Abstract
Because hematopoietic cells derived from Fanconi anemia (FA) patients of the C-complementation group (FA-C) are hypersensitive to the inhibitory effects of interferon γ (IFNγ), the products of certain IFNγ-inducible genes known to influence hematopoietic cell survival were quantified. High constitutive expression of the IFNγ-inducible genes, IFN-stimulated gene factor 3 gamma subunit (ISGF3γ), IFN regulatory factor-1 (IRF-1), and the cyclin-dependent kinase inhibitor p21WAF1 was found inFANCC mutant B lymphoblasts, low-density bone marrow cells, and murine embryonic fibroblasts. Paradoxically, these cells do not activate signal transducer and activator of transcription (STAT) 1 properly. In an attempt to clarify mechanisms by which FA-C cells overexpress IFNγ-inducible genes in the face of defective STAT1 phosphorylation, it was reasoned that decreased levels of activated STAT1 might result in reduced expression of a hematopoietic IFNγ-responsive protein that normally modulates expression of other IFNγ-responsive genes. Levels of the IFNγ-inducible factor IFN consensus sequence binding protein (ICSBP), a negative trans-acting regulator of some IFNγ-inducible genes, were quantified. ICSBP levels were reduced in FA-C B lymphoblasts and MEFs. However, enforced expression of ICSBP failed to down-regulate IRF-1, ISGF3γ, and p21WAF1. Thus, the FANCC protein functions to modulate expression of a family of genes that in normal cells are inducible only by specific environmental cues for apoptosis or mitogenic inhibition, but it does so independently of the classic IFN-STAT1 pathway and is not the direct result of reduced ICSBP expression.
Introduction
Fanconi anemia (FA) is an autosomal recessive disease characterized by progressive bone marrow failure, multiple congenital anomalies, and a high incidence of acute myelogenous leukemia.1-4 Cells from FA patients are hypersensitive to the effects of DNA cross-linking agents, such as mitomycin C and diepoxybutane.5,6 The disorder is genetically heterogeneous, with at least 7 different complementation groups.7,8 The genes corresponding to group A (FANCA), C (FANCC), E (FANCE), F (FANCF), and G (FANCG) have been cloned.9-13 Fanconi gene sequences show no significant homology to any known genes, and the functions of their gene products are unknown.14 15
Hematopoietic precursor cells from children with FA of the C complementation group (FA-C) are excessively apoptotic and hypersensitive to a variety of cytokines known to induce apoptosis, including interferon γ (IFNγ). IFNγ is a cytokine involved in host defense against viral infections and regulation of cell growth.16 IFNγ clearly plays a role in hematopoietic suppression and has been implicated in the pathogenesis of acquired aplastic anemia.16-22 Cells with inactivatingFANCC mutations are hypersensitive to the apoptotic effects of IFNγ, an effect that depends in part on an intactfas/fas-ligand pathway.23-26Specifically, exposure to low doses of IFNγ in vitro primes a substantial fraction of FA-C progenitor cells to undergo apoptosis after subsequent exposure to fas ligand. Such doses of IFNγ have no effect on normal hematopoietic progenitor cells.23 24 Because of the influence of IFNγ on thefas pathway, we sought to determine whether other IFNγ-inducible genes are hyperactive in FA-C cells. We carried out studies designed to analyze expression of IFNγ-inducible genes known to promote apoptosis and/or mitogenic inhibition in hematopoietic progenitor cells. We describe here studies revealing that the transactivators interferon regulatory factor-1 (IRF-1) and IFN-stimulated gene factor 3 gamma subunit (ISGF3γ) and the cyclin-dependent kinase (cdk) inhibitor p21WAF1 are expressed at constitutively higher levels in FA-C B lymphoblasts, low-density bone marrow cells (LDBMCs), and murine embryonic fibroblasts (MEFs), compared with cells expressing a normalFANCC. However, these proteins are expressed at normal levels in mature mutant fibroblasts, suggesting some degree of tissue or developmental stage specificity of FANCC function in control of these genes.
IFNγ exerts its effects, at least in part, through the Jak/signal transducer and activator of transcription (STAT) signaling pathway,27,28 However, FA-C cells are defective in their ability to properly activate STAT1.29 We show here that up-regulation of IFNγ-inducible genes occurs precisely in those cell types that fail to properly activate STAT1. For this reason, we hypothesized that loss of a STAT1-inducible transcriptional repressor may result in up-regulation of IFNγ-inducible genes in FA-C cells. Indeed, we found expression of the transcriptional repressor IFN consensus sequence binding protein (ICSBP) is decreased in both FA-C B lymphoblasts and murine embryonic fibroblasts. However, enforced expression of ICSBP failed to suppress IRF-1, ISGF3γ, or p21WAF1.
We conclude that in FA-C hematopoietic and embryonic cells, expression of IFNγ-inducible genes is increased even in the face of defective STAT1 phosphorylation but is not linked directly with reduced levels of ICSBP. We suggest that FANCC functions to indirectly suppress expression of IFNγ-inducible genes in a cell-type–specific, STAT1-independent manner.
Materials and methods
Cell culture and IFNγ stimulation
Epstein-Barr virus (EBV)–transformed human lymphoblasts were maintained in RPMI media 1640 (Life Technologies, Rockville, MD) supplemented with 15% heat-inactivated fetal calf serum and grown in a humidified 5% CO2-containing atmosphere at 37°C. The FA-A B-lymphoblast lines HSC72 and HSC72/FANCC were a generous gift from Dr Walsh (University of North Carolina, Chapel Hill). The lymphoblast lines JY (normal), HSC536N (FANCCmutant), HSC536N/FANCC, and HSC536N/neo were described previously.24 Briefly, HSC536N/FANCC was derived by transducing the HSC536N with a retrovirus encoding bothFANCC and neomycin phosphotransferase (neo). The HSC536N/neo line was derived by transducing the HSC536N with a vector encoding for neomycin phosphotransferase alone.
MEFs were established from FancC knockout and wild-type mice as previously described23 and grown in Dulbecco's modified Eagle's medium (Life Technologies) supplemented with 10% heat-inactivated fetal bovine serum. MEFs were transformed and immortalized by SV40 large T antigen.30
The following Fanconi fibroblast lines were received from the Oregon Health Sciences University Fanconi Anemia Cell Repository: PD134 (FA-C); PD426/FANCA (FA-C–expressing FANCA); PD426/FANCC (FA-C corrected); PD331.T (FA-C); PD720/FANCC (FA-A–expressing FANCC); PD720/FANCA (FA-A corrected); ML7334 (FA-G); and ML7334/FANCG (FA-G corrected). Human primary fibroblasts cell lines were established by means of standard procedures.31 The PD331.T line was immortalized as previously described.31 Cells were grown in MEM-α (Life Technologies) supplemented with 20% heat-inactivated fetal bovine serum. Correction of PD134 and PD331 was done by means of a retroviral vector carrying the humanFANCC complementary DNA (cDNA) as previously described.24 Cells were incubated with virus for 3 hours twice daily over a 5-day period and then selected with G418.
ICSBP-expressing HSC536N cells were made by means of a PLXSN-based retroviral vector expressing the human ICSBP cDNA. The pLXSN was a generous gift of Dr A. D. Miller (Fred Hutchinson Cancer Research Center, Seattle, WA). The pTarget/ICSBP was a generous gift supplied by Dr Ben-Zion Levi (Technion-Israel Institute of Technology, Haifa, Israel) and contains the entire human ICSBP cDNA. The pLXSN/ICSBP was made as follows: pTarget/ICSBP was digested with EcoRI; a 1.3-kilobase insert was isolated and cloned into pLXSN. Proper orientation was determined by means of a KpnI/NotI digest. Viruses were made as previously described.24 HSC536N were incubated with pLXSN/ICSBP or pLXSN virus plus 8 μg/mL polybrene for 3 hours twice daily for 5 days. Cells were then selected with G418.
IFNγ treatment
MEFs were serum starved for 24 hours and treated with the indicated amount of recombinant murine IFNγ (R&D Systems, Minneapolis, MN) for the indicated times in 5% CO2 at at 37°C. B lymphoblasts and human primary fibroblasts were treated with the indicated amounts of recombinant human IFNγ (R&D Systems) in 5% CO2 at 37°C.
Preparation of total cell lysates and nuclear extracts
Total cell lysates were made by washing cells twice with phosphate-buffered saline (PBS), and cell pellets were solubilized in RIPA: 10 mM Tris-HCl (pH 7.6), 150 mM NaCl, 1% (wt/vol) sodium dodecyl sulfate (SDS), 1% (vol/vol) aprotinin, 2 mM Na3VO4, 1 μg/mL leupeptin, 1 μg/mL pepstatin, and 1 mM phenylmethylsulfonylfluoride (PMSF). Lysates were centrifuged at 16 000g for 15 minutes at 4°C, and protein concentrations were determined by means of protein micro-assay of the Bradford method (BioRad, Hercules, CA). Nuclear extracts were prepared as previously described.32 Briefly, cells were washed in ice-cold PBS and resuspended in cold hypotonic buffer: 10 mM Hepes, pH 7.9, 1.5 mM MgCl2, 10 mM KCl, 0.2 mM PMSF, 0.5 mM dithiothreitol (DTT), 2 μg/mL aprotinin, and 2 μg/mL leupeptin. Cells were allowed to swell on ice 10 minutes and then lysed with a Dounce B homogenizer (Bellco Glass, Vineland, NJ). Nuclei were pelleted at 3300g for 20 minutes at 4°C and then resuspended in low-salt buffer: 20 mM Hepes, pH 7.9, 25% glycerol, 1.5 mM MgCl2, 20 mM KCl, 0.2 mM EDTA, 0.2 mM PMSF, 0.5 mM DTT, 2 μg/mL aprotinin, and 2 μg/mL leupeptin. High-salt buffer (0.6 M KCl) was added dropwise, and nuclear proteins were extracted by incubation at 4°C for 30 minutes and centrifuged at 13 000g for 30 minutes at 4°C. Supernatants were dialyzed at 4°C for 3 to 5 hours against a 250-fold volume excess of 20 mM Hepes, pH7.9, 20% glycerol, 100 mM KCl, 0.2 mM EDTA, 0.2 mM PMSF, 0.5 mM DTT, 2μg/mL aprotinin, and 2 μg/mL leupeptin.
Immunoblot analysis
Total cell lysates and nuclear extracts were mixed with Laemmli sample buffer,33 heated at 94°C for 5 minutes. Equivalent amounts of protein were subjected to SDS-polyacrylamide gel electrophoresis and electroblotted onto Bio-Blot (Costar, Cambridge, MA) as previously described.34 Blots were then subjected to Ponceau S. staining to demonstrate equivalent loading. Nonspecific binding was blocked for 1 hour at room temperature in Tris-buffered saline plus 0.05% Tween 20 (TBS-T) containing 5% milk. Blots were incubated with primary antibody, washed with TBS-T, then incubated with secondary antibody, and washed again with TBS-T. Antibody-reactive proteins were detected by means of Enhanced Chemiluminescence (Amersham, Arlington Heights, IL). IRF-1 antibody (1:1000), p21WAF1 antibody (1:500), ISGF3γ (BD Pharmingen) (1:250), p27KIP1 (BD Pharmingen) (1:500), p53 (1:1000), IRF-2 (1:1000), and total STAT1 (1:2000) were used at the indicated dilutions and incubated with the blots for 1 hour at room temperature. Donkey antirabbit immunoglobulin G horseradish peroxidase (HRP) secondary antibody (1:5000) (Amersham) was used against these primary antibodies. ICSBP antibody was diluted 1:500 for 1 hour at room temperature. Rabbit antigoat HRP secondary antibody (1:2000) (Pierce, Rockford, IL) was used against ICSBP primary antibody. Phospho-specific STAT1 (New England Biolabs, Beverly, MA) (1:500) was incubated at 4°C overnight. Donkey antirabbit HRP (New England Biolabs) (1:2000) secondary antibody was used against phosphorylated STAT1 primary antibody. Antibodies were used at the indicated dilutions and purchased from Santa Cruz Biotechnology (CA) unless otherwise indicated.
Electromobility shift assay
Sequences for oligonucleotides used in binding reactions were as follows: human IRF-1 gamma activation sequence (GAS), 5′-ACAACAGCCTGATTTCCCCGAA-3′; murine IRF-1 GAS, 5′-CCTGATTTCCCCGAAATGATG-3′; murine ICSBP GAS, 5′-AGTGATTTCTCGGAAAGAGAG-3′. Oligonucleotides were synthesized at the Molecular Biology Core Laboratory (Portland, OR, Veterans Affairs Medical Center), then labeled with [γ32P-ATP] to 25 000 cpm/ng by means of T4 polynucleotide kinase (Roche Molecular Biochemical, Indianapolis, IN). Binding reactions (20 μL) contained 5.0 to 7.5 μg nuclear extracts, 0.2 ng labeled oligo, 2 μg poly (dI-dC), and 10 μg bovine serum albumin in 10 mM Tris-Cl, pH 7.4, 50 mM NaCl, 1 mM dithiothreitol, 1 mM EDTA, and 10% glycerol. Reactions were incubated at room temperature for 30 minutes and then resolved on a 4% polyacrylamide gel in 25 mM Tris, 190 mM glycine, and 1 mM EDTA. Gels were dried and autoradiographed with intensifying screens at −80°C.
RNA isolation, cDNA synthesis, and real-time reverse-transcriptase polymerase chain reaction
Total RNA was isolated from cells with TriReagent (Molecular Research Center, Cincinnati, OH) according to the manufacturer's instructions. We performed cDNA synthesis with 1 μg total RNA, 200 ng random hexamers (Life Technologies), and Superscript II reverse transcriptase (Life Technologies) according to the manufacturer's instructions. The cDNAs were then diluted to an approximate concentration of 20 ng/mL before performing the amplification reactions. Amplifications were performed in an ABI Prism 7700 Sequence Detection System (PE Applied Biosystems, Foster City, CA) by means of 100 ng cDNA, 7.5 pmol each target primer, 2.5 pmol target probe, 2.5 pmol each housekeeping primer, and 2.5 pmol housekeeping probe (h18S). As recommended by the manufacturer, the TaqMan polymerase chain reaction (PCR) kit (PE Applied Biosystems) was used as directed. The primer and probe sequences, designed by means of ABI Primer Express software (PE Applied Biosystems) and synthesized by Integrated DNA Technologies (Coralville, IA), are as follows: hMxA (forward primer) 5′-TGGTGGTGGTCCCCAGTAAT-3′; hMxA (reverse primer) 5′-CGTCAAGATTCCGATGGTCC-3′; hMxA (dual labeled probe) 5′-FAM-CCACCACAGAGGCTCTCAGCATGG-TAMRA-3′; h18S (forward primer) 5′-CGGCTACCACATCCAAGGAA-3′; h18S (reverse primer) 5′-GGGCCTCGA AAGAGTCCTGT-3′; and 18S Probe 5′-VIC-CA GCAGGCGCGCAAATTACCCA-TAMRA-3′.
Results
Constitutive overexpression of IFNγ-inducible genes in mutant FA-C cell lines
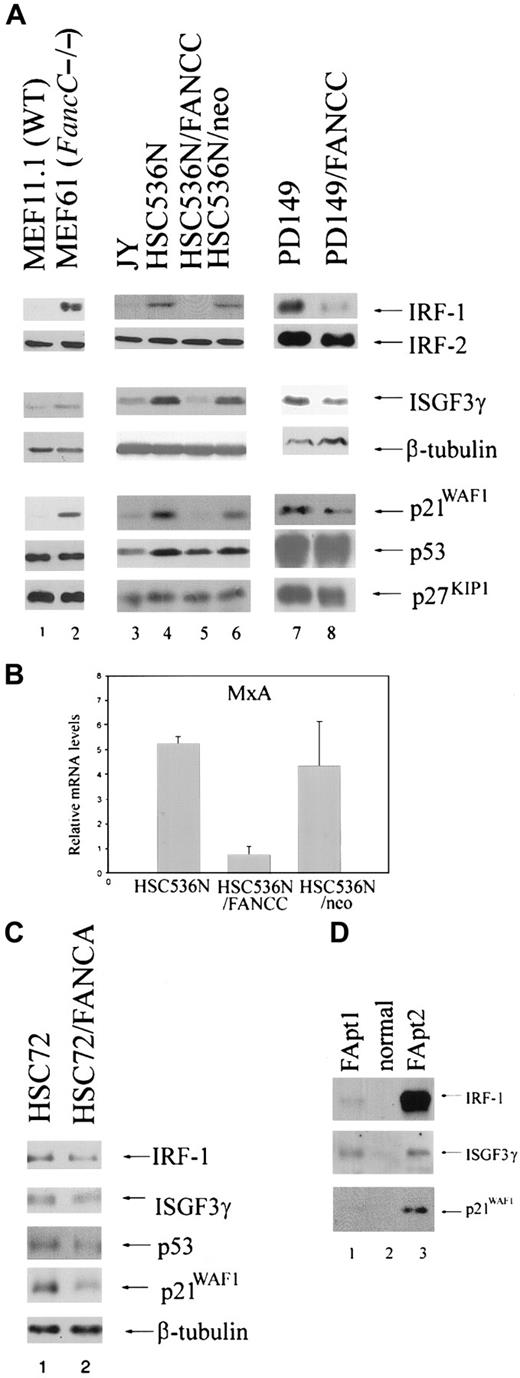
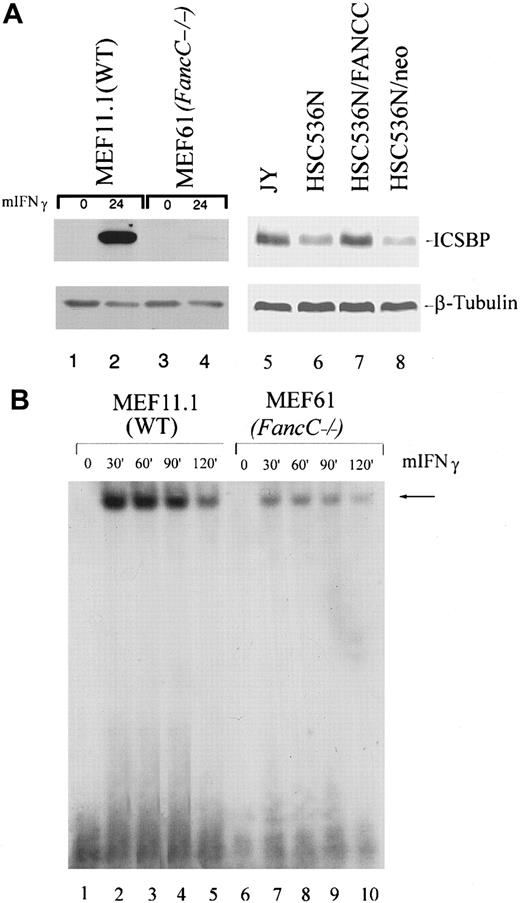
Because FA-C cells are hypersensitive to the inhibitory effects of IFNγ,23,24 we quantified protein levels of transcriptional activators known to be downstream of the IFNγ/Jak/STAT pathway by immunoblot analysis of total cell lysates or nuclear extracts. Analysis of the IFNγ-inducible transactivating factors IRF-1 and ΙSGF3γ consistently revealed high constitutive expression levels in nuclear extracts and whole cell lysates derived from the FancC mutant MEF cell line (MEF61) compared with the wild-type (WT) MEF cell line (MEF11.1). We observed the same phenomenon using (1) whole cell lysates and nuclear extracts from the EBV-transformed FANCC mutant human B-lymphoblast line HSC536N; (2) nuclear extracts from the EBV-transformed B-lymphoblast line PD149, relative to levels in those cells corrected for the defect by retroviral transduction of the normal FANCC cDNA (Figure1A); and (3) whole cell lysates from LDBMCs derived from FA patients compared with LDBMCs derived from a normal person (Figure 1D). In addition, we have previously found constitutively high expression of IRF-1 messenger RNA (mRNA) also occurs in diploid bone marrow cells from children with FA-C but not in normal bone marrow cells.24 However, no differences in IRF-2 expression levels were found between FA-C cells and normal cells (Figure 1A). In addition, mRNA levels of the IFNγ-inducible guanosine triphosphatase (GTPase) MxA are reduced in the HSC536N B-lymphoblast line compared with corrected cells (Figure1B). We found no differences in IRF-1 and ISGF3γ expression in the FA-A lymphoblast line HSC72 (Figure 1C).
Expression of IRF-1, ISGF3γ, and p21WAF1 is reduced in FA-C MEFs, B lymphoblasts, and FA patient LDBMCs.
(A) Immunoblot analysis of the FancC mutant MEF cell line (MEF61) shows increased expression levels of IRF-1, ISGF3g, and p21WAF1 compared with the WT MEF cell line (MEF1.1) (lane 2 vs lane 1). The FANCC mutant B-lymphoblast line HSC536N, the HSC536N line transduced with a vector only (HSC536N/neo), and PD149 (lanes 4, 6, and 7, respectively) show increased IRF-1, ISGF3γ, and p21WAF1 expression compared with the normal cell line JY (lane 3) and with cells corrected for the defect by retroviral transduction of the FANCC cDNA (HSC536N/FANCC) (lane 5) or PD149/FANCC (lane 8). However, FA-C cells express normal levels of IRF-2, p53, and p27KIP1. To demonstrate equivalent loading of the ISGF3γ blots, β-tubulin is used. (B) Real-time reverse transcriptase PCR demonstrates up-regulation of MxA in HSC536N and HSC536N/neo compared with the corrected cells HSC536N/FANCC. This is a representative experiment. Error bars are based on duplicate samples. (C) The FA-C cell line HSC72 expresses equivalent levels of IRF-1 and ISGF3γ compared with cells corrected for the defect. The cells do express slightly more p21WAF1, but equivalent p53. To demonstrate equivalent loading of the p21 blot, β-tubulin is used. (D) LDBMCs derived from 2 FA patients show up-regulation of IRF-1, ISGF3γ, and p21WAF1 compared with cells derived from normal individual (lanes 1 and 3 vs lane 2).
Expression of IRF-1, ISGF3γ, and p21WAF1 is reduced in FA-C MEFs, B lymphoblasts, and FA patient LDBMCs.
(A) Immunoblot analysis of the FancC mutant MEF cell line (MEF61) shows increased expression levels of IRF-1, ISGF3g, and p21WAF1 compared with the WT MEF cell line (MEF1.1) (lane 2 vs lane 1). The FANCC mutant B-lymphoblast line HSC536N, the HSC536N line transduced with a vector only (HSC536N/neo), and PD149 (lanes 4, 6, and 7, respectively) show increased IRF-1, ISGF3γ, and p21WAF1 expression compared with the normal cell line JY (lane 3) and with cells corrected for the defect by retroviral transduction of the FANCC cDNA (HSC536N/FANCC) (lane 5) or PD149/FANCC (lane 8). However, FA-C cells express normal levels of IRF-2, p53, and p27KIP1. To demonstrate equivalent loading of the ISGF3γ blots, β-tubulin is used. (B) Real-time reverse transcriptase PCR demonstrates up-regulation of MxA in HSC536N and HSC536N/neo compared with the corrected cells HSC536N/FANCC. This is a representative experiment. Error bars are based on duplicate samples. (C) The FA-C cell line HSC72 expresses equivalent levels of IRF-1 and ISGF3γ compared with cells corrected for the defect. The cells do express slightly more p21WAF1, but equivalent p53. To demonstrate equivalent loading of the p21 blot, β-tubulin is used. (D) LDBMCs derived from 2 FA patients show up-regulation of IRF-1, ISGF3γ, and p21WAF1 compared with cells derived from normal individual (lanes 1 and 3 vs lane 2).
We next quantified levels of the cell cycle modulator p21WAF1, which is known to be induced by IRF-1 and IFNγ.35 36 FA-C B lymphoblasts, MEFs, and LDBMCs express high levels of p21WAF1 compared with normal or corrected cells (Figure 1A,D). The FA-A cell line HSC72 also expresses higher levels of p21WAF1 compared with corrected cells, but does not overexpress IRF-1 or ISGF3γ (Figure 1C). This suggests that overexpression of the transactivators IRF-1 and ISGF3γ and of the cell cycle inhibitor p21WAF1 may be due to abnormalities in different pathways, one of which is influenced by both FANCA and FANCC and the other by FANCC alone. We found no differences in p53 binding to its p21 binding site (data not shown) and minimal differences in p53 expression levels, indicating that p21WAF1 overexpression is p53-independent (Figure 1A). No differences in expression levels of the cdk inhibitor p27KIP1 were found (Figure 1A).
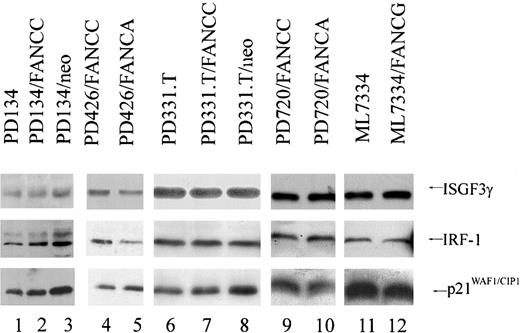
We found no differences in protein levels of IRF-1, ISGF3γ, and p21WAF1 in mature primary human FA fibroblasts (Figure2). Finding molecular differences between mature fibroblasts and hematopoietic cells is not surprising given that hematopoietic abnormalities represent the dominant phenotype in FA patients. In addition, finding that a normal FANCC is required for proper regulation of IRF-1, p21WAF1, and ISGF3γ in hematopoietic cells and embryonic fibroblasts but not in mature fibroblasts suggests that MEFs have retained more plasticity and may exhibit responses more reflective of hematopoietic and germ cells than adult fibroblasts.
Human FA fibroblasts express equivalent levels of IRF-1, ISGF3γ, and p21WAF1.
Immunoblot analysis of human FA-C fibroblasts PD134 (lane 1), PD134/neo (lane 3), PD426/FANCA (lane5), PD331.T (lane 6), and PD331.T/neo (lane 8); the FA-A fibroblasts PD720/FANCC (lane 9); and the FA-G fibroblast line ML7334 (lane 11) express equivalent levels of ISGF3γ, IRF-1, and p21WAF1 compared with those cells corrected for the defect by retroviral transduction of the appropriate FA cDNA: PD134/FANCC (lane 2), PD426/FANCC (lane 4), PD331.T/FANCC (lane 7), PD720/FANCA (lane 10), ML7334/FANCG (lane 12).
Human FA fibroblasts express equivalent levels of IRF-1, ISGF3γ, and p21WAF1.
Immunoblot analysis of human FA-C fibroblasts PD134 (lane 1), PD134/neo (lane 3), PD426/FANCA (lane5), PD331.T (lane 6), and PD331.T/neo (lane 8); the FA-A fibroblasts PD720/FANCC (lane 9); and the FA-G fibroblast line ML7334 (lane 11) express equivalent levels of ISGF3γ, IRF-1, and p21WAF1 compared with those cells corrected for the defect by retroviral transduction of the appropriate FA cDNA: PD134/FANCC (lane 2), PD426/FANCC (lane 4), PD331.T/FANCC (lane 7), PD720/FANCA (lane 10), ML7334/FANCG (lane 12).
FA-C cells overexpressing IFNγ-inducible genes are deficient in STAT1 phosphorylation
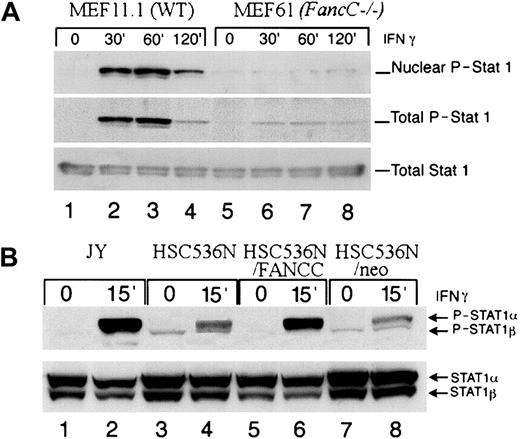
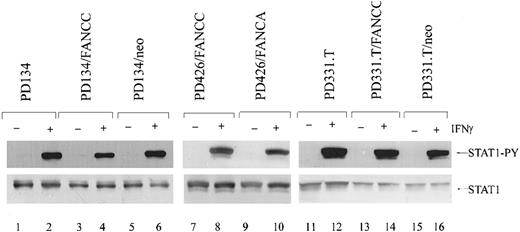
FA-C cells have been shown to be defective in STAT1 phosphorylation, despite having normal activation of the IFNγ receptor, JAK1, and JAK2.29 We sought to determine if this occurred in cells that overexpress IFNγ-inducible genes and those that do not. We analyzed the levels of phosphorylated STAT1 in IFNγ-induced cells. MEFs were serum starved for 24 hours and then incubated at 37°C with 1 ng/mL of murine IFNγ for the indicated time periods. Using immunoblot analysis, we determined that in both total cell lysates and nuclear extracts from MEFs, the level of phosphorylated STAT1 was significantly decreased in FancCmutant cells compared with the WT cells. When these blots were stripped and reprobed for total STAT1, levels of total STAT1 were equivalent (Figure 3A). In addition, analysis of total cell lysates from the mutant FANCC B-cell lines HSC536N and HSC536N/neo demonstrated that the amount of phosphorylated STAT1 was diminished when compared with the normal cell line JY and the FANCC-transduced cell line HSC536N/FANCC. When the same blots were stripped and reprobed for STAT1, total STAT1 was slightly increased in the mutant cells (but phosphorylated levels were decreased) (Figure 3B). However, STAT1 phosphorylation was found to be normal in all mature human FA fibroblast lines tested (Figure4). Thus, in FA cells, STAT1 phosphorylation defects are specific to certain cell types, and paradoxically, those cell types defective in STAT1 activation have high constitutive expression of several IFNγ-inducible genes.
STAT1 phosphorylation at tyrosine 701 is reduced in MEFs and B lymphoblasts.
(A) Immunoblot analysis demonstrates that phosphorylation of STAT1 is reduced in both nuclear extracts and total cell extracts (upper and middle panel respectively) from FancC−/− MEFs (MEF61) (lanes 5 through 8) compared with WT cells (MEF11.1) (lanes 1 though 4) following stimulation with 1 ng/mL IFNγ for the indicated times. Total STAT1 levels (lower panel) were equivalent (lanes 1 through 4 compared with lanes 5 through 8). (B) Phosphorylation of STAT1 (upper panel) is reduced in total cell extracts from HSC536N (lanes 3 and 4) and HSC536N/neo (lanes 7 and 8) compared with JY (lanes 1 and 2) and HSC536N/FANCC (lanes 5 and 6) following stimulation with 1 ng/mL IFNγ at 37°C for 15 minutes (lanes 2, 4, 6, and 8). Total STAT1 levels were slightly higher in the mutant cells (but phosphorylation levels are reduced) (lower panel).
STAT1 phosphorylation at tyrosine 701 is reduced in MEFs and B lymphoblasts.
(A) Immunoblot analysis demonstrates that phosphorylation of STAT1 is reduced in both nuclear extracts and total cell extracts (upper and middle panel respectively) from FancC−/− MEFs (MEF61) (lanes 5 through 8) compared with WT cells (MEF11.1) (lanes 1 though 4) following stimulation with 1 ng/mL IFNγ for the indicated times. Total STAT1 levels (lower panel) were equivalent (lanes 1 through 4 compared with lanes 5 through 8). (B) Phosphorylation of STAT1 (upper panel) is reduced in total cell extracts from HSC536N (lanes 3 and 4) and HSC536N/neo (lanes 7 and 8) compared with JY (lanes 1 and 2) and HSC536N/FANCC (lanes 5 and 6) following stimulation with 1 ng/mL IFNγ at 37°C for 15 minutes (lanes 2, 4, 6, and 8). Total STAT1 levels were slightly higher in the mutant cells (but phosphorylation levels are reduced) (lower panel).
Normal phosphorylation of STAT1 in mature FA-C fibroblasts.
Immunoblot analysis of human FA-C fibroblasts by means of an antibody specific for phosphorylated STAT1 demonstrates that STAT1 is phosphorylated equivalently between mutant and corrected IFNγ-stimulated cells (1 ng/mL IFNγ) (top panel). In addition, total STAT1 is expressed at approximately equal levels (bottom panel).
Normal phosphorylation of STAT1 in mature FA-C fibroblasts.
Immunoblot analysis of human FA-C fibroblasts by means of an antibody specific for phosphorylated STAT1 demonstrates that STAT1 is phosphorylated equivalently between mutant and corrected IFNγ-stimulated cells (1 ng/mL IFNγ) (top panel). In addition, total STAT1 is expressed at approximately equal levels (bottom panel).
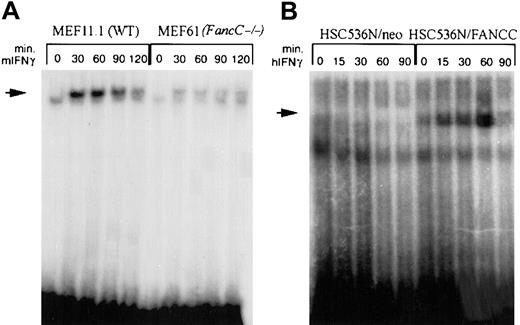
Reduced binding to the GAS element in FA-C cells
It was theoretically possible that a more complete and rapid nuclear translocation of STAT1 in FA-C cells might have given an appearance of reduced STAT1 activation. Consequently, we asked whether apparently reduced levels of STAT1 phosphorylation were associated with increased binding to the STAT1 response element, GAS. Using an oligonucleotide derived from the GAS element of murine IRF-1 for electromobility shift assay (EMSA) analysis of murine MEF lines, we found that IFNγ-inducible binding to the GAS element was significantly reduced in theFancC−/− MEF line compared with the WT MEF line (Figure5A). Using oligonucleotides representing the human IRF-1 GAS element, we found that IFNγ-inducible binding was not detectable in mutant FANCC human B cells, whereas normal inducible binding was observed in the FANCC-complemented cell line (Figure 5B). These results indicate that the diminished ability to phosphorylate STAT1 in FA-C cells results in decreased activated STAT1 nuclear translocation and binding to GAS.
STAT1 binding to the IRF-1 GAS element is reduced in FA-C cell lines.
EMSA of STAT1 binding to oligonucleotides corresponding to the GAS element in the IRF-1 promoter. (A) IFNγ-inducible STAT1 binding is diminished in FancC−/− cells (MEF61) (lanes 6 through 10) compared with WT MEFs (MEF11.1) (lanes 1 through 5). Murine IFNγ was used at 1 ng/mL at 37°C. (B) No STAT1 binding occurs in HSC536N/neo (lanes 1 through 5), whereas in HSC536N/FANCC, STAT1 binds in an IFNγ-inducible manner (lanes 6 through 10). Cells were treated with 1 ng/mL human IFNγ at 37°C.
STAT1 binding to the IRF-1 GAS element is reduced in FA-C cell lines.
EMSA of STAT1 binding to oligonucleotides corresponding to the GAS element in the IRF-1 promoter. (A) IFNγ-inducible STAT1 binding is diminished in FancC−/− cells (MEF61) (lanes 6 through 10) compared with WT MEFs (MEF11.1) (lanes 1 through 5). Murine IFNγ was used at 1 ng/mL at 37°C. (B) No STAT1 binding occurs in HSC536N/neo (lanes 1 through 5), whereas in HSC536N/FANCC, STAT1 binds in an IFNγ-inducible manner (lanes 6 through 10). Cells were treated with 1 ng/mL human IFNγ at 37°C.
Expression of the IFNγ response modulator ICSBP is reduced in the FA-C cell lines
Seeking to identify potential molecular points of control involved in the aberrant expression of IRF-1, p21WAF1, and ISGF3γ in FA-C cells, we quantified ICSBP, an interferon-responsive transcription factor known to modulate IFN responses in hematopoietic cells.37-39 ICSBP levels were constitutively low in HSC536N and HSC536N/neo cell lines compared with the normal cell line JY and the complemented line HSC536N/FANCC (Figure6A). In addition, although ICSBP is reported to be expressed mainly in hematopoietic cells, we found ICSBP expression to be consistently inducible in WT MEFs as well. Expression of ICSBP is significantly decreased in FANCC mutant MEFs. Therefore a functional FANCC is necessary for optimal constitutive expression of ICSBP. In addition, we find reduced binding to the GAS element derived from murine ICSBP (Figure 6B), suggesting that decreased ICSBP expression may be a direct result of reduced STAT1 phosphorylation.
ICSBP is expressed at reduced levels in FA-C cells.
(A) Immunoblot analysis demonstrates that ICSBP is expressed inducibly (50 ng/mL IFNγ for 24 hours) in wild-type MEFs (MEF11.1) (lane 2) at high levels. ICSBP expression in the FancC−/−MEF line (MEF61) is significantly reduced compared with the wild-type cells (lane 4 vs lane 2). Constitutive expression of ICSBP is reduced in FA-C B lymphoblasts compared with normal and corrected cells (lanes 6 and 8 vs lanes 5 and 7). The same blot was stripped and reprobed with β-tubulin to demonstrate equivalent loading (lower panel). (B) EMSA of the GAS element of murine ICSBP demonstrates reduced IFNγ-stimulated binding in FancC−/− MEFs compared with WT MEFs. Cells were stimulated with 1 ng/mL murine IFNγ for the indicated times.
ICSBP is expressed at reduced levels in FA-C cells.
(A) Immunoblot analysis demonstrates that ICSBP is expressed inducibly (50 ng/mL IFNγ for 24 hours) in wild-type MEFs (MEF11.1) (lane 2) at high levels. ICSBP expression in the FancC−/−MEF line (MEF61) is significantly reduced compared with the wild-type cells (lane 4 vs lane 2). Constitutive expression of ICSBP is reduced in FA-C B lymphoblasts compared with normal and corrected cells (lanes 6 and 8 vs lanes 5 and 7). The same blot was stripped and reprobed with β-tubulin to demonstrate equivalent loading (lower panel). (B) EMSA of the GAS element of murine ICSBP demonstrates reduced IFNγ-stimulated binding in FancC−/− MEFs compared with WT MEFs. Cells were stimulated with 1 ng/mL murine IFNγ for the indicated times.
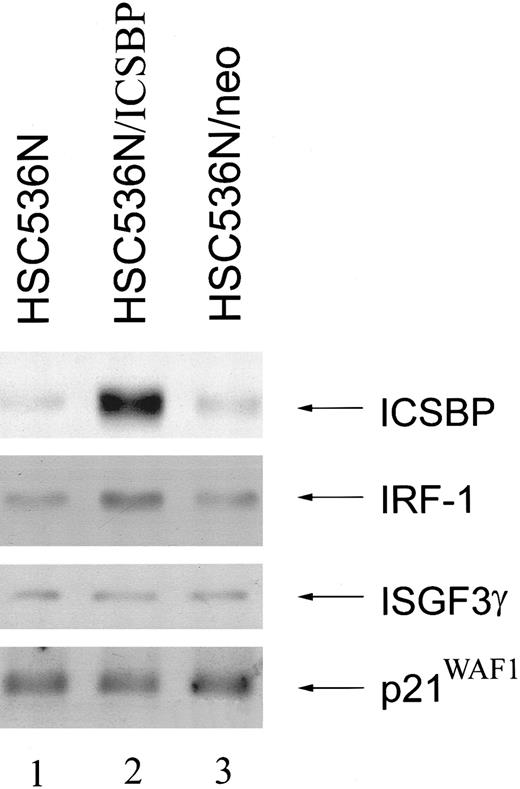
Enforced expression of ICSBP does not suppress IRF-1, p21WAF1/CIP1, or ISGF3γ
To examine the linkage of reduced ICSBP with overexpression of IRF-1, ISGF3γ, or p21WAF1, we transduced the HSC536N B-lymphoblast cell line with a retroviral vector carrying the humanICSBP cDNA. Although these cells express high levels of ICSBP, ICSBP expression failed to suppress the expression of IRF-1, p21WAF1, or ISGF3γ (Figure7). In fact; expression of IRF-1 was slightly increased in cells expressing ICSBP. Therefore, up-regulation of IRF-1, ISGF3γ, and p21WAF1 is not directly linked to reduced ICSBP expression.
Enforced expression of ICSBP fails to down-regulate IRF-1 ISGF3γ or p21WAF1.
Immunoblot analysis of FA-C B lymphoblasts HSC536N (lane1), HSC536N transduced with the human ICSBP cDNA (HSC536N/ICSBP) (lane2), and HSC536N transduced with vector alone (HSC536N/neo) (lane 3) demonstrates that HSC536N/ICSBP are expressing high constitutive levels of ICSBP, but this expression fails to suppress IRF-1, ISGF3γ, or p21WAF1.
Enforced expression of ICSBP fails to down-regulate IRF-1 ISGF3γ or p21WAF1.
Immunoblot analysis of FA-C B lymphoblasts HSC536N (lane1), HSC536N transduced with the human ICSBP cDNA (HSC536N/ICSBP) (lane2), and HSC536N transduced with vector alone (HSC536N/neo) (lane 3) demonstrates that HSC536N/ICSBP are expressing high constitutive levels of ICSBP, but this expression fails to suppress IRF-1, ISGF3γ, or p21WAF1.
Discussion
Interferons are involved in a broad range of mammalian biological functions, including immunity, inflammation, hematopoiesis, cell proliferation, and differentiation.16,17 In hematopoietic progenitor cells, IFNγ induces apoptosis,21,26,40,41 and there is compelling indirect evidence that it plays a pathophysiologically significant role in acquired aplastic anemias.17-19,21,41 Because FA-C cells are hypersensitive to IFNγ,23 24 we believe that IFNγ hypersensitivity and bone marrow failure may be linked in FA. Accordingly, we have focused our attention on the IFNγ signaling pathway in FA-C cells as a way of deciphering the function of the FANCC protein in modulating the apoptotic response to extracellular cues.
The effects of IFNγ are mediated, at least in part, through transcriptional regulation of downstream genes, which include members of the IRF family of transcription factors. We find that certain IRF family members are constitutively overexpressed in specific FA-C cell types compared with normal cells. ISGF3γ, which is constitutively expressed at higher levels in FANCC mutant B lymphoblasts, MEFs, and patient LDBMCs, is of particular interest because it is the only protein of the IRF family known to complex with STAT molecules.42 ISGF3γ exerts its transcriptional activation only in conjunction with STAT1 and STAT243 and plays a crucial role in the regulation of IFN-β gene expression,44-46 a finding of relevance because some researchers have suggested that the induced expression of IFN-β is required for the hematopoietic suppressive effects of IFNγ.47 IRF-1 is required for carrying out the apoptotic effects of IFNγ signaling. The ratio of IRF1 to IRF2 expression is important for proper cell cycle control.48,49 IRF-1 expression is significantly increased in FA-C hematopoietic and embryonic cells, but no differences in IRF-2 expression levels were found (Figure 1), upsetting the proper balance of IRF-1 and IRF-2 levels in FA-C cells. IRF-1 also cooperates with p53 to induce expression of the cdk inhibitor p21WAF1.35 50Although equivalent p53 expression levels were found betweenFancC−/− MEFs andFancC+/+ MEFs and only minimal differences in p53 expression were found in human FA-C B lymphoblasts compared with normal cells, FA-C cells expressed significantly greater amounts of p21WAF1 (Figure 1). There was no difference in p53 binding to its p21WAF1-binding site (data not shown), and expression of the IRF-1 independent cdk inhibitor p27KIP1was equivalent (Figure 1). Thus, it is possible that IRF-1 up-regulation contributes to the overexpression of p21WAF1. However, given that we find high constitutive p21WAF1 in FA-A cells and that these cells do not overexpress IRF-1, it is more likely that up-regulation of p21WAF1 and the IRF transactivators may be due to abnormalities in different pathways, one of which is influenced by both FANCA and FANCC or the entire FA complex and the other by FANCC alone.
In addition to IRF-1, ISGF3γ, and p21WAF1, others have found that IFNγ-inducible GTPase, MxA, which confers intracellular antiviral activity in IFN-induced cells,51,52 is also constitutively expressed in FA cells,53 a finding we confirm here (Figure 1B). Thus, several IFNγ-inducible factors that are important in regulation of apoptosis and mitogenic inhibition are constitutively upregulated in FA-C cells in a cell-type–specific manner.
Surprisingly, though FA-C hematopoietic and embryonic cells (1) are hypersensitive to the mitotic inhibitory effects of IFNγ and (2) express constitutively high levels of several IFNγ-inducible genes (effects abrogated by transduction of normal FANCC cDNA), activation of STAT1 is impaired.29 We sought to determine if loss of STAT1 activation occurs only in those cell types exhibiting high constitutive expression of IFNγ-inducible genes. We found STAT1 phosphorylation to be defective in FA-C B lymphoblasts and MEFs, but not in mature human fibroblasts (Figures 3, 4). Because mature human fibroblasts do not overexpress these genes, we reason that the overexpression of IRF-1, ISGF3γ, and p21WAF1 occurs only in FA-C cells defective in their ability to activate STAT1. Up-regulation of these genes in these cells appears to require that (1) FANCC be inactivated and (2) the particular FANCC mutation suppress STAT1 activation. Therefore, constitutive expression of IFNγ-responsive genes in FANCC mutant cells is due to either ground-state activation of a pathway that does not require STAT1 phosphorylation or loss of a hematopoietic IFNγ-responsive protein that normally modulates expression of other IFN-responsive genes. One IFNγ-responsive modulator of interest to us was the transcriptional repressor ICSBP, a member of the IRF family expressed mainly in hematopoietic cells.38
ICSBP interacts with both IRF-1 and IRF-2 in vitro and in vivo and inhibits the DNA-binding activity of ISGF3γ.37 ICSBP binds to IRF-1 and suppresses some IFNγ-induced gene responses.38 Stable expression of the DNA binding domain of ICSBP blocks IRF-1 transactivation and also inhibits binding of STAT1 to GAS.54 In FANCC mutant B lymphoblasts we found less ICSBP expression than normal cells (Figure 6). In fact, ICSBP is the only IFNγ-inducible protein of those we have tested that is reduced in FA-C cells. Surprisingly, ICSBP is also expressed in an inducible manner in MEFs, and this expression is significantly reduced in FancC mutant MEFs. We first hypothesized that low expression levels of ICSBP in FA-C cells may be a direct result of a deficiency in STAT1 phosphorylation in FA-C cells. Indeed, STAT1 binding to the GAS element from ICSBP is significantly reduced in FA-C cells (Figure 6). Because ICSBP is decreased in those cells with up-regulated IFNγ-inducible genes, we next reasoned that loss of ICSBP contributes to the up-regulation of these genes. However, enforced expression of ICSBP into these cells failed to down-regulate IRF-1, ISGF3γ, or p21WAF1 (Figure 7). Therefore, up-regulation of these IFNγ-inducible factors in FA cells is not directly linked with reduced ICSBP expression levels.
FA cells, both in vitro and in vivo, are not hardy. They grow slowly and have a high apoptotic fraction.21,24,26 Studies in our laboratory have focused, in large part, on clarifying molecular determinants of the death pathways in FA cells and on beginning to establish an ordered relationship of these death pathways to discrete FA proteins. We have identified roles for the fas/fas-ligand pathway,24 caspases 8 and 3,55 and the double-stranded RNA-dependent protein kinase.56 In this work, although we cannot yet clarify the cause, it is clear that even without being exposed to environmental cues for apoptosis or mitotic arrest, at a gene-expression level, FA-C cells act as if they have been stimulated by such factors. This is similar to another chromosomal instability syndrome, ataxia telangiectasia (AT). AT cells are hypersensitive to γ irradiation and constitutively overexpress several γ-irradiation–inducible factors.57 58 In view of the apoptotic and mitogenic inhibitory activities of these proteins, we suggest that they contribute to the characteristic apoptotic phenotype in FA bone marrow cells and may contribute to the IFN-hypersensitive phenotype by specifically setting ground-state levels of these proteins that are higher than those in normal cells. Clarification of the signaling pathways by which aberrantly expressed genes are controlled in FA-C cells should lead to an understanding of why such cells are hypersensitive to both biological and chemical inducers of apoptosis and may reveal clearer functions for theFANCC gene product.
The authors thank Dr Ben-Zion Levi for kindly providing the human ICSBP cDNA; Dr Markus Grompe, Dr Petra Jakobs, Dr Yasmine Akarri, and Dr Barbara Cox for providing the human FA fibroblasts; Dr Manuel Buchwald for the HSC536N cell line; Dr Chris Walsh for the HSC72 and HSC72/FANCA cell lines; Tara Koretsky and Keaney Rathbun for technical support; and Dr Qishen Pang for experimental help and advice.
Supported by grant HL48546 from the National Institutes of Health.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Grover C. Bagby, Oregon Cancer Center, Oregon Health Sciences University, CR-145, 3181 SW Sam Jackson Park Rd, Portland, OR 97201; e-mail: grover@ohsu.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal