The reverse transcriptase–polymerase chain reaction (RT-PCR) has become widely used for monitoring minimal residual disease after allogeneic stem cell transplantation (SCT) for chronic myeloid leukemia (CML). However, most of these studies were performed using qualitative RT-PCR, and the interpretation of the results obtained has been conflicting. The correlation of a quantitative RT-PCR test performed early after SCT (at 3 to 5 months) and long-term outcome of CML patients surviving for more than 6 months was studied. Between January 1991 and June 1999, data from 138 CML patients who received allografts were evaluated. Early RT-PCR results were classified as (1) negative if there were no BCR-ABLtranscripts detected (n = 61), (2) positive at low level if the total number of BCR-ABL transcripts was less than 100 per μg RNA and/or the BCR-ABL/ABL ratio was less than 0.02% (n = 14), or (3) positive at high level if transcript levels exceeded the thresholds defined above (n = 63). Three years after SCT the cumulative incidence of relapse was 16.7%, 42.9%, and 86.4%, respectively (P = .0001). The relationship betweenBCR-ABL transcript level and probability of relapse was apparent whether patients had received sibling or unrelated donor SCT and also whether or not the transplantation was T cell depleted. The results suggest that quantitative RT-PCR performed early after SCT is useful for predicting outcome and may help to define the need for further treatment.
Introduction
At present, allogeneic stem cell transplantation (SCT) is the only curative treatment for patients with chronic myeloid leukemia (CML). Although allogeneic SCT often results in the disappearance of the Ph chromosome, this Ph negativity does not exclude the presence of residual leukemic cells because the sensitivity of cytogenetic analysis is low. The chimeric BCR-ABL gene is a tumor-specific marker that allows detection of minimal residual disease (MRD) using a reverse transcriptase–polymerase chain reaction (RT-PCR) assay.1-4
Several studies have attempted to assess the clinical significance and predictive value of detecting BCR-ABL transcripts by RT-PCR assay after SCT. However, most studies were performed using qualitative RT-PCR, and the results have been conflicting.1-8Moreover, the sensitivity of the RT-PCR assay varies considerably in different laboratories, and it is difficult to compare and interpret results reported by different investigators in the absence of internal quantitative controls. When the sensitivity of the assay is increased, only a few patients remain RT-PCR− early after SCT, thus rendering RT-PCR “too sensitive” to serve a prognostic purpose.9 10
Because nonquantitative RT-PCR analysis gives only limited information, several groups have developed quantitative or semiquantitative RT-PCR assays that enable the kinetics of residual disease to be monitored over time.11-15 If a threshold of residual disease can be established above which a patient is likely to relapse, then monitoring residual disease using quantitative RT-PCR would have clinical value. We have previously shown that in patients treated with interferon, low levels or absence of MRD is associated with continuing remission.16,17 After allogeneic SCT, serial quantitative RT-PCR analysis of peripheral blood specimens can effectively distinguish those patients who will remain in remission from those patients who are destined to relapse.11 13 Moreover, early recognition of relapse at the molecular level provides a window for therapeutic intervention while the burden of disease is still relatively low. In this study we have tested the value of a quantitative RT-PCR assay in predicting relapse when performed early (at 3 to 5 months) after SCT.
Patients, materials, and methods
Patients
Between January 1991 and June 1999 a total of 238 consecutive patients with Ph+ chromosome and/orBCR-ABL+ CML in chronic phase (CP) or accelerated phase (AP) underwent allogeneic SCT at the Hammersmith Hospital in London, England, and survived for more than 6 months after SCT. All patients gave informed consent according to the Declaration of Helsinki. The study was approved by the Hammersmith Hospital research ethics committee. Data from early RT-PCR, performed within 3-5 months after SCT, were available for 138 of these patients. Characteristics of these patients are summarized in Table 1. The overall median age was 34.0 years (range, 6.5-56.7 years); the median interval from diagnosis to SCT was 14 months (range, 3-73 months). Of those patients who underwent an SCT, 67 patients received cells from a human leukocyte antigen (HLA)–identical sibling (SIB), and 71 patients received cells from an unrelated donor (URD). Where the donor was unrelated, iso-electric focusing and molecular typing for DRB1 were employed to confirm HLA identity at class I and II loci, respectively. Where a choice of unrelated donor existed, the cytotoxic T-lymphocyte precursor (CTLp) assay was used to aid donor selection.18
Clinical characteristics of 138 chronic myeloid leukemia patients according to early reverse transcriptase–polymerase chain reaction results after allogeneic stem cell transplantation
| . | Negative . | Low-level positive . | High-level positive . | P . |
|---|---|---|---|---|
| Sex, no. (%) | ||||
| Male | 30 (49) | 6 (43) | 38 (60) | NS |
| Female | 31 (51) | 8 (57) | 25 (40) | |
| Age, y*(range) | 33.5 | 35.5 | 33.9 | NS |
| (6.5-55.4) | (22.6-54.2) | (9.7-56.7) | ||
| Type of donor, no. (%) | ||||
| SIB | 36 (59) | 7 (50) | 24 (38) | .01 |
| URD | 25 (41) | 7 (50) | 39 (62) | |
| Interval from diagnosis to SCT, mo*(range) | 11 (4-73) | 15 (4-36) | 15 (3-64) | NS |
| Disease status at SCT, no. (%) | ||||
| Chronic phase | 51 (84) | 10 (71) | 52 (83) | NS |
| Accelerated phase | 10 (16) | 4 (29) | 11 (17) | |
| GVHD prophylaxis, no. (%) | ||||
| Non-TCD | 33 (54) | 6 (43) | 20 (32) | .02 |
| TCD | 28 (46) | 8 (57) | 43 (68) | |
| CMV seropositive, no. (%) | 26 (43) | 8 (57) | 27 (42) | NS |
| Acute GVHD after SCT, no. (%) | ||||
| 0-I | 23 (38) | 7 (50) | 33 (52) | |
| II | 32 (52) | 5 (36) | 27 (43) | NS |
| III-IV | 6 (10) | 2 (14) | 3 (5) |
| . | Negative . | Low-level positive . | High-level positive . | P . |
|---|---|---|---|---|
| Sex, no. (%) | ||||
| Male | 30 (49) | 6 (43) | 38 (60) | NS |
| Female | 31 (51) | 8 (57) | 25 (40) | |
| Age, y*(range) | 33.5 | 35.5 | 33.9 | NS |
| (6.5-55.4) | (22.6-54.2) | (9.7-56.7) | ||
| Type of donor, no. (%) | ||||
| SIB | 36 (59) | 7 (50) | 24 (38) | .01 |
| URD | 25 (41) | 7 (50) | 39 (62) | |
| Interval from diagnosis to SCT, mo*(range) | 11 (4-73) | 15 (4-36) | 15 (3-64) | NS |
| Disease status at SCT, no. (%) | ||||
| Chronic phase | 51 (84) | 10 (71) | 52 (83) | NS |
| Accelerated phase | 10 (16) | 4 (29) | 11 (17) | |
| GVHD prophylaxis, no. (%) | ||||
| Non-TCD | 33 (54) | 6 (43) | 20 (32) | .02 |
| TCD | 28 (46) | 8 (57) | 43 (68) | |
| CMV seropositive, no. (%) | 26 (43) | 8 (57) | 27 (42) | NS |
| Acute GVHD after SCT, no. (%) | ||||
| 0-I | 23 (38) | 7 (50) | 33 (52) | |
| II | 32 (52) | 5 (36) | 27 (43) | NS |
| III-IV | 6 (10) | 2 (14) | 3 (5) |
NS indicates not sufficient data; SIB, sibling donor; URD, unrelated donor; SCT, stem cell transplantation; GVHD, graft-versus-host disease; TCD, T-cell depletion; CMV, cytomegalovirus.
Median values.
The transplantation conditioning and prophylaxis against graft-versus-host disease (GVHD) were performed according to our standard protocols as previously described.18 19 All patients were conditioned with 120 mg/kg cyclophosphamide and 1200-1440 cGy total body irradiation in 6 fractions for 3 days. In addition, some patients received busulfan, daunorubicin, cytarabine, thiotepa, and/or splenic irradiation. Patients receiving stem cells from an unrelated donor were given intravenous anti-CD52 monoclonal antibody (mAb) (Campath 1G or 1H; Therapeutic Antibody Center–TAC, Oxford, United Kingdom) from day −5 to day +4 as a method of T-cell depletion (TCD). In addition, Campath was used in vitro to treat the bone marrow of 8 SIB donors. All patients received cyclosporine and “short” methotrexate as GVHD prophylaxis. Four patients received 10 mg/kg antithymocyte globulin (ATG) instead of Campath antibody in 1992 because Campath was temporarily unavailable.
Molecular monitoring
Peripheral blood samples in all cases were studied every 3 months after SCT for the presence of BCR-ABL transcripts using a double-step, nested, semiquantitative RT-PCR assay as previously described.3,11 Bone marrow samples for cytogenetic analysis were performed when clinically indicated. The frequency of cytogenetic and RT-PCR monitoring after transplantation was similar in the SIB and URD transplantations. If a sample was found positive by RT-PCR testing, quantitation was performed by competitive RT-PCR, and further samples were analyzed at monthly intervals. Until 1996 results were expressed as the number of BCR-ABLtranscripts per μg RNA. Subsequently, results were expressed as theBCR-ABL/ABL ratio, the number of total ABLtranscripts (including BCR-ABL) serving as an internal control for sample quality.11 13
Classification of “early” RT-PCR results
Early RT-PCR was defined as a quantitative RT-PCR performed between 3 and 5 months after SCT. For patients with more than one sample analyzed, the highest level of RT-PCR positivity was considered. In a previous study we showed that for patients who in sequential sampling remained below a ratio of 0.02%, the probability of relapse was extremely low.13 Therefore we have used the 0.02% cutoff for classifying early RT-PCR samples. The results of early RT-PCR were classified as (1) negative if there were noBCR-ABL transcripts detected; (2) low-level positive if the total number of BCR-ABL transcripts was less than 100 per μg RNA and/or the BCR-ABL/ABL ratio was less than 0.02%; or (3) high-level positive if transcript levels exceeded the thresholds defined above.
Definition of relapse
Currently we classify a patient as having molecular relapse if (1) the BCR-ABL/ABL ratio is more than 0.02% on 3 consecutive occasions at least one month apart; (2) the ratio is more than 0.05% on 2 consecutive samples; or (3) the transcript number is rising in 3 consecutive samples, with the last 2 samples greater than 0.02%. This molecular relapse was dated from the time of the first detection of BCR-ABL positivity, including the results of the early RT-PCR. Cytogenetic relapse is considered to be present if one or more Ph+ metaphases were detected on 2 consecutive analyses without evidence of hematologic relapse. Hematologic relapse is defined as peripheral blood leukocytosis usually with the presence of myelocytes, basophils, and an excess of neutrophils in the differential count with a hypercellular bone marrow. A functional definition of disease recurrence after SCT was used in this study to calculate the incidence of relapse after SCT. This disease recurrence was defined by the detection of any type of relapse (molecular, cytogenetic, or hematologic) and the need for antileukemia therapy.1
Statistical analysis
The Fisher exact test, χ2 test, or χ2 trend test, and the Mann-Whitney test were employed to compare groups where appropriate. Survival and leukemia-free probabilities were calculated by the Kaplan-Meier method.20 The probability of relapse was calculated by the cumulative incidence procedure.21 The “failure of interest” was molecular, cytogenetic, or hematologic relapse, whichever was recognized first, and death without relapse was the competing risk. The log-rank test was used to compare survival curves. All quoted P values are 2-sided, and confidence intervals (CI) refer to 95% boundaries.
Results
Early RT-PCR
Of the 138 patients with early RT-PCR tests, 63 (46%) patients were classified as high-level positive, 14 (10%) patients were low-level positive, and 61 (44%) patients were negative. In the high-level positive group, 24 (38%) patients received SIB transplantations and 39 (62%) patients received URD transplantations. In the negative group, 36 (59%) patients received SIB transplantations, and 25 (41%) patients received URD transplantations. In the low-level positive group, the distribution was equally balanced; 7 patients received SIB transplantations, and 7 patients received URD transplantations. Clinical characteristics and transplantation-related factors were distributed homogeneously between all 3 groups, with the exception of a significantly greater proportion of patients undergoing TCD and URD SCT in the high-level positive cohort (Table 1).
Association of early RT-PCR positivity and subsequent relapse
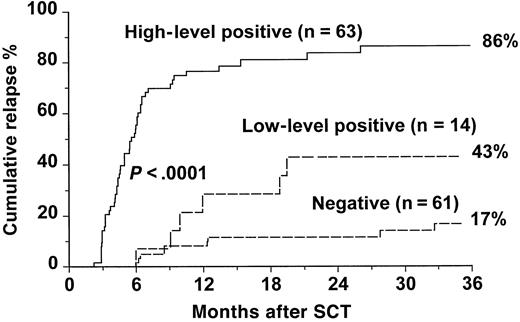
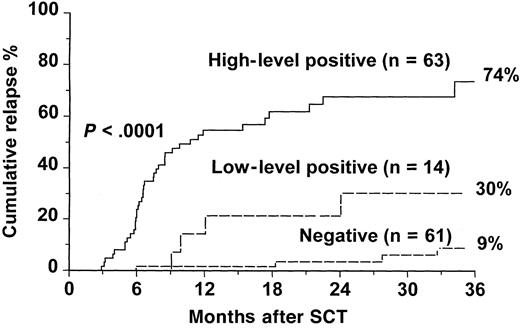
Of the 63 patients who had high-level positive RT-PCR, 52 (83%) patients relapsed at the molecular, cytogenetic, or hematologic level, and 11 (17%) patients have remained in continuous remission, with a median follow-up of 16 months (range, 7-92 months) (Table2). In the low-level positive group, 8 (57%) of 14 patients relapsed. In contrast, of the 61 patients who had negative RT-PCR results, only 14 (23%) patients relapsed, and 47 (77%) patients remained in remission after a median follow-up of 36 months (range, 7-103 months). The cumulative incidence of relapse for patients with high-level positive RT-PCR was 86.4% at 3 years compared with 42.9% for patients with low-level positive RT-PCR and 16.7% for patients with negative RT-PCR results (P < .0001) (Figure1). A further analysis was performed in which relapse was defined only on cytogenetic or hematologic criteria, and patients whose disease did not progress beyond molecular relapse were classified as “nonrelapsed” patients (Figure2). In this analysis, the cumulative incidence of relapse at 3 years was 74% for the high-level positive group versus 30% for the low-level group and 9% for the negative group (P < .0001).
Association between incidence of relapse and early reverse transcriptase–polymerase chain reaction result
| . | Patients relapsed, no. . | Patients in continuous remission, no. . | Total patients, no. . |
|---|---|---|---|
| RT-PCR result | |||
| Negative | 14 | 47 | 61 |
| Low-level positive | 8 | 6 | 14 |
| High-level positive | 52 | 11 | 63 |
| Total patients, no. | 74 | 64 | 138 |
| . | Patients relapsed, no. . | Patients in continuous remission, no. . | Total patients, no. . |
|---|---|---|---|
| RT-PCR result | |||
| Negative | 14 | 47 | 61 |
| Low-level positive | 8 | 6 | 14 |
| High-level positive | 52 | 11 | 63 |
| Total patients, no. | 74 | 64 | 138 |
RT-PCR indicates reverse transcriptase–polymerase chain reaction.
Cumulative incidence of relapse after SCT for CML according to results of an early RT-PCR test.
Cumulative incidence of disease recurrence at any (molecular, cytogenetic, or hematologic) level for patients with negative, low-level positive, and high-level positive RT-PCR at 3-5 months after SCT, respectively. The P value shown has been pooled over strata. The pairwise over strata P values are negative versus low positive, P = .01; negative versus high positive, P < .0001; and low positive versus high positive, P = .001.
Cumulative incidence of relapse after SCT for CML according to results of an early RT-PCR test.
Cumulative incidence of disease recurrence at any (molecular, cytogenetic, or hematologic) level for patients with negative, low-level positive, and high-level positive RT-PCR at 3-5 months after SCT, respectively. The P value shown has been pooled over strata. The pairwise over strata P values are negative versus low positive, P = .01; negative versus high positive, P < .0001; and low positive versus high positive, P = .001.
Cumulative incidence of relapse at the cytogenetic or hematologic level after SCT for CML according to results of an early RT-PCR test.
Relapse is defined only on cytogenetic or hematologic criteria, and patients whose disease did not progress beyond molecular relapse are classified as “nonrelapsed” patients. The P value shown has been pooled over strata. The pairwise over strata Pvalues are negative versus low positive, P = .01; negative versus high positive, P < .0001; and low positive versus high positive, P = .001.
Cumulative incidence of relapse at the cytogenetic or hematologic level after SCT for CML according to results of an early RT-PCR test.
Relapse is defined only on cytogenetic or hematologic criteria, and patients whose disease did not progress beyond molecular relapse are classified as “nonrelapsed” patients. The P value shown has been pooled over strata. The pairwise over strata Pvalues are negative versus low positive, P = .01; negative versus high positive, P < .0001; and low positive versus high positive, P = .001.
When patients were subdivided by donor type, the cumulative incidence of relapse for the SIB and URD subgroups remained significantly higher for patients with a high-level positive result (Table3). In SIB recipients, patients with high-level positive RT-PCR had a cumulative incidence of relapse at 3 years of 70.6% compared with 28.6% in the low-level group and 10.5% in the negative group (P < .0001). In URD SCT, the cumulative incidence of relapse at 3 years in patients with high-level positive RT-PCR was 95% compared with 57% in the low-level group and 25% in the negative group (P < .0001).
Association between cumulative incidence of relapse at 3 years and early reverse transcriptase–polymerase chain reaction result according to different clinical variables
| RT-PCR result . | Total patients, % . | Type of donor, % . | GVHD prophylaxis, % . | Disease stage, % . | |||
|---|---|---|---|---|---|---|---|
| SIB . | URD . | TCD . | Non-TCD . | CP . | AP . | ||
| Negative | 16.7 | 10.5 | 25 | 26.4 | 6.2 | 14.8 | 28.7 |
| Low-level positive | 42.9 | 28.6 | 57 | 62.5 | 16.7 | 50 | 3-150 |
| High-level positive | 86.4 | 70.6 | 95 | 95.3 | 64.7 | 84.5 | 90.9 |
| RT-PCR result . | Total patients, % . | Type of donor, % . | GVHD prophylaxis, % . | Disease stage, % . | |||
|---|---|---|---|---|---|---|---|
| SIB . | URD . | TCD . | Non-TCD . | CP . | AP . | ||
| Negative | 16.7 | 10.5 | 25 | 26.4 | 6.2 | 14.8 | 28.7 |
| Low-level positive | 42.9 | 28.6 | 57 | 62.5 | 16.7 | 50 | 3-150 |
| High-level positive | 86.4 | 70.6 | 95 | 95.3 | 64.7 | 84.5 | 90.9 |
Cumulative incidence of relapse is expressed as probability (%) at 3 years after SCT.
Indicates less than 5 patients.
The probability of relapse remained significantly higher for patients with high-level positive RT-PCR when patients were subdivided according to GVHD prophylaxis (TCD versus non-TCD) and disease stage (CP vs AP) at SCT (Table 3). Patients who had received a TCD and URD SCT and had a high-level positive result in the early RT-PCR test showed a strong tendency to relapse rapidly after SCT. More than 89% of these patients relapsed within 9 months of SCT. Overall, for patients who have relapsed, the median time from SCT to relapse was 6 months (range, 3-92 months) for the high-level positive group compared with 19 months (range, 6-90 months) for the low-level positive group and 28 months (range, 6-99 months) for patients with a negative RT-PCR.
Management of relapse
Following SCT, 74 patients developed disease recurrence. Of these patients, 5 received no treatment or were treated with hydroxyurea, interferon-alpha, and/or other chemotherapy agents at the time of hematologic relapse (Table 4). The other 49 patients received donor lymphocyte infusion (DLI). The same proportion of patients received DLI in all 3 groups. The median time from first detection of relapse until DLI was identical in all 3 groups (10 months). Reasons for not having DLI included GVHD (5 patients), disease progression (5 patients), and withdrawal of immunosuppression (9 patients); in 9 cases, DLI had been planned but not yet implemented. At the time of DLI, 75% of patients in the negative RT-PCR group were in molecular or cytogenetic relapse compared with 43% and 35% in the low- and high-level groups, respectively.
Clinical evolution after relapse
| . | RT-PCR result . | ||
|---|---|---|---|
| Negative, no. (%) . | Low-level positive, no. (%) . | High-level positive, no. (%) . | |
| Treatment of relapse | |||
| DLI | 8 (57) | 7 (62) | 34 (65) |
| Other | 6 (43) | 1 (38) | 18 (35) |
| Disease status at DLI | |||
| Molecular/cytogenetic | 6 (75) | 3 (43) | 12 (35) |
| Hematologic | 2 (25) | 4 (57) | 22 (65) |
| Response to DLI | |||
| Molecular remission | 4 (50) | 5 (72) | 14 (41) |
| No response | 2 (25) | 1 (14) | 13 (38) |
| Not yet evaluable | 2 (25) | 1 (14) | 7 (21) |
| . | RT-PCR result . | ||
|---|---|---|---|
| Negative, no. (%) . | Low-level positive, no. (%) . | High-level positive, no. (%) . | |
| Treatment of relapse | |||
| DLI | 8 (57) | 7 (62) | 34 (65) |
| Other | 6 (43) | 1 (38) | 18 (35) |
| Disease status at DLI | |||
| Molecular/cytogenetic | 6 (75) | 3 (43) | 12 (35) |
| Hematologic | 2 (25) | 4 (57) | 22 (65) |
| Response to DLI | |||
| Molecular remission | 4 (50) | 5 (72) | 14 (41) |
| No response | 2 (25) | 1 (14) | 13 (38) |
| Not yet evaluable | 2 (25) | 1 (14) | 7 (21) |
RT-PCR indicates reverse transcriptase–polymerase chain reaction; DLI, donor lymphocyte infusion.
Of the 49 patients treated with DLI, 10 patients are still receiving escalating doses of lymphocytes, and 39 patients have completed this therapy and are evaluable for response. Overall, 23 of 39 patients have achieved a molecular remission after DLI. The probability of achieving a molecular remission at 2 years seemed higher (although not statistically significant) among patients with negative or low-level positive RT-PCR (83%; n = 12) compared to patients with high-level positive RT-PCR (60%; n = 27).
Survival and disease-free survival
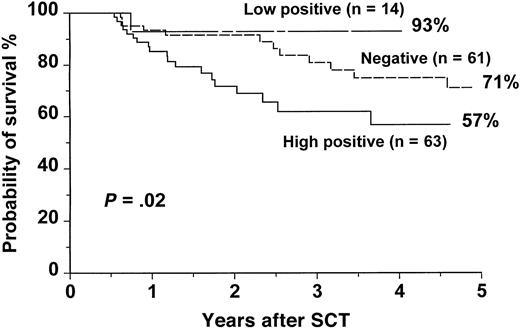
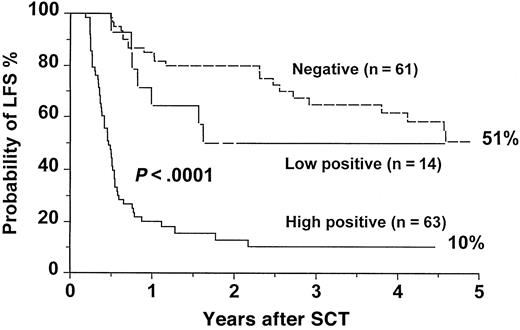
The actuarial overall survival (OS) for the entire group was 67.4% (range, 61% to 73%) at 5 years. For patients with a high-level positivity, the OS was 56.7% (range, 40% to 72%) at 5 years compared with 92.9% (range, 68% to 99%) and 71% (range, 56% to 83%) for patients with low-level positive and negative RT-PCR, respectively (P = .02) (Figure3). The leukemia-free survival (LFS) for patients with high-level positive RT-PCR was 10.4% (range, 5% to 22%) at 5 years compared with 50% (range, 27% to 74%) and 50.7% (range, 34% to 67%) for patients with low-level positive and negative RT-PCR, respectively (P < .0001) (Figure4). There were no statistically significant differences in OS and LFS between patients with negative and low-positive RT-PCR results. There were no statistically significant differences in OS and LFS between patients who had an early RT-PCR test (n = 138) and those who did not (n = 100) (data not shown).
OS after SCT for CML according to the results of an early RT-PCR test.
The P value shown has been pooled over strata. The pairwise over strata P values are: negative versus low positive,P = .1; negative versus high positive,P = .04; and low positive versus high positive,P = .02.
OS after SCT for CML according to the results of an early RT-PCR test.
The P value shown has been pooled over strata. The pairwise over strata P values are: negative versus low positive,P = .1; negative versus high positive,P = .04; and low positive versus high positive,P = .02.
LFS after SCT for CML according to the results of an early RT-PCR test.
The P value shown has been pooled over strata. The pairwise over strata P values are: negative versus low positive,P = .1; negative versus high positive,P < .0001; and low positive versus high positive,P = .001.
LFS after SCT for CML according to the results of an early RT-PCR test.
The P value shown has been pooled over strata. The pairwise over strata P values are: negative versus low positive,P = .1; negative versus high positive,P < .0001; and low positive versus high positive,P = .001.
Discussion
RT-PCR is by far the most sensitive method to detect residual disease in CML and can enable a single leukemia cell to be detected in a background of 105 to 106 normal cells. This is approximately 1000-fold greater than the sensitivity of routine cytogenetic analysis.1 Many investigators (including ourselves) have used nonquantitative RT-PCR techniques to detect MRD in patients who have undergone allogeneic SCT for CML. Using a nested primer technique, Roth et al7 analyzed 64 CML patients after allogeneic SCT and detected BCR-ABL transcripts at one time point in 37 patients. They concluded that nested RT-PCR could define subgroups of patients in apparent clinical remission, but with an increased risk of disease recurrence. Others,22 also using nested RT-PCR, found no association between RT-PCR positivity and subsequent relapse. In an early study5 we showed that RT-PCR positivity within 6 months after transplantation did not predict a worse outcome, whereas RT-PCR positivity later than 6 months after transplantation did. Radich et al6 presented a comprehensive multivariate analysis of 346 patients after allogeneic SCT. They identified RT-PCR positivity at 6-12 months after SCT as one independent variable influencing subsequent relapse. The significance of the presence of BCR-ABL transcripts in predicting disease recurrence was, however, lost in patients who tested positive more than 36 months after SCT. More recently, Serrano et al23 have used a combination of lineage-specific chimerism and detection of p190BCR-ABL transcripts to predict relapse.
All these studies employed a nonquantitative PCR technique, which could only give a positive or negative answer. A quantitative technique that allows one to follow the kinetics of low-level PCR positivity may, however, be more valuable in the clinical setting. For example, a proportion of CML patients are BCR-ABL+ for prolonged periods after allogeneic SCT, but they do not satisfy conventional criteria for relapse; for such patients one or more positive RT-PCR results would be difficult to interpret. Conversely, serial quantitative RT-PCR techniques can distinguish those RT-PCR+ patients who have low or falling BCR-ABLlevels from those whose levels are increasing.13 Patients destined not to relapse after SCT have persistently undetectable, low, or falling BCR-ABL levels on sequential analysis; after 6 to 12 months, BCR-ABL transcripts are usually undetectable and remain so indefinitely. In contrast, increasing or persistently high levels of BCR-ABL messenger RNA (mRNA) precede relapse, often several months before the cytogenetic detection of the Ph+ chromosome marrow metaphases.1 13 Provided assays are performed with sufficient frequency, rising or persistently high numbers of BCR-ABL transcripts can be detected prior to frank relapse, and this information may be used for early therapeutic intervention.
This study has focused on the potential association between a quantitative RT-PCR test performed early after SCT (at 3 to 5 months) and subsequent outcome. For the purposes of this investigation patients were classified into 3 groups (high-level positive, low-level positive, and negative) according to the level of quantitation (see “Patients, materials, and methods”). The cutoff between low-level and high-level transcript numbers was based on our previous experience. In our laboratory, patients who have a BCR-ABL/ABL ratio of less than 0.02% fall below the detection of relapse by cytogenetic analysis. Moreover, on sequential analysis, some patients with a ratio above 0.02% remained continuously RT-PCR+ in long-term follow-up, and some patients below such threshold were likely to fluctuate between positive and negative RT-PCR results.13This level correlates with a burden of CML which is approximately 50-fold lower than that seen in early cytogenetic relapse.1 13 Before 1996, quantitative results were expressed in numbers of BCR-ABL transcripts per μg RNA. Assuming a good quality sample, 100 transcripts per μg RNA corresponds to a BCR-ABL/ABL ratio of 0.02%.
We have found that 63 of the 138 patients had a high-level of RT-PCR positivity early after SCT. Early RT-PCR was able to differentiate between 3 well-defined groups, with different probabilities of developing disease recurrence. Those with high-level positivity had an extremely high risk of relapse; those with a negative RT-PCR result had a low probability, and those with a low-level RT-PCR positivity had an intermediate risk of disease recurrence 3 years after SCT. This correlation remained significant when only patients who had progressed to or beyond cytogenetic relapse were considered. Patients with negative RT-PCR results soon after SCT had more than a 95% probability of remaining free of leukemia at the chromosomal level 2 years after SCT.
The association between early RT-PCR and relapse was particularly strong in patients who underwent a TCD and URD SCT. This finding may represent a reduced graft-versus-leukemia effect in this type of transplantation.24 When rigorous TCD is performed, the antileukemic efficacy of the SCT must be due largely to the chemoradiotherapy of the conditioning. If this fails to eradicate the leukemia, as measured by a positive early RT-PCR, then relapse is the likely outcome. The kinetics of disappearance of residual disease may prove informative, as has been shown in acute lymphoblastic leukemia and acute promyelocytic leukemia.25-28
The overall relapse rate in this study was relatively high. This may, in part, have reflected the use of Campath as an in vivo method of TCD. An additional factor could also have been prolonged use of post-SCT immunosuppression. Withdrawal of immunosuppression can result in an enhanced graft-versus-leukemia effect in some patients.29 30 Early RT-PCR could be a useful tool in deciding when to stop cyclosporine or other immunosuppressive agents and thus reduce the risk of relapse. In this series, more than 70% of patients with high-level positive RT-PCR relapsed within 9 months after SCT, and more than 48% of patients had relapsed at the cytogenetic level. Thus many patients were still on immunosuppressive therapy at the time of relapse.
DLI has become a standard treatment for patients who relapse after allogeneic SCT. Responses to DLI are higher in patients with molecular or cytogenetic relapse compared with hematologic relapse.31-34 Furthermore, DLI is much less effective in patients with advanced disease (accelerated or blastic phase).33 Patients in the low- and high-level positive groups were more likely to receive DLI in hematologic relapse compared with the negative RT-PCR group. At our center, the median interval between first recognition of relapse and institution of DLI is more than 9 months. Because this is partly due to the delay in diagnosing relapse, early RT-PCR could reduce this delay.
In summary, this study has shown the usefulness of a quantitative RT-PCR performed early after SCT. Patients who remain RT-PCR+ at a high level soon after SCT have an increased risk of relapse and are more likely to progress in the short-term, thereby limiting the efficacy of post-SCT therapies. These patients may represent a subgroup with biologically distinct characteristics and a different response to DLI. Conversely, patients with a negative RT-PCR have an excellent prognosis. An intermediate subgroup of patients was identified with an intermediate risk of relapse, although after relapse the disease was less aggressive. Patients in this intermediate group may resemble patients treated by “nonmyeloablative” SCT, and the kinetics of response to treatment with DLI may be similar in the 2 groups.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Eduardo Olavarria, Department of Haematology, Hammersmith Hospital, ICSM, Du Cane Rd, London W12 0NN, England; e-mail: e.olavarria@ic.ac.uk.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal