, a bicistronic vector for the long terminal repeat (LTR)-driven expression of HOXA10 and GFP was constructed in the MSCV viral backbone (A10-GFP virus, Figure1A). The analogous vector containing GFP alone (GFP virus) was used as a control. Full-length proviral integration and transcription were confirmed by Southern (Figure 1B) and Northern blot (Figure 1C) analyses, respectively, of stable PG13 producer cells, transduced hematopoietic target cell lines, K562, MO7E, and transduced primary cord blood cells. Fifty-four percent ( ± 17%) and 70% ( ± 12%) of low-density Lin−cord blood cells exposed to the A10-GFP vector or the GFP vector, respectively, were found to be GFP+ when examined by FACS, demonstrating the high efficiency of gene transfer by both vectors.
Structure and expression of the
HOXA10 retrovirus. (A) Diagrammatic representation of the integrated MSCV-HOXA10-IRES-GFP (A10-GFP) construct and the MIG (GFP) control vector. The expected sizes of the full-length viral transcripts are shown. (B) Southern blot analysis and (C) Northern blot analysis of total RNA of transduced cell lines and primary cord blood cells after 4 weeks in vitro culture. Membranes were hybridized with probes specific for GFP (Southern blot) or HOXA10 (Northern blot).
Structure and expression of the
HOXA10 retrovirus. (A) Diagrammatic representation of the integrated MSCV-HOXA10-IRES-GFP (A10-GFP) construct and the MIG (GFP) control vector. The expected sizes of the full-length viral transcripts are shown. (B) Southern blot analysis and (C) Northern blot analysis of total RNA of transduced cell lines and primary cord blood cells after 4 weeks in vitro culture. Membranes were hybridized with probes specific for GFP (Southern blot) or HOXA10 (Northern blot).
HOXA10-transduced cord blood cells produce increased numbers of primitive cells in vitro
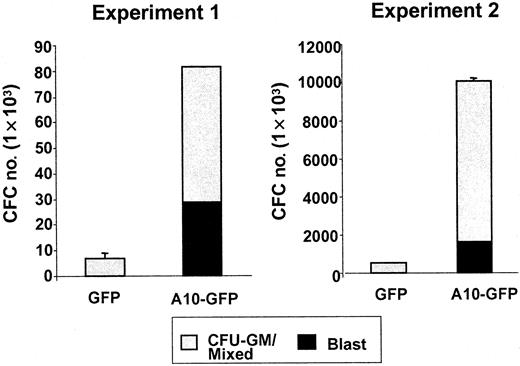
An immediate effect of HOXA10 overexpression on transduced cord blood cells was revealed by comparing the numbers and morphology of cells generated from CD34+ GFP+cells isolated by FACS 48 hours after their transduction with the A10-GFP or GFP vectors and placed in serum-free liquid cultures containing fetal liver, SF, IL-3, IL-6, and G-CSF. One week later, the total number of cells present in cultures initiated withHOXA10-transduced cord blood cells was approximately 1.7-fold higher than in parallel cultures initiated with GFP-transduced cord blood cells. Mean expansion (range) over input was 49 (17 to 109)-fold versus 33 (6.5 to 77)-fold, respectively; n = 4). This increased expansion was associated with an even greater (2.5-fold) increased production of blast cells. The mean proportion was 52% ± 18% vs 30% ± 11%, resulting in absolute numbers of 6.8 × 105 (0.5 × 105 to 18.6 × 105) versus 2.7 × 105(0.06 × 105 to 7.1 × 105) blasts, respectively, per 1 × 104CD34+GFP+ cells initially placed in culture. Analysis of the hematopoietic progenitor content of serum-free suspension cultures after 2 weeks showed that the mean absolute number of CFCs was 21-fold higher. The number of blast colonies was 125-fold higher in the cultures initiated with HOXA10-transduced cells than in the control cultures (n = 2; HOXA10, 29 and 1600 blast colonies, respectively; GFP, 0 and 13 blast colonies, respectively) (Figure 2). These results suggested that HOXA10 overexpression delayed myeloid differentiation.
Absolute numbers of CFU-blast and CFU-GM/mixed generated by A10-GFP or GFP-transduced progenitors cultured 2 weeks in serum-free medium supplemented with IL-3, IL-6, G-CSF, FLT3-ligand (FL), and SF.
Colonies were scored on day 14.
Absolute numbers of CFU-blast and CFU-GM/mixed generated by A10-GFP or GFP-transduced progenitors cultured 2 weeks in serum-free medium supplemented with IL-3, IL-6, G-CSF, FLT3-ligand (FL), and SF.
Colonies were scored on day 14.
HOXA10 induces alterations in cord blood progenitor cell differentiation in vitro
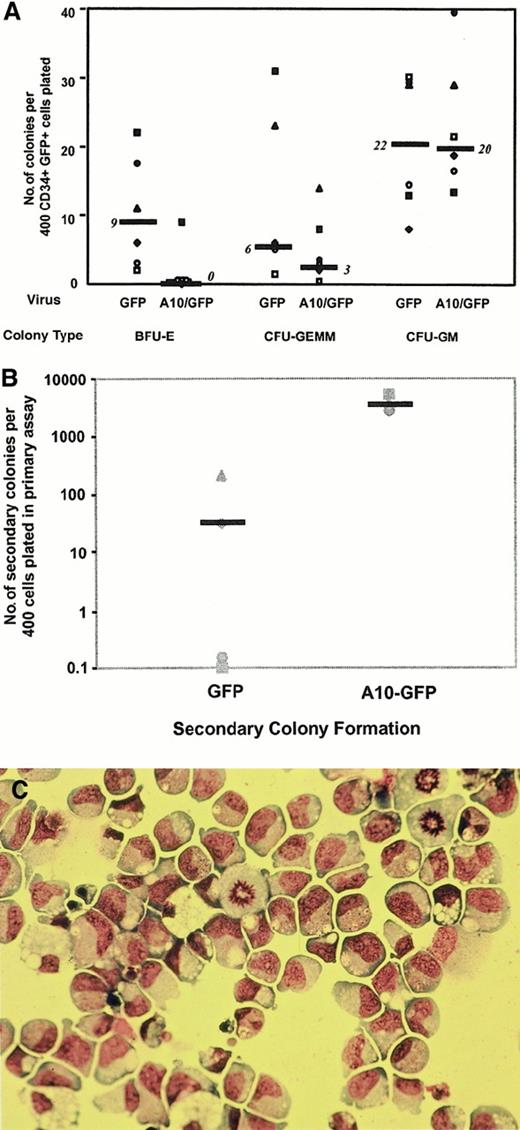
To determine whether dysregulated HOXA10 expression would also affect the proliferation and differentiation activity of progenitors detectable in other assay systems, CD34+/GFP+ cord blood cells isolated by FACS 48 hours after transduction were either plated directly into methylcellulose cultures or first cultured for 6 weeks on murine stromal fibroblast feeder layers engineered to produce human SF, IL-3, and G-CSF. Colony numbers obtained from direct CFC assays of cells from 6 independent transduction experiments are summarized in Figure3A. The total number of CFCs detected immediately after transduction was reduced for theHOXA10-transduced cells compared to the control GFP-transduced cells. This was because of an almost complete block of erythroid differentiation in the colonies generated byHOXA10-transduced cells (more than 85% reduction in the number of “pure” erythroid colonies; n = 6;P = .017; 54% reduction of mixed erythroid–“myeloid” colonies by comparison to the control GFP-transduced cells). Replating studies showed that, as expected, only small numbers of colonies were obtained when control primary CFC cultures were assayed in secondary cultures (median, 30 secondary colonies per 400 primary CD34+ GFP+ cells plated into the primary CFC assays) (Figure 3B). In contrast, more than 100-fold higher numbers of secondary colonies were generated on the replating of cells from primary CFC assays of HOXA10-transduced cells (median, 3700 colonies per 400 initially plated CD34+GFP+;P < .01). Moreover, Wright-Giemsa staining showed approximately 80% of these HOXA10-derived secondary colonies to consist morphologically of blasts (Figure 3C). In 2 of 4 experiments, HOXA10-transduced cells obtained from the secondary colonies were able to form tertiary colonies, though at somewhat reduced numbers, and all tertiary colonies consisted of blast cells as assessed by morphology.
Colonies derived from A10–GFP- and GFP-transduced progenitors.
(A) Comparison of the frequency and distribution of colonies derived from A10–GFP- and GFP-transduced progenitors in methylcellulose scored on day 14. Median frequencies of 6 independent experiments performed with 6 different cord blood samples are indicated by bars. (B) Methylcellulose dishes of primary CFC cultures derived from A10–GFP- and GFP-transduced progenitors were killed on day 14, and progenitor cells resuspended in IMDM 2% FCS were replated in secondary methylcellulose cultures. Secondary colony formation was assessed on day 14. Medians of secondary colony formation of 4 independent experiments performed with 4 different cord blood samples are indicated by bars. (C) Morphology of blast colonies was confirmed by staining individually plucked colonies with Wright-Giemsa. CFU-GEMM indicates colony-forming unit–granulocyte erythroid macrophage megakaryocyte; BFU-E, burst-forming unit–erythroid.
Colonies derived from A10–GFP- and GFP-transduced progenitors.
(A) Comparison of the frequency and distribution of colonies derived from A10–GFP- and GFP-transduced progenitors in methylcellulose scored on day 14. Median frequencies of 6 independent experiments performed with 6 different cord blood samples are indicated by bars. (B) Methylcellulose dishes of primary CFC cultures derived from A10–GFP- and GFP-transduced progenitors were killed on day 14, and progenitor cells resuspended in IMDM 2% FCS were replated in secondary methylcellulose cultures. Secondary colony formation was assessed on day 14. Medians of secondary colony formation of 4 independent experiments performed with 4 different cord blood samples are indicated by bars. (C) Morphology of blast colonies was confirmed by staining individually plucked colonies with Wright-Giemsa. CFU-GEMM indicates colony-forming unit–granulocyte erythroid macrophage megakaryocyte; BFU-E, burst-forming unit–erythroid.
When the initially transduced CD34+GFP+ cells were cultured under LTC-IC assay conditions, the number of CFCs present 6 weeks later was similar for both HOXA10 and control GFP+ input populations. However, most (68% ± 8%) of colonies generated by HOXA10-transduced cells maintained under these conditions again consisted of blasts or granulocyte precursors, whereas the CFCs obtained from LTC-IC assays of control cells produced mainly mature granulocyte-macrophage colonies (99.5% ± 0.25%). These studies indicate the ability ofHOXA10 overexpression to significantly impair myeloid differentiation, particularly the terminal aspects of erythroid differentiation.
HOXA10 perturbs the output of lymphoid cells by in vivo repopulating human progenitor cells
To determine whether overexpression of HOXA10 would affect the proliferation and differentiation of cord blood cells able to engraft the marrow of NOD/SCID mice, cord blood or fetal liver cells were injected into mice immediately after infection, without any preselection of CD34+ GFP+ cells. Each mouse was injected with the progeny of an original input of Lin−cells containing 105 CD34+ cells. Engraftment with human cells was first assessed by FACS analysis of bone marrow cells obtained by femoral bone marrow aspirate 8 weeks after transplantation and again 12 weeks after, when the mice were killed. At 8 weeks after transplantation, both human lymphoid (CD34−CD19+) and human myeloid (CD15+) cells could be detected in 13 of 14 animals injected with A10-GFP cells and in 6 of 14 mice injected with control GFP cells. Moreover, in all cases, human lymphomyeloid engraftment was evident among both the GFP+ and the GFP−cells. Four weeks later, 11 of 14 animals in the HOXA10group and 7 of 14 in the GFP control group showed lymphomyeloid engraftment, with a median frequency in human cells of 18.5% (0.7%-66%) and 23% (5%-66%), respectively. Twelve percent (2%-48%) of the engrafted human cells were transduced in the mice transplanted with A10-GFP cells versus 12% (2%-89%) in the mice transplanted with control GFP cells.
Although overexpression of HOXA10 in human NOD/SCID repopulating cells was compatible with lymphomyeloid engraftment, the proportion and absolute yield of human lymphoid and myeloid cells produced in the recipients of HOXA10-transduced cells were significantly altered. In both the GFP+ and the GFP− populations in mice transplanted with control cells, the size of the human lymphoid compartment was approximately 4-fold greater than the size of the human myeloid compartment, as previously reported.22 27 An almost identical approximately 4-fold predominance of human B lymphoid cells was observed in the GFP− (nontransduced) compartment of human cells in mice injected with HOXA10-transduced cells (Figure4A,B). In contrast, in the same mice, the number of GFP+ lymphoid cells was greatly reduced, by 70%, compared with the number of GFP+ lymphoid cells in the control mice—3.7 × 106 CD19+ cells (±2.7) versus 11.9 × 106 cells (±9.1) per mouse, respectively, with a proportional predominance of human myeloid cells in this compartment (P < .02) (Table1). These alterations in the proportion and absolute yield of human lymphoid and myeloid cells were the same in mice transplanted with cord blood cells or fetal liver cells. Although on average the number of transduced human myeloid cells in the GFP+ compartment in mice injected withHOXA10-transduced cells or control cells was similar at both time points assessed—1.7 × 106 (±0.7) vs 1.4 × 106 cells (±0.7), respectively—closer examination of individual mice revealed substantial effects ofHOXA10 on human myelopoiesis. In 3 of 11 mice, the transduced myeloid cells had a clear, competitive growth advantage, with a 2.6 (±0.5)-fold higher absolute number of human myeloid cells in the GFP+ compartment than in the GFP−compartment. In these mice, 72% (±3.9%) of the human myeloid cells were GFP+, whereas only 46.6% (±25.6%) of the human cells injected at the time point of transplantation were GFP+. In contrast, only 15.6% (±6.7%) of the human lymphoid compartment was GFP+, indicating a myeloproliferative effect of HOXA10 overexpression in a subgroup of NOD/SCID mice (Table 2). Although we did not observe an increase in the CD34+/GFP+ compartment in the HOXA10mice compared with the control, more detailed analysis of the progenitor content of this cell population revealed that in all animals analyzed (n = 6) HOXA10 overexpression perturbed normal myelopoiesis, as reflected by an increase in the frequency of CFCs and an impairment of terminal myeloid differentiation ex vivo. As shown in Table 3, the frequency of CFCs in the GFP+ fraction of CD34+ cells isolated from HOX-A10 mice (HOXA10+) 12 weeks after transplantation was consistently higher (1.2- to 12.9-fold;P = .04) than in the GFP− fraction of these mice. Moreover, a large proportion of those derived from GFP+ precursors formed blast cell colonies, whereas this was rarely seen in the assays of the GFP− cells. Sufficient CD34+ cells were isolated from 2 of these mice to permit their growth in LTC to be evaluated. In both cases, significantly more (34- and 44-fold) CFCs were detected in the 6-week-old LTCs initiated with the GFP+ than with GFP− cells.
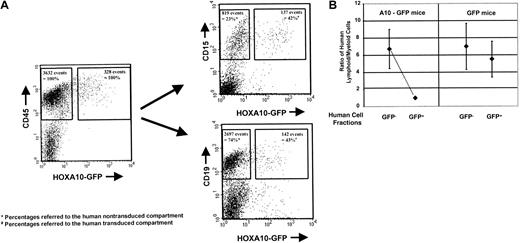
Engraftment of human cells in NOD/SCID mice: A10-GFP virus GFP.
A10–GFP- or GFP-transduced progenitors and the nontransduced cell fraction were injected into NOD/SCID mice directly after finishing transduction. Engraftment of human lymphoid and myeloid cells was assessed by quantifying CD19+/CD34− or CD15+12 weeks after transplantation by FACS analysis. The FACS profile of the BM of a representative mouse transplanted withHOXA10-transduced and nontransduced fetal liver cells is shown at 12 weeks after transplantation. Percentages of GFP+ and GFP− human lymphoid and myeloid cells are presented as proportions of the human GFP+ and GFP− CD45+ cell populations, respectively (A). Ratios of human lymphoid and myeloid cells were calculated for the individual mice in the GFP and A10-GFP experimental groups (n = 7 and n = 11, respectively) for both the GFP+ and the GFP− compartments. Median ratios are indicated (B).
Engraftment of human cells in NOD/SCID mice: A10-GFP virus GFP.
A10–GFP- or GFP-transduced progenitors and the nontransduced cell fraction were injected into NOD/SCID mice directly after finishing transduction. Engraftment of human lymphoid and myeloid cells was assessed by quantifying CD19+/CD34− or CD15+12 weeks after transplantation by FACS analysis. The FACS profile of the BM of a representative mouse transplanted withHOXA10-transduced and nontransduced fetal liver cells is shown at 12 weeks after transplantation. Percentages of GFP+ and GFP− human lymphoid and myeloid cells are presented as proportions of the human GFP+ and GFP− CD45+ cell populations, respectively (A). Ratios of human lymphoid and myeloid cells were calculated for the individual mice in the GFP and A10-GFP experimental groups (n = 7 and n = 11, respectively) for both the GFP+ and the GFP− compartments. Median ratios are indicated (B).
Proportions of lymphoid and myeloid engraftment in NOD/SCID mice
| . | Proportions of human cells . | |
|---|---|---|
| CD34−/CD19+(lymphoid) . | CD15+ (myeloid) . | |
| Median % (range) | Median % (range) | |
| A10-GFP mice (n = 11) | ||
| GFP+ compartment | 27.6 (4-71) | 31 (8-81.3) |
| GFP− compartment | 65.5 (43-90) | 14.7 (3-28) |
| GFP mice (n = 7) | ||
| GFP+ compartment | 72.7 (35-87.5) | 15.3 (7.5-64) |
| GFP−compartment | 61.5 (53-81) | 16 (4-32) |
| . | Proportions of human cells . | |
|---|---|---|
| CD34−/CD19+(lymphoid) . | CD15+ (myeloid) . | |
| Median % (range) | Median % (range) | |
| A10-GFP mice (n = 11) | ||
| GFP+ compartment | 27.6 (4-71) | 31 (8-81.3) |
| GFP− compartment | 65.5 (43-90) | 14.7 (3-28) |
| GFP mice (n = 7) | ||
| GFP+ compartment | 72.7 (35-87.5) | 15.3 (7.5-64) |
| GFP−compartment | 61.5 (53-81) | 16 (4-32) |
Number of engrafted myeloid and lymphoid human cells in the transduced versus nontransduced compartment of HOXA10mice
| HOXA10 mice . | 5 A10#3 . | 6 A10#1 . | 7 A10#2 . |
|---|---|---|---|
| Cell no./mouse (× 105)* . | |||
| CD15+GFP+ | 37 | 0.73 | 14.6 |
| CD15+GFP− | 13.7 | 0.24 | 7 |
| CD19+GFP+ | 34 | 0.6 | 7.2 |
| CD19+GFP− | 165 | 6.6 | 26.3 |
| HOXA10 mice . | 5 A10#3 . | 6 A10#1 . | 7 A10#2 . |
|---|---|---|---|
| Cell no./mouse (× 105)* . | |||
| CD15+GFP+ | 37 | 0.73 | 14.6 |
| CD15+GFP− | 13.7 | 0.24 | 7 |
| CD19+GFP+ | 34 | 0.6 | 7.2 |
| CD19+GFP− | 165 | 6.6 | 26.3 |
Assessed 12 weeks after transplantation.
Ex vivo colony formation of A10–GFP-transduced human cells 12 weeks after transplantation into NOD/SCID mice
| Experiment no. . | Ex vivo CFC assay . | ||
|---|---|---|---|
| . | Total colony no./1.5 × 103 CD34+ cells . | % Blasts . | |
| 5 A10#1 | GFP+ | 5.5 | 100 |
| GFP− | 2.5 | 0 | |
| 6 A10#3 | GFP+ | 83 | 53 |
| GFP− | 32 | 9 | |
| 7 A10#1 | GFP+ | 97 | 45 |
| GFP− | 7.5 | 0 | |
| 7 A10#2 | GFP+ | 99.5 | 0 |
| GFP− | 84.5 | 0 | |
| 7 A10#4 | GFP+ | 96 | 43 |
| GFP− | 55 | 7 | |
| 7 A10#5 | GFP+ | 72.5 | 23 |
| GFP− | 54 | 13 | |
| P value | .04 | .04 | |
| GFP+ vs GFP− | |||
| Experiment no. . | Ex vivo CFC assay . | ||
|---|---|---|---|
| . | Total colony no./1.5 × 103 CD34+ cells . | % Blasts . | |
| 5 A10#1 | GFP+ | 5.5 | 100 |
| GFP− | 2.5 | 0 | |
| 6 A10#3 | GFP+ | 83 | 53 |
| GFP− | 32 | 9 | |
| 7 A10#1 | GFP+ | 97 | 45 |
| GFP− | 7.5 | 0 | |
| 7 A10#2 | GFP+ | 99.5 | 0 |
| GFP− | 84.5 | 0 | |
| 7 A10#4 | GFP+ | 96 | 43 |
| GFP− | 55 | 7 | |
| 7 A10#5 | GFP+ | 72.5 | 23 |
| GFP− | 54 | 13 | |
| P value | .04 | .04 | |
| GFP+ vs GFP− | |||
Discussion
Several lines of evidence have shown that the homeobox family plays a critical role in normal hematopoietic development and that their imbalanced expression is linked to leukemogenesis. In particular, the 5′-located A cluster HOX genes, such asHOXA9 and HOXA10, are associated with the development of leukemia by the molecular characterization of leukemia-specific fusion genes or by studies in murine transplantation models, using bone marrow cells engineered to overexpress theHOXA10 or HOXA9 gene.3,4,12,13 The precise regulatory role of these Abd-like A clusterHOX genes in human hematopoietic development has not yet been dissected. To test the hypothesis that HOXA10 is critically involved in early hematopoietic differentiation in humans, we aberrantly expressed this gene in human hematopoietic progenitor cells and studied the effect of misexpression on hematopoietic development in vitro and in vivo. For this, we exploited recent advances in the technique of retroviral gene transfer that allow the transduction of human hematopoietic progenitor cells, including those that repopulate NOD/SCID mice with high efficiency.21Thus, it was possible to analyze the effect of deregulatedHOX gene expression in vivo for the first time in the human system.
In vitro studies revealed striking effects of HOXA10overexpresssion on cell expansion in liquid culture or cell proliferation of clonogenic progenitors in methylcellulose. In parallel, HOXA10 profoundly altered the normal differentiation program of early progenitors and impaired myeloid differentiation: it increased the yield of blast cells in liquid culture, induced a greater than 100-fold enhanced generation of blast colonies after in vitro expansion or after replating of methylcellulose colony assays, and led to an almost complete block of erythroid colony formation. Furthermore, it perturbed the differentiation of LTC-IC such that most of their clonogenic progeny detected after 6 weeks in culture were CFU-blast. These observations probably highlight key regulatory features of this gene—to enhance proliferation and, in parallel, to impair differentiation of early hematopoietic progenitors. Thus, high-expression levels of HOXA10 in concert with otherHOX genes may be necessary to maintain CD34+cells in an undifferentiated state with high proliferative potential, while subsequent silencing of the gene may be permissive for maturation and a parallel decrease in proliferative capacity. These predictions are consistent with the high expression of HOXA10 in CD34+ progenitors and its down-regulation in the differentiating CD34− fraction.19Furthermore, our model of retrovirally transduced progenitor cells, with an increased and extended HOXA10 gene expression, mimics the deregulation of HOXA10 demonstrated in blasts of patients with AML, which lack the normal down-regulation ofHOXA10 expression in the more differentiated CD34− cell population.9 Interestingly, the impact of HOXA10 dysregulation on cell proliferation and replating efficiency were comparable to the in vitro effects observed with engineered overexpression of the leukemia-specific fusion geneTLS-ERG, which is associated with poor-prognosis AML.28 Comparable to HOXA10 effects, overexpression of HOXA5 was shown to inhibit erythroid colony formation, whereas overexpression of HOXB7 orHOXC4 induced unchanged or increased BFU-E colony formation, respectively.29-32
In our study we also analyzed the impact of HOXA10deregulation on the differentiation of human progenitors in vivo in sublethally irradiated NOD/SCID mice. These experiments demonstrated a substantial impairment in B-cell development in mice transplanted with cells overexpressing the gene. So far, there are no data on the effect of other HOX genes on B-cell differentiation in the human system. However, data from murine transplantation models demonstrated that both HOXA10 andHOXA9 are detrimental to B-cell development in vivo.12,13 There are no precise data concerning at which stage of B-cell development the block occurs, but pre-B colony formation assays suggest that the development of murine IL-7–sensitive pre-B cells is impaired by HOXA10overexpression.12 The impact of HOXA10 on early B-cell progenitors was confirmed by recent results suggesting thatHOXA10 overexpression critically impairs the transition from the pro-B to the pre–B-cell stage, as defined by Hardy inHOXA10 transgenic mice.33 34
In addition, the in vivo studies confirmed that HOXA10overexpression perturbed normal myeloid development: misexpression of the gene had a myeloproliferative effect with an increased frequency of CFCs in the human CD34+ compartment, an impairment of terminal myeloid differentiation ex vivo, and a competitive growth advantage of transduced myeloid cells in a subgroup of mice. However, we did not observe frank signs of a myeloproliferative syndrome or of leukemia in the animals. It may be that other HOXcofactors are required for accelerated leukemogenesis. This was shown for HOXA9 by a shortened latency period to the development of leukemia in mice transplanted with cells overexpressing both HOXA9 and its cofactor, MEIS1, as compared with the control animals transplanted with cells overexpressingHOXA9 alone. In addition, structure-function analysis has demonstrated the importance of the MEIS- and PBX-interacting domains for HOXA9-mediated immortalization.13 35However, it is likely that the limited proliferative potential demonstrated by most human hematopoietic cells in the NOD/SCID mouse model have made it difficult to detect a HOXA10-induced myeloproliferative process in these animals. With regard to the impact of HOXA10 on erythroid differentiation in vivo, we noticed a substantial decrease in the median proportion of erythroid glycophorin A+ cells in the HOXA10-transduced human cell population compared with the GFP+ cell population in the control mice (HOXA10, 0.5%; GFP, 12.9%; data not shown). However, the frequency of glycophorin A+cells was low, and xenotransplant models, which allow a more efficient erythroid differentiation of human engrafted cells, will help to analyze more precisely the effect of HOXA10 on erythropoiesis in vivo.
In summary, these data demonstrate that regulated HOXA10expression may be pivotal for normal progenitor development and that aberrant expression of this gene results in increased proliferation and impaired differentiation of normal human hematopoietic cells. When combined with additional genetic events, these changes may be key factors in HOXA cluster-linked leukemogenesis. This in vitro and in vivo study now allows the study of these aspects in detail. It opens the door to assess more precisely the role of HOXcofactors, such as PBX1 or MEIS1, in HOXA10-induced perturbations of hematopoiesis and to perform structure-function analysis of putative HOXA10 regulatory domains in the experimental human systems described here.
Supported by the National Cancer Institute of Canada, with funds from the Canadian Cancer Society and the Terry Fox Run, and by the National Institutes of Health (grant DK48642). C.B. is supported by a grant from the Deutsche Forschungsgesellschaft (Bonn, Germany). M.F.-B. is supported by a grant from the Deutsche Krebshilfe (Bonn, Germany).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
R. Keith Humphries, The Terry Fox Laboratory, British Columbia Cancer Agency, 10th Ave West, Vancouver, BC, Canada V5ZIL3; e-mail: keith@terryfox.ubc.ca.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal