Abstract
TEL-AML1 fusion resulting from the t(12;21)(p13;q22) is one of the most common genetic abnormalities in childhood acute lymphoblastic leukemia. Recent findings that site-specific cleavage of the MLL gene can be induced by chemotherapeutic agents such as topoisomerase-II inhibitors suggest that apoptogenic agents can cause chromosomal translocations in hematopoietic cells. This study demonstrates a possible relationship between exposure to apoptogenic stimuli, TEL breaks, and the formation ofTEL-AML1 fusion in immature B lymphocytes. Short-term culture of immature B cell lines in the presence of apoptogenic stimuli such as serum starvation, etoposide, or salicylic acid induced double-strand breaks (DSBs) in intron 5 of the TEL gene and intron 1 of the AML1 gene. TEL-AML1fusion transcripts were also identified by reverse transcriptase–polymerase chain reaction (RT-PCR) analysis in cell lines treated by serum starvation or aminophylline. DSBs within theTEL gene were also associated with fusion to other unknown genes, presumably as a result of chromosomal translocation. We also examined 67 cord blood and 147 normal peripheral blood samples for the existence of in-frame TEL-AML1 fusion transcripts. One cord blood sample (1.5%) and 13 normal peripheral blood samples (8.8%) were positive as detected by nested RT-PCR. These data suggest that breakage and fusion of TEL andAML1 may be relatively common events and that sublethal apoptotic signals could play a role in initiating leukemogenesis via the promotion of DNA damage.
Introduction
The t(12;21)(p13;q22) is one of the most common chromosome translocations in childhood acute lymphoblastic leukemia (ALL).1-3 This translocation is associated with rearrangement of the TEL (ETV6) gene on chromosome 12 and the AML1(CBFA2) gene on chromosome 21 resulting inTEL-AML1 fusion. Although many studies on theTEL-AML1 fusion gene focused on the biologic significance of leukemogenesis, little is known about the mechanisms involved in the cleavage of TEL and AML1 and subsequent fusion of the TEL-AML1 gene. Chemotherapy-related secondary leukemia is frequently associated with rearrangement of the MLL(ALL1) gene. Several clinicoepidemiologic studies have indicated that topoisomerase-II (topo-II) inhibitors cause rearrangement of the MLLgene.4 These findings are supported by in vitro studies that have demonstrated that MLL rearrangement could be induced by topo-II inhibitors in human hematopoietic cells.5 Site-specific cleavage within the MLLgene can also be induced by nongenotoxic stimuli involved in apoptotic cell death, suggesting that this cleavage is not specific to topo-II inhibitors but is rather part of a chromatin fragmentation similar to the generalized cellular response to apoptotic stimuli.6Based on these early findings, we speculated that general apoptotic stimuli on hematopoietic cells could play a key role in chromosomal translocation involving not only MLL but alsoTEL, another gene frequently altered in hematopoietic malignancies. To investigate whether apoptotic stimuli could trigger a cascade of events that ultimately leads to the formation of theTEL-AML1 fusion gene, we studied the in vitro conditions inducing the formation of DNA double-strand break (DSB) ends within TEL and AML1 genes. Our study indicates that DSBs of TEL and AML1 can be induced by apoptotic stimuli and this DNA cleavage appears to be the first step in the generation of TEL-AML1 fusion in immature B lymphocytes, which is considered to be an early or initiating event in childhood ALL.7 8
Materials and methods
Cell lines
RS4;11 is an ALL cell line and has MLL-AF4 fusion gene as result of t(4;11)(q21;q23).9 The TS-2 cell line was established from childhood pre-B ALL10 and carries t(1;19)(q23;p13) without the E2A gene rearrangement. REH, a t(12;21)-positive ALL cell line,11 was used as a positive control for reverse transcriptase–polymerase chain reaction (RT-PCR) analysis for TEL-AML1 fusion transcript. Epstein-Barr virus (EBV)–immortalized lymphoblastoid cell lines (EB-LCLs) were established from peripheral blood lymphocytes infected with the EBV strain B95-8, as described previously.12 Cell lines were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS) at 37°C in 5% CO2 in air.
Peripheral blood and cord blood samples
Peripheral blood samples of nonleukemic individuals were obtained from outpatients consulting several hospitals during December 1997 to April 1998 with informed consent. These individuals had no history of hematopoietic malignancy or administration of any chemotherapeutic or immunosuppressive agents. A total of 147 peripheral blood samples were collected. In each sample, mononuclear cells were separated by Ficoll-Hypaque gradient centrifugation and total RNA was extracted for RT-PCR analysis. Cord blood samples were collected at the time of birth of full-term newborns after informed consent was obtained from the mothers.
Serum starvation and chemicals
The ALL cell lines at exponential growth phase were centrifuged and washed twice with phosphate-buffered saline (PBS). Cells were resuspended in RPMI 1640 without FBS at a concentration of 5 × 105 cells/mL. As primary hematopoietic cells, mononuclear cells were separated from fresh cord blood samples and suspended in RPMI 1640 without FBS. VP-16 (10 μM), H2O2 (5 μM), salicylic acid (2 mM, 2-hydroxybenzoic acid, Sigma Co, St Louis, MO), caffeine (2 mM, Sigma), or aminophylline (2 mM, Neophyllin; Eisai Pharmaceuticals, Tokyo, Japan) was added to RPMI 1640 supplemented with 10% FBS. Cells were cultured at 37°C in 5% CO2 in air for indicated times and collected for further analysis.
Ligation-mediated PCR for detection of broken-ended DNA
To detect DSB ends of DNA, ligation-mediated PCR (LM-PCR) was performed as described previously.13-15 BW linker was produced by annealing the BW-1 and BW-2 oligonucleotides (Table1A). Two micrograms of high-molecular-weight DNA was ligated to the BW linker with T4 DNA ligase (Takara Shuzo, Otsu, Japan), precipitated with ethanol, and dissolved in 20 μL distilled water. One microliter of the ligated sample was analyzed by PCR with Ex Taq (Takara) using BW-1 linker primer (0.02 μM) and gene-specific primers (0.4 μM) of target genes (Figure 1, Table 1A). Seminested PCR was performed with a specific primer and BW-1 primer for first round PCR, and with internal specific primer and BW-1 for the second round PCR. The conditions for PCR were as follows: 94°C for 2 minutes, followed by 30 cycles of 94°C for 1 minute, 65°C for 1 minute, 72°C for 1 minute, and final extension at 72°C for 10 minutes. PCR products were purified using a PCR purification kit (Qiagen, Santa Clarita, CA) and subcloned to pGEM-T easy vector with a TA-cloning kit (Promega, Madison, WI). Plasmid DNA was extracted using the Qiaprep Spin Plasmid system (Qiagen) and insert DNAs were sequenced with 373A Autosequencer (Applied Biosystems, Urayasu, Japan). Obtained nucleotide sequences were compared with the sequences of intron 5 of TEL and intron 1 of AML1 (GenBank accession number: HSU6137516 and AJ229043, respectively). DSB ofBCR, β-globin, andglyceraldehyde-3-phosphate dehydrogenase (GAPDH) genes (GenBank accession number U07000, AF007546, and J04038, respectively) were also studied with LM-PCR analysis.
Polymerase chain reaction primers
| For LM-PCR and inverse PCR . | For RT-PCR . | ||
|---|---|---|---|
| BW-1 | 5′-GCGGTGACCCGGGAGATCTGAATTC-3′ | TEL940f | 5′-CTCTCTCATCGGGAAGACCTGGC-3′ |
| BW-2 | 5′-GAATTCAGATC-3′ | TEL981f | 5′-GGTCTCTGTCTCCCCGCCTGAAG-3′ |
| E1F (TEL) | 5′-GGATACCCCTGTGTGTACTCCACTCT-3′ | AML1-r41 | 5′-ATTGCCAGCCATCACAGTGAC-3′ |
| E2F (TEL) | 5′-CTGATGTCTTTCTGGACTCCTCGATC-3′ | AML1-r42 | 5′-CAGAGTGCCATCTGGAACAT-3′ |
| E3F (TEL) | 5′-CTGAGGTGGGAGAATCGCTTGAAC-3′ | AML1-r31 | 5′-AGCGGCAACGCCTCGCTCAT-3′ |
| E4F (TEL) | 5′-GGAAGGTGGAGGTTGTAATGAGCC-3′ | AML1-r32 | 5′-TCATCTTGCCTGGGCTCAGCG-3′ |
| E1R (TEL) | 5′-ACTACAGACTGGCAGAGCTGGAAGG-3′ | GAPDHf | 5′-CCATGGAGAAGGTGGGG-3′ |
| E2R (TEL) | 5′-GGATGATTCAGTCCTACCTTCTTCGG-3′ | GAPDHr | 5′-CAAAGTTGTCATGGATGACC-3′ |
| A1R (AML1) | 5′-CCTAGATCCCTGGACATGCCTTTACC-3′ | ||
| A2R (AML1) | 5′-GTCCTCTCTTCCCTTGGCTAGATACC-3′ | ||
| A3R (AML1) | 5′-TGCCTTAGCGATGTCACTCCTGG-3′ | ||
| A4R (AML1) | 5′-GGATGGGGTAAGGTAAGATGTAGGG-3′ | ||
| B1F (BCR) | 5′-GGAAGGAGAGTCTGAAAGTGCAGGGG-3′ | ||
| B2F (BCR) | 5′-AGCATAGGATACTCAGGTGGCTCTGG-3′ | ||
| BG1F (β-globin) | 5′-GCAACCTCAAACAGACACCATGG-3′ | ||
| BG2F (β-globin) | 5′-GGTGAACGTGGATGAAGTTGGTG-3′ | ||
| G1F (GAPDH) | 5′-GTTCGACAGTCAGCCGCATCTTC-3′ | ||
| G2F (GAPDH) | 5′-ATCGCTCAGACACCATGGGGAAG-3′ | ||
| For LM-PCR and inverse PCR . | For RT-PCR . | ||
|---|---|---|---|
| BW-1 | 5′-GCGGTGACCCGGGAGATCTGAATTC-3′ | TEL940f | 5′-CTCTCTCATCGGGAAGACCTGGC-3′ |
| BW-2 | 5′-GAATTCAGATC-3′ | TEL981f | 5′-GGTCTCTGTCTCCCCGCCTGAAG-3′ |
| E1F (TEL) | 5′-GGATACCCCTGTGTGTACTCCACTCT-3′ | AML1-r41 | 5′-ATTGCCAGCCATCACAGTGAC-3′ |
| E2F (TEL) | 5′-CTGATGTCTTTCTGGACTCCTCGATC-3′ | AML1-r42 | 5′-CAGAGTGCCATCTGGAACAT-3′ |
| E3F (TEL) | 5′-CTGAGGTGGGAGAATCGCTTGAAC-3′ | AML1-r31 | 5′-AGCGGCAACGCCTCGCTCAT-3′ |
| E4F (TEL) | 5′-GGAAGGTGGAGGTTGTAATGAGCC-3′ | AML1-r32 | 5′-TCATCTTGCCTGGGCTCAGCG-3′ |
| E1R (TEL) | 5′-ACTACAGACTGGCAGAGCTGGAAGG-3′ | GAPDHf | 5′-CCATGGAGAAGGTGGGG-3′ |
| E2R (TEL) | 5′-GGATGATTCAGTCCTACCTTCTTCGG-3′ | GAPDHr | 5′-CAAAGTTGTCATGGATGACC-3′ |
| A1R (AML1) | 5′-CCTAGATCCCTGGACATGCCTTTACC-3′ | ||
| A2R (AML1) | 5′-GTCCTCTCTTCCCTTGGCTAGATACC-3′ | ||
| A3R (AML1) | 5′-TGCCTTAGCGATGTCACTCCTGG-3′ | ||
| A4R (AML1) | 5′-GGATGGGGTAAGGTAAGATGTAGGG-3′ | ||
| B1F (BCR) | 5′-GGAAGGAGAGTCTGAAAGTGCAGGGG-3′ | ||
| B2F (BCR) | 5′-AGCATAGGATACTCAGGTGGCTCTGG-3′ | ||
| BG1F (β-globin) | 5′-GCAACCTCAAACAGACACCATGG-3′ | ||
| BG2F (β-globin) | 5′-GGTGAACGTGGATGAAGTTGGTG-3′ | ||
| G1F (GAPDH) | 5′-GTTCGACAGTCAGCCGCATCTTC-3′ | ||
| G2F (GAPDH) | 5′-ATCGCTCAGACACCATGGGGAAG-3′ | ||
LM-PCR indicates ligation-mediated polymerase chain reaction; RT-PCR, reverse transcriptase-PCR.
Location of PCR primers in intron 5 of
TEL and intron 1 of AML1.Arrows indicate the location and direction of primers used in this study. In TEL gene all primers were located within the 4.2-kb HindIII fragment of intron 5. E1F, E2F, and E3F were used for LM-PCR analysis and E3F, E4F, E1R, and E2R were used for inverse PCR analysis. In AML1 gene, 2 sets of primers for LM-PCR analysis (A1R, A2R, A3R, and A4R) were located within 2 kb of one long purine-pyrimidine repeat (about 55 kb downstream of exon 1) in intron 1. Numbered solid boxes indicate the exons of each gene. Hatched boxes (Pu/Py) represent alternative purine-pyrimidine tract located on each gene. H: HindIII, E: EcoRI.
Location of PCR primers in intron 5 of
TEL and intron 1 of AML1.Arrows indicate the location and direction of primers used in this study. In TEL gene all primers were located within the 4.2-kb HindIII fragment of intron 5. E1F, E2F, and E3F were used for LM-PCR analysis and E3F, E4F, E1R, and E2R were used for inverse PCR analysis. In AML1 gene, 2 sets of primers for LM-PCR analysis (A1R, A2R, A3R, and A4R) were located within 2 kb of one long purine-pyrimidine repeat (about 55 kb downstream of exon 1) in intron 1. Numbered solid boxes indicate the exons of each gene. Hatched boxes (Pu/Py) represent alternative purine-pyrimidine tract located on each gene. H: HindIII, E: EcoRI.
Inverse PCR for detection of rearrangement of theTEL gene
Inverse PCR strategy was performed as previously reported.17 High-molecular-weight DNA (2 μg) was digested with HindIII and ligated at 15°C for 30 minutes in a total volume of 250 μL with 5 U T4 DNA ligase (Takara). The ligated DNA was then purified using the Geneclean (BIO 101 Inc, La Jolla, CA) and eluted in a final volume of 50 μL distilled water. Ligated DNA (4 μL) was used as the template and amplified by nested PCR using LA Taq (Takara). First-step PCR amplification was performed with E3F as the sense primer and E1R as the antisense primer. Second-step PCR was performed with E4F and E2R (Figure 1, Table 1A). The PCR amplification condition was as follows: 94°C for 2 minutes, followed by 30 cycles of 98°C for 20 seconds and 68°C for 7 minutes and terminal extension at 72°C for 10 minutes. PCR products were electrophoresed onto 0.8% agarose gel and positive bands were eluted from the gel, purified with Geneclean and subcloned into pGEM-T easy vector. The extracted plasmid DNAs were sequenced using 373A Autosequencer. Obtained nucleotide sequences were compared with the sequence of intron 5 of TEL and unknown sequences fused toTEL were analyzed by NCBI BLAST server.
Fluorescence in situ hybridization analysis
Mononuclear cells separated from peripheral blood of healthy volunteers were cultured with phytohemagglutinin (PHA) for 72 hours and used for the normal metaphase sample for fluorescence in situ hybridization (FISH) analysis. DNA fragments for chromosome mapping were labeled with digoxigenin-11-dUTP (Boehringer Mannheim, Mannheim, Germany) by PCR after sequence-independent amplification method18 and were hybridized to metaphase samples as described previously.19
RT-PCR
Nested RT-PCR amplification for theTEL-AML1 fusion gene was performed using the specific primers (Table 1B) as described previously.20GAPDH was used for control expression to confirm RNA integrity. Strict precautions were followed to prevent cross-contamination of samples and negative controls were included in each PCR amplification step. RNA for positive control experiments (extracted from the REH cell line) were handled only in the first experiment for setting of PCR conditions and sensitivity assay of the RT-PCR analysis. Positive RNA was never handled in whole step while analyzing RNA samples from nonleukemic volunteers.
Results
DSBs of TEL and AML1 after serum starvation
Serum starvation induced apoptosis in TS-2 and RS4;11 cell lines and the remaining surviving cells showed growth arrest within 48 hours. Cell lines cultured for 12 hours under serum starvation were subjected to detection of DSBs by LM-PCR analysis. About 15% and 30% of TS-2 and RS4;11 cells were identified to have undergone apoptosis, respectively (data not shown). Representative results of LM-PCR are shown in Figure 2A. A clear but weak band representing DSB of intron 5 of TEL was detected with E1F and E2F primers in the RS4;11 cell line. By PCR with E1F and E3F primers both leukemic cell lines showed smearing bands ranging in size from 400 base pairs (bp) to 1 kilobase (kb). As for AML1, the 2 cell lines showed a clear band with A1R and A2R primers and a broad 1- to 2-kb band with A3R and A4R primers, both indicating DNA break in intron 1. Sequence analysis revealed that 5 independent clones obtained from E1F/E2F product in the RS4;11 cell line included the same DSB DNA fragment, which corresponded to intron 5 of TEL (BP1 in Figure 3A). As for AML1, 5 and 6 independent clones from RS4;11 and TS-2, respectively, were obtained from A1R/A2R products and 4 different sites of breakpoints in intron 1 were isolated (BP2-5 in Figure 3A). Interestingly, sequences of the DNA break ends from these immature B lymphocytes showed nontemplated base addition to the break ends. HL60 cell line was also examined with the same primer sets and DSBs of TEL andAML1 were also detected after 12 hours of serum starvation. The sequences of DNA break ends from HL60 cell line showed no addition of nontemplated bases to the break ends (data not shown). We also studied DSB formation of the BCR gene in both ALL cell lines, which is also frequently involved in pathogenesis of leukemia. As a result DSB ends of BCR were detected after 12 hours of serum starvation. DSBs of the β-globin andGAPDH genes were also studied and only GAPDHshowed DSB ends in the same condition (Figure 2B). To confirm whether the formation of DSB ends of TEL and AML1 genes is a general event that occurs in not only immortal cell lines but also primary hematopoietic cells, we applied LM-PCR analysis to mononuclear cells of 3 cord blood samples. Although we could not obtain clear bands of the LM-PCR products, subcloning and sequencing of the LM-PCR products revealed that DSBs occurred in TEL andAML1 also in the cord blood after 24 hours of serum starvation (Figure 3C).
DSBs of
TEL and AML1 genes as detected by LM-PCR. Representative results of LM-PCR are shown. PCR products were electrophoresed on 2% agarose gels and stained with ethidium bromide. (A) DSBs of TEL and AML1 genes in TS-2 and RS4;11 cell lines. Gene-specific primers used for seminested PCR are indicated in each panel. FBS(−) indicates cells treated with serum starvation; salicylic acid, cells treated with salicylic acid as described in “Materials and methods.” (B) Representative results of LM-PCR for BCR,β-globin, and GAPDH genes in TS-2 and RS4;11 cell lines. Primers used for LM-PCR are listed in Table 1. (C) DSBs ofTEL and AML1 genes in EB-LCLs after treatment with various chemical agents. Primers used for seminested PCR were indicated in parentheses. H2O represents negative control.
DSBs of
TEL and AML1 genes as detected by LM-PCR. Representative results of LM-PCR are shown. PCR products were electrophoresed on 2% agarose gels and stained with ethidium bromide. (A) DSBs of TEL and AML1 genes in TS-2 and RS4;11 cell lines. Gene-specific primers used for seminested PCR are indicated in each panel. FBS(−) indicates cells treated with serum starvation; salicylic acid, cells treated with salicylic acid as described in “Materials and methods.” (B) Representative results of LM-PCR for BCR,β-globin, and GAPDH genes in TS-2 and RS4;11 cell lines. Primers used for LM-PCR are listed in Table 1. (C) DSBs ofTEL and AML1 genes in EB-LCLs after treatment with various chemical agents. Primers used for seminested PCR were indicated in parentheses. H2O represents negative control.
Nucleotide sequences of DSB ends of the
TEL and AML1 genes. (A) Nucleotide sequences of DNA break ends of the TEL andAML1 genes obtained from the RS4;11 or TS-2 cell line. BW1 represents the sequence of BW linker connected to each DNA break end. Underlined letters indicate nontemplated nucleotide inserts between BW linker and DNA break ends with no homology to germline sequence. Arrows indicate the breakpoint of genomic sequence observed in DSBs. One identical breakpoint in intron 5 of TEL gene and 4 different breakpoints in intron 1 of AML1 gene were isolated. (B) Nucleotide sequences of DSB ends of AML1 gene obtained from EB-LCL induced by chemical agents. (C) Nucleotide sequences of DSB ends of the TEL and AML1 gene obtained from cord blood samples after 24 hours of serum starvation.
Nucleotide sequences of DSB ends of the
TEL and AML1 genes. (A) Nucleotide sequences of DNA break ends of the TEL andAML1 genes obtained from the RS4;11 or TS-2 cell line. BW1 represents the sequence of BW linker connected to each DNA break end. Underlined letters indicate nontemplated nucleotide inserts between BW linker and DNA break ends with no homology to germline sequence. Arrows indicate the breakpoint of genomic sequence observed in DSBs. One identical breakpoint in intron 5 of TEL gene and 4 different breakpoints in intron 1 of AML1 gene were isolated. (B) Nucleotide sequences of DSB ends of AML1 gene obtained from EB-LCL induced by chemical agents. (C) Nucleotide sequences of DSB ends of the TEL and AML1 gene obtained from cord blood samples after 24 hours of serum starvation.
DSBs of TEL and AML1 are induced by chemicals
Formation of DSB by topo-II inhibitor (VP-16), H2O2, and salicylic acid was also examined to determine whether these agents could induce DNA breaks inTEL and AML1 genes. After 24 hours' culture with VP-16 or H2O2, DSBs of AML1 were observed by LM-PCR analysis (data not shown). In addition DSBs of theAML1 gene could be induced by 24 hours' culture of RS4;11 with salicylic acid (Figure 2A). Nucleotide sequences of the DSB ends induced by salicylic acid were determined and DNA breaks in intron 1 ofAML1 were confirmed in all examined clones.
The DSBs of TEL and AML1 were tested in 4 EB-LCLs after apoptogenic stimuli with salicylic acid, caffeine, or H2O2. After 24 hours of culture with the chemical agents, cells were harvested and formation of DSBs was analyzed by LM-PCR (Figure 2C). DSBs of TEL were observed as weak bands in EB-LCL4 after salicylic acid treatment and in EB-LCL1 after H2O2 treatment. DSBs of AML1were also detected as clear bands in EB-LCL1 and 3 after salicylic acid treatment, in EB-LCL3 and 4 after caffeine treatment, and as smearing band in all 4 EB-LCLs after H2O2 treatment. When cells were cultured without the addition of any chemical agent, all 4 EB-LCLs showed no positive band by LM-PCR analysis (data not shown). Representative nucleotide sequences of DSB ends ofAML1 are shown in Figure 3B. As with HL60, the addition of nontemplated bases to the break ends was not observed in DSB ends detected in EB-LCLs.
Fusion gene formation of the TEL gene after serum starvation
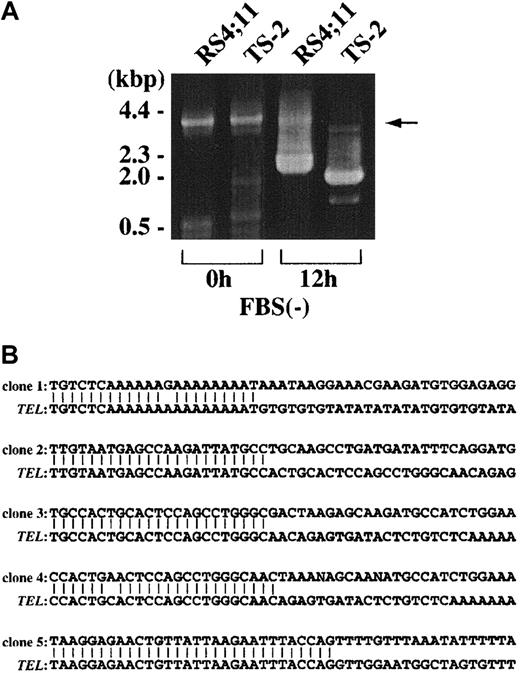
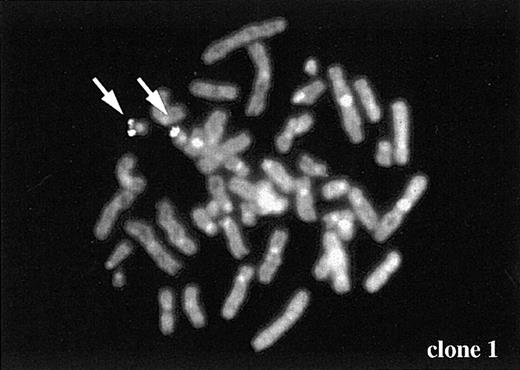
Because DSBs of TEL and AML1 were frequently detected in ALL cell line after serum starvation, we investigated whether formation of broken DNA ends was followed by chromosome translocation after repair of broken DNA ends. For this purpose, we applied inverse PCR strategy to detect rearrangement of theTEL gene. DNA samples extracted from cells before serum starvation were used for negative control samples (0 hour). As shown in Figure 4A, only the 4-kb germline band was detected in these control samples. However, both cell lines cultured for 12 hours under serum starvation showed PCR fragments with smaller size than the germline band. By subcloning the PCR fragments to pGEM-T easy vector, 17 clones were isolated, which exhibited rearrangement within the 4-kb HindIII fragment of intron 5. Twelve of 17 clones had interstitial deletion of about 100 to 200 bp within the 4-kb germline HindIII fragment. The other 5 clones showed fusion of TEL genomic fragment to unknown genes whose length ranged from 30 bp to 2 kb. These 5 fusion partner genes showed no homology to AML1 or to any published sequences based on BLAST homology search. Sequences of the 5 clones are shown in Figure 4B. Clone 1 was the only one that was long enough for chromosome mapping by FISH. This clone was mapped to chromosome 19 (Figure 5), indicating that chromosome translocation had occurred between TEL gene and clone 1 on chromosome 19.
Inverse PCR analysis of rearranged
TEL gene after serum starvation. (A) Representative results of inverse PCR-amplified fragments of the rearranged TEL gene after serum starvation. PCR products were electrophoresed on 0.8% agarose gels and stained with ethidium bromide. Control samples obtained before serum starvation (0 hour) show only normal products derived from the 4-kb HindIII fragment (arrow). Samples from 12 hours of serum starvation culture show rearranged products with smaller size than the germline band in RS4;11 and TS-2. (B) Nucleotide sequences of the breakpoints of theTEL gene and unknown sequences fused to DNA break ends in 5 rearranged clones.
Inverse PCR analysis of rearranged
TEL gene after serum starvation. (A) Representative results of inverse PCR-amplified fragments of the rearranged TEL gene after serum starvation. PCR products were electrophoresed on 0.8% agarose gels and stained with ethidium bromide. Control samples obtained before serum starvation (0 hour) show only normal products derived from the 4-kb HindIII fragment (arrow). Samples from 12 hours of serum starvation culture show rearranged products with smaller size than the germline band in RS4;11 and TS-2. (B) Nucleotide sequences of the breakpoints of theTEL gene and unknown sequences fused to DNA break ends in 5 rearranged clones.
Chromosome mapping of a partner gene fused to the
TEL gene. One of the partner genes (clone 1 in Figure 4B) was determined to be located on chromosome 19 by FISH. Arrows indicate FISH signal of the gene.
Chromosome mapping of a partner gene fused to the
TEL gene. One of the partner genes (clone 1 in Figure 4B) was determined to be located on chromosome 19 by FISH. Arrows indicate FISH signal of the gene.
TEL-AML1 fusion transcript is detected after serum starvation
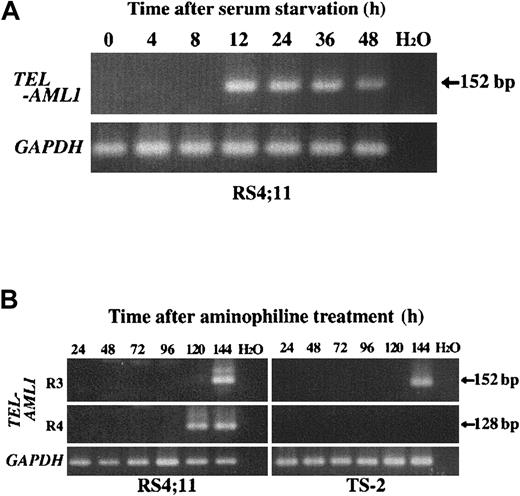
AML1 was not included as the fusion partner in isolated 5 TEL-rearranged clones. Therefore, we performed RT-PCR analysis to detect possible TEL-AML1 fusion transcript in the RS4;11 and TS-2 cell lines after serum starvation. Sensitivity analysis of RT-PCR was performed using RNA extracted from the REH cell line mixed with the HL60 cell line at different ratios, which showed that a single cell with TEL-AML1fusion transcript in 105 cells could be detected (data not shown). Representative results of RT-PCR analysis are shown in Figure6A. Samples obtained from cells before serum starvation showed no amplified product using combination ofTEL exon 5 forward primer and AML1 exon 3 or exon 4 reverse primer. Twelve hours after serum deprivation RS4;11 showed positive TEL-AML1 fusion transcript with combination of TEL exon 5 forward primer and AML1exon 3 reverse primer. Sampling of cell lines before and after serum starvation, RNA extraction, and RT-PCR analysis was repeated 3 times and only RS4;11 showed positive results. Amplified PCR products were directly sequenced and confirmed to be identical to the sequence of fusion transcript in ALL with TEL-AML1rearrangement.21
TEL-AML1 fusion transcript as detected after serum starvation and aminophylline treatment by RT-PCR.
(A) Representative results of RT-PCR of RS4;11 cell line at various time points after serum starvation. TEL exon 5 forward primers (TEL940f and TEL981f) and AML1 exon 3 reverse primers (AML1-r31 and AML1-r32) were used in nested PCR analysis for TEL-AML1 fusion transcript. (B) Representative results of RT-PCR of RS4;11 and TS-2 cell lines at various time points after treatment with aminophylline. TELexon 5 forward primers (TEL940f and TEL981f) and AML1 exon 3 or exon 4 reverse primers (R3: AML1-r31 and AML1-r32, R4: AML1-r41 and AML1-r42, respectively) were used in nested PCR for TEL-AML1fusion transcript. H2O represents negative control in both panels A and B.
TEL-AML1 fusion transcript as detected after serum starvation and aminophylline treatment by RT-PCR.
(A) Representative results of RT-PCR of RS4;11 cell line at various time points after serum starvation. TEL exon 5 forward primers (TEL940f and TEL981f) and AML1 exon 3 reverse primers (AML1-r31 and AML1-r32) were used in nested PCR analysis for TEL-AML1 fusion transcript. (B) Representative results of RT-PCR of RS4;11 and TS-2 cell lines at various time points after treatment with aminophylline. TELexon 5 forward primers (TEL940f and TEL981f) and AML1 exon 3 or exon 4 reverse primers (R3: AML1-r31 and AML1-r32, R4: AML1-r41 and AML1-r42, respectively) were used in nested PCR for TEL-AML1fusion transcript. H2O represents negative control in both panels A and B.
TEL-AML1 fusion transcript is detected after aminophylline treatment
It is important to study whether chemicals that can induce apoptosis of immature B lymphocytes could also induce the formation ofTEL-AML1 fusion as serum starvation. Aminophylline was added to the culture, which resulted in growth impairment of both ALL cell lines. At a concentration of 3 mM, aminophylline caused apoptosis of almost all the cell populations after 72 hours of culture. However, after the addition of 2 mM aminophylline, cells were still alive and proliferated slowly up to day 7. Total RNA was extracted every 24 hours after the addition of 2 mM aminophylline and TEL-AML1 fusion transcripts were analyzed by RT-PCR. As shown in Figure 6B, TEL-AML1 fusion transcripts were detected in both cell lines at 5 and 6 days after addition of aminophylline. Nucleotide sequences of these products showed that all positive bands corresponded to one of the 4 isoforms ofTEL-AML1 fusion transcript observed in ALL.21 Although TS-2 cell line showed only one isoform, RS4;11 cell line showed 2 isoforms depending on the time points analyzed.
TEL-AML1 fusion transcripts are present in normal blood
We examined the presence of TEL-AML1 fusion transcript in lymphocytes from peripheral blood of nonleukemic individuals to assess whether the fusion gene is frequently generated in vivo (Table 2). Surprisingly, 13 of 147 samples (8.8%) showed at least one type ofTEL-AML1 fusion transcript as detected by nested RT-PCR. Specifically, 11 of 99 samples (11.1%) obtained from individuals younger than 20 years of age and 2 of 48 samples (4.2%) from subjects older than 20 years of age were positive for the fusion transcript. Representative results of RT-PCR analysis are shown in Figure 7. All positive products were directly sequenced, which confirmed that fusion of TEL toAML1 was identical to that of leukemic patients (data not shown). All the 4 isoforms of TEL-AML1 fusion transcript were identified by DNA sequence analysis. Different types of isoforms were observed in each of the positive individuals and some of them had more than one type of fusion transcripts (data not shown), thus making contamination of the tested samples unlikely. We also examined 67 cord blood samples for the existence ofTEL-AML1 fusion transcripts and one sample was positive.
TEL-AML1 fusion transcripts in nonleukemic individuals
| Sample . | No. tested . | TEL-AML1 positive . |
|---|---|---|
| Peripheral blood | 147 | 13 (8.8%) |
| <20 y of age | 99 | 11 (11.1%) |
| >20 y of age | 48 | 2 (4.2%) |
| Cord blood | 67 | 1 (1.5%) |
| Sample . | No. tested . | TEL-AML1 positive . |
|---|---|---|
| Peripheral blood | 147 | 13 (8.8%) |
| <20 y of age | 99 | 11 (11.1%) |
| >20 y of age | 48 | 2 (4.2%) |
| Cord blood | 67 | 1 (1.5%) |
TEL-AML1 fusion transcript in normal peripheral blood samples.
Representative results of nested RT-PCR for TEL-AML1 fusion transcript of normal peripheral blood samples. TEL exon 5 forward primers (TEL940f and TEL981f) and AML1 exon 4 reverse primers (AML1-r41 and AML1-r42) were used. H2O represents negative control. Arrow indicates positive band indicatingTEL-AML1 fusion transcript.
TEL-AML1 fusion transcript in normal peripheral blood samples.
Representative results of nested RT-PCR for TEL-AML1 fusion transcript of normal peripheral blood samples. TEL exon 5 forward primers (TEL940f and TEL981f) and AML1 exon 4 reverse primers (AML1-r41 and AML1-r42) were used. H2O represents negative control. Arrow indicates positive band indicatingTEL-AML1 fusion transcript.
Discussion
Chromosome translocation and consequent fusion gene formation with acquired oncogenic function is a frequent event in hematopoietic malignancy. The mechanism of chromosome translocation and fusion with each other is an important issue in leukemogenesis. The general mechanism of gene fusion appears to be error-prone DNA repair of DSBs via nonhomologous recombination.17,22,23MLL gene is considered to be highly susceptible to topo-II inhibitors, and DSB of MLL gene after chemotherapy may produce translocations and gene fusion resulting in secondary leukemia.4 Previous studies have shown that site-specific cleavage of MLL is induced not only by topo-II inhibitors but also by more general nongenotoxic, apoptogenic stimuli such as serum starvation.5 6 These findings suggest that apoptogenic stimuli in general can cause DSB in leukemogenic genes. In our study, we hypothesized that in immature B lymphocytes, apoptogenic stimuli might trigger DSBs of TEL and AML1 genes and that the TEL-AML1 fusion gene could result from aberrant or error-prone DSB repair.
In 12;21 translocation, the breakpoint of the TEL gene is located at intron 5, and more than half of the cases have breakpoints in the 4-kb HindIII fragment of intron 5 (Figure1).17,24 As shown in this study, this region is indeed susceptible to DNA cleavage and produces DSB ends after exposure to several apoptogenic stimuli. In contrast to TEL, breakpoints in AML1 of leukemic cells are dispersed throughout more than one region of the large intron 1.17 Because some previous reports indicated the relationship between DNA cleavage/chromosome translocation and alternating purine-pyrimidine tract25 26that is also present in the 4-kb HindIII fragment ofTEL gene, we tentatively focused on the purine-pyrimidine tracts in intron 1 of AML1 as the target region for LM-PCR analysis. Our data indicated that this region and, we assume, other regions of AML1 intron 1 included in TEL-AML1fusion might produce DNA break ends under apoptotic stress.
Because efficiency of detecting DSB ends by LM-PCR analysis may depend on the selection and location of gene-specific primers, we have no insight into the relative frequency of DSB formation between analyzed genes with our LM-PCR strategy. The level of DSB formation may depend on the level and duration of apoptogenic stress. We have no grounds for suspecting that TEL and AML1genes are unusually susceptible to breakage and recombination. Vulnerability to DSB may be associated with some general features of the gene including open or accessible chromatin and transcriptional activity. We found that like TEL and AML1, other genes that are widely expressed, BCR andGAPDH also underwent DSB in response to apoptotic signals in contrast to transcriptionally silent (in lymphoid cells)β-globin in which we could observe no DSBs. We assume that the selectivity of TEL-AML1 fusion to B-cell precursor ALL has some link to the cell type selectivity of the fusion protein function in B-cell precursor compared with other cell types.27
As reported by Richardson and Jasin,28 the presence of DSB ends of both the TEL and AML1 genes simultaneously in a single cell might be expected to increase substantially the frequency of TEL-AML1 fusion gene formation. Our results showed that DSBs of the TEL gene under serum starvation were repaired and rearrangement and fusion to other genes occurred. Although genomic fusion of TEL toAML1 gene was not detected by inverse PCR, RT-PCR analysis demonstrated TEL-AML1 fusion transcripts were formed after 12 hours of serum starvation or aminophylline treatment. These findings suggest that the DSB end of the TEL gene may fuse to the break end of AML1 gene during the repair process. Like TEL and AML1 genes, apoptogenic stimuli on normal hematopoietic cells could induce DSB ends of some other genes leading to misjoining of the leukemia-associated genes. Several studies have previously shown that fusion genes specific to hematopoietic malignancy, such as BCR-ABL,29IGH-BCL2,30-33 and tandem duplication ofMLL,34 could be detected in peripheral blood of healthy volunteers at a relatively high frequency. Based on the results of our study, we have added TEL-AML1 to the catalog of leukemic fusion genes that can be detected in normal blood samples. A limitation of this finding on normal blood is that the cell type harboring the fusion gene is unidentified. It is therefore unclear if the presence of a fusion protein is associated with a transformation event (or is neutral in an “irrelevant” cell). Even if these fusion genes are functionally active in some samples it is likely that they are insufficient for leukemogenesis. Studies on identical twins with ALL and TEL-AML1 fusion provide a strong argument for the fusion gene being an initiating event (prenatally) but insufficient to generate ALL.8 35 The failure of a TEL-AML1transgene to induce leukemia in mice (A. M. Ford and M. F. Greaves, personal communication, June 2000) leads to a similar conclusion.
Recently, observations on identical twins with ALL and retrospective PCR analysis of neonatal blood spots of patients with ALL have established an in utero origin of leukemic cells with t(12;21) in most cases of childhood ALL.7,8,35,36 Considered together with our findings, it is possible that exposure to apoptogenic stimuli to immature B lymphocytes in utero can trigger the initial molecular change in childhood ALL. Whether this mechanism is dependent on exposure of the pregnant mother and fetus to exogenous agents (eg, pesticides, organic solvents or other chemicals, viruses) or the endogenous “physiologic” apoptotic stress of development36 is unknown but may be clarified by ongoing case/cohort epidemiologic studies.
In summary, we have demonstrated that DSB of TEL andAML1 genes occurs after exposure of immature B lymphocytes to apoptotic stimuli and thatTEL-AML1 fusion gene can be generated during the apoptotic response in these cells. Based on these results we speculate that apoptotic stimuli triggering DSB with subsequent fusion gene formation might play a role in initiating childhood ALL.
Acknowledgments
We thank Prof Kiyoshi Miyagawa (Department of Pathology, Research Institute for Radiation Biology and Medicine, Hiroshima University, Hiroshima, Japan) for providing adult peripheral blood samples. We also thank Dr Masataka Imaizumi (Department of Pediatrics, Tohoku University School of Medicine, Sendai, Japan) for providing the TS-2 cell line, Dr Yoshinobu Matsuo (Fujisaki Cell Center, Hayashibara Institute, Okayama, Japan) for providing the REH cell line, and Prof Mel Greaves (Leukaemia Research Fund Centre, Institute of Cancer Research, London) for helpful advice.
Supported by a Grant-in-Aid for Pediatric Research from the Ministry of Health and Welfare, Japan; by a Grant-in-Aid for Cancer Research from the Ministry of Health and Welfare, Japan; by a Grant-in-Aid from the Ministry of Health and Welfare, Japan, as part of a comprehensive 10-year strategy for cancer control; by a Grant-in-Aid from the Ministry of Education, Science and Culture, Japan; by Health Science Research Grants; and by Organized Research Combination System.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Minenori Eguchi-Ishimae, Leukaemia Research Fund Centre, Institute of Cancer Research, Chester Beatty Laboratories, 237 Fulham Rd, London SW3 6JB, United Kingdom; e-mail: mieguchi@icr.ac.uk.





This feature is available to Subscribers Only
Sign In or Create an Account Close Modal