Abstract
In a search for key molecules that prevent murine M1 leukemia cells from undergoing interleukin (IL)-6–induced differentiation into macrophages, we isolated an antisense complementary DNA (cDNA) that encodes full-length mouse MgcRac-GTPase-activating protein (GAP) through functional cloning. Forced expression of this antisense cDNA profoundly inhibited IL-6–induced differentiation of M1 cells into macrophage lineages. We also isolated a full-length human MgcRacGAP cDNA, which encodes an additional N-terminal polypeptide of 105 amino acid residues compared with the previously published human MgcRacGAP. In human HL-60 leukemic cells, overexpression of the full-length form of human MgcRacGAP alone induced growth suppression and macrophage differentiation associated with hypervacuolization and de novo expression of the myelomonocytic marker CD14. Analyses using a GAP-inactive mutant and 2 deletion mutants of MgcRacGAP indicated that the GAP activity was dispensable, but the myosin-like domain and the cysteine-rich domain were indispensable for growth suppression and macrophage differentiation. The present results indicated that MgcRacGAP plays key roles in controlling growth and differentiation of hematopoietic cells through mechanisms other than regulating Rac GTPase activity.
Introduction
Rho-related guanosine triphosphate (GTP)-binding proteins constitute a functionally distinct group within the small G protein family, which includes Rho A, B, C, and G, Rac1 and Rac2, Cdc42, and TC10.1 These proteins are 30% identical to Ras and 50% to 60% identical to each other at the amino acid level. Rho family members have been linked to a variety of cellular functions, including changes in cytoskeletal dynamics (actin polymerization and reorganization), gene expression (p38/Jun NH2-terminal kinase and serum response factor activity), G1 cell cycle progression, endocytosis, exocytosis, and superoxide production.2-5GTPase-activating proteins (GAPs) for Rho GTPases constitute a class of regulatory proteins that can bind GTP-bound active forms of small G proteins and stimulate GTP hydrolysis.6,7 Harboring this catalytic function, RhoGAPs negatively regulate Rho-mediated signals. However, certain GAPs, including p120RasGAP, n-chimaerin, and phospholipase C (a GAP for heterotrimeric G proteins) simultaneously function as effectors downstream of the GTPases.8-10 More specifically, n-chimaerin was shown to cooperate with Rac1 and Cdc42 in inducing specific changes in cytoskeletal morphology, ie, the formation of lamellipodia and filopodia, respectively.8 This effect ofn-chimaerin requires G protein binding capacity and the non-GAP N-terminal extension but not GAP activity.8 A conventional myosin, myosin-IXb, harboring a chimaerin-like Rho/Rac GAP domain in its tail colocalizes with F-actin in the cell periphery in undifferentiated human HL-60 leukemia cells, while in differentiated cells the localization becomes more cytoplasmic, with the highest levels being seen in the perinuclear region.11Regulation of the cytoskeleton by Rho family members seems to depend on a cascade of events within the acto-myosin system.8 Thus, the view is favored that RhoGAPs control a variety of cellular functions through Rho family proteins as well as other signaling molecules.
We report here functional cloning of murine MgcRacGAP (mMgcRacGAP) as a differentiation-regulating gene of murine M1 leukemia cells. Overexpression of the full-length human MgcRacGAP (hMgcRacGAP) suppressed cell growth and induced macrophage differentiation of HL-60 cells, indicating that MgcRacGAP is involved in the control of growth and differentiation of hematopoietic cells.
Materials and methods
Cell lines
The murine myeloid leukemia cell line M1 was grown in Dulbecco's modified Eagle's medium (DMEM; GIBCO, Rockville, MD) containing 10% fetal calf serum (FCS). A human myeloid leukemia cell line HL-60 was grown in RPMI 1640 medium (GIBCO) containing 10% FCS. An ecotropic retrovirus packaging cell line BOSC23 12was maintained in DMEM containing 10% FCS and GPT selection reagents (Specialty Media, Lavallette, NJ). Two days before transfection, the cells were transferred into DMEM-10% FCS without GPT selection reagents.
Retrovirus vectors
A bicistronic retroviral vector pMX-IRES-EGFP was constructed as described.13 pMX-MgcRacGAP-IRES-EGFP was constructed by inserting a 1.9-kb EcoRI-NotI fragment of the Flag-tagged full-length form of hMgcRacGAP gene into EcoRI and NotI sites of pMX-IRES-EGFP, upstream of the IRES (internal ribosomal entry site) sequence so that both MgcRacGAP and EGFP (enhanced green fluorescent protein) are expressed from a single messenger RNA (mRNA) in the same cells.
Production of retroviruses and infection with them
High-titer retroviruses carrying the hMgcRacGAP-IRES-EGFP were produced with a transient retrovirus packaging cell line BOSC23 12 as described previously.13 We first established a stable transfectant expressing the ecotropic virus receptor.14,15 Infection was performed as described.16 Briefly, cells were incubated with 10 mL of the retroviruses in the presence of 10-μg/mL hexadimethrine bromide (Sigma, St Louis, MO). Twenty-four hours after infection, cells were washed, refed with growth medium, and left for one more day before cell sorting with EGFP.
Cell sorting and flow cytometry
EGFP+ cells were sorted using a modification of the technique described earlier.17 Briefly, 2 days after virus infection, the infected cells were washed twice with phosphate buffered saline (PBS), suspended in PBS containing 1% bovine serum albumin (BSA), and then sorted based on EGFP expression on a FACSVantage (Becton Dickinson, San Jose, CA). The sorted cells (40 per well) were then expanded in growth medium. Flow cytometric analysis was carried out to quantify morphologic changes and to confirm the expression of EGFP in the HL-60 transfectants on a FACSCalibur flow cytometer (Becton Dickinson). The cells were also stained with a phycoerythrin (PE)-conjugated mouse antihuman CD14 monoclonal antibody (PharMingen, San Diego, CA) on ice for 30 minutes after blocking with 100-fold excess of mouse immunoglobulin (Ig) G, and analyzed on a FACSCalibur flow cytometer. PE-conjugated mouse IgG2a was used as an isotype-matched negative control.
Immunoprecipitation and Western blotting
Immunoprecipitation, gel electrophoresis, and immunoblotting were performed as described18 but with minor modifications. Exponentially growing cells were washed with PBS, lysed in lysis buffer (50-mmol/L Tris-HCl, pH 7.5; 150-mmol/L NaCl; 1% Triton X-100; 1-mmol/L ethylenediaminetetraacetic acid [EDTA]; 0.2-mmol/L Na3VO4; 2-mmol/L phenylmethylsulfonyl fluoride; 2-μg/mL leupeptin; 10-μg/mL aprotinin) (5 × 106cells/mL), and incubated on ice for 30 minutes. Cell lysates were clarified by centrifugation for 15 minutes at 12 000g prior to incubation with the anti-Flag M2 monoclonal antibody (Eastman Kodak, Kingsport, TN) or the control whole-mouse IgG and protein A-Sepharose at 4°C overnight. The immunoprecipitates were washed 3 times with lysis buffer, subjected to sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis, and electrophoretically transferred onto Immobilon filters (Millipore, Bedford, MA). After blocking in a solution containing 3% BSA, the filter was probed with the anti-Flag M2 monoclonal antibody. The filter-bound antibody was detected using the enhanced chemiluminescence system (Amersham, Buckinghamshire, UK).
Northern blot analysis
Poly(A)+ RNA was isolated from cells using FastTrack 2.0 kit (Invitrogen, San Diego, CA). Two micrograms of Poly(A)+ RNA was denatured in 50% formamide at 60°C, electrophoresed on a 1% agarose formaldehyde gel, and blotted to Hybond-N nylon filter (Amersham). The filter was incubated at 42°C in 50% formamide, 3 × Denhardt's reagent (0.06% polyvinylpyrrolidone, 0.06% BSA, 0.06% Ficoll), 5 × standard saline citrate (SSC; 1 × SSC = 0.15-mol/L NaCl, 0.015-mol/L trisodium citrate), 1% SDS, 200-μg/mL denatured salmon sperm DNA, and a 32P-labeled complementary DNA (cDNA) probe, prepared using Random Prime Kit (Stratagene, La Jolla, CA). After hybridization, the filter was washed in 0.1 × SSC, 0.1% SDS at room temperature, and autoradiographed. Reprobing was carried out after washing the filter in a stripping buffer of 10-mmol/L Tris-HCl (pH 7.5), 1-mmol/L EDTA, 0.1% SDS, and 0.3 × Denhardt's reagent at 90°C for 20 minutes.
GTP hydrolysis assays
The [γ-32P] GTP-bound form of small GTPases was prepared by incubating 10 pmol of protein with 74 kBq of [γ-32P] GTP (1.11 TBq/mmol, NEN Life Science Products, Boston, MA) in a 50-μL volume of 25-mmol/L Tris-HCl (pH 7.5), 5-mmol/L EDTA, 0.2-mmol/L MgCl2, 0.1-mg/ml BSA, 1-mmol/L dithiotreitol, and 4-μmol/L GTP for 15 minutes at room temperature. GTP hydrolysis was initiated by raising MgCl2 and GTP to a final concentration of 20 mmol/L and 200 μmol/L, respectively, and was stopped after 3 minutes by adding 2 mL of ice-cold 50-mmol/L Tris-HCl (pH 8), 35-mmol/L MgCl2, 1-mmol/L dithiotreitol, and 150-mmol/L NaCl. Amounts of [γ-32P] GTP bound to GTPases were determined by radioactivity counting after rapid vacuum filtration of the samples on BA 85 nitrocellulose (Schleicher and Schuell, Dassel, Germany). Typically, 100% of [γ-32P] GTP bound to GTPases is around 25 000 cpm in our hands. The GAP assay was conducted in the presence of the GAP domain (20 pmol) of hMgcRacGAP or a GAP inactive mutant of hMgcRacGAP (R385A*MgcRacGAP) during the step of GTP hydrolysis. GTP exchange reactions during the GTP hydrolysis step were examined using [α-32P] GTP-preloaded GTPases under similar conditions and were not significantly affected by the presence of hMgcRacGAP.
Mutagenesis and production of retroviruses
The mutation R385A was introduced by overlap-extension polymerase chain reaction (PCR) mutagenesis.19 DNA encoding the conserved arginine residue was amplified by PCR using primer pairs as follows:
Upstream primer: 5′-AGCTGCGGAATTCCGGAATT-3′
Mutagenic primer, sense: 5′-AGGCCTGTATGCGATCTCTGGCT-3′
Downstream primer: 5′-TTCACCAACAGCTTGGTACAT-3′
Mutagenic primer, antisense: 5′-AGCCAGAGATCGCATACAGGCCT-3′.
PCR products were joined and amplified by the upstream primer and the downstream primer and cloned into EcoRI-BstXI sites of pMX-MgcRacGAP-IRES-EGFP. Mutagenic sequence was confirmed by automated sequencing by an ABI PRISM 310 Genetic Analyzer (Perkin-Elmer, Branchburg, NJ). To construct a mutant MgcRacGAP lacking the myosin-like domain (ΔMyo-MgcRacGAP), the deletion constructs were generated by PCR with a 5′ primer that contained an EcoRI site (5′-GAAAGAATTCCGAGAGATGCTCATGTGTGA-3′) and a 3′ primer that is located downstream of the BstXI site of hMgcRacGAP (5′-TTCACCAACAGCTTGGTACAT-3′). The resulting PCR fragment was digested with EcoRI and BstXI and subcloned into the corresponding cloning sites of pMX-MgcRacGAP-IRES-EGFP. A mutant MgcRacGAP lacking the cysteine-rich domain (ΔCys-MgcRacGAP) was generated by overlapping extension using PCR. In brief, complementary oligonucleotide primers were used for PCR to generate 2 DNA fragments that have overlapping ends. To construct the N-terminal half of ΔCys-MgcRacGAP, primer pairs 5′-AGCTGCGGAATTCCGGAATT-3′ (primer A) and 5′-GGTGTTCCTATCAGGGTAGGCATCCCTCCATTACTCTGT-3′ (primer B) were used for PCR with the full-length hMgcRacGAP cDNA as a template. Primer pairs 5′-ACAGAGTAATGGAGGGATGCCTACCCTGATAGGAACACC-3′ (primer C) and 5′-TTCACCAACAGCTTGGTACAT-3′ (primer D) were used to generate the C-terminal half. These fragments were combined in a subsequent reaction in which the overlapping strands were annealed, allowing the 3′ overlap of each strand to serve as a primer for the extension of the complementary strand. The resulting fusion product was further amplified by PCR using primers A and D. The product was digested byEcoRI and BstXI, separated by agarose gel electropheresis, purified, and cloned into pMX-MgcRacGAP-IRES-EGFP digested by EcoRI and BstXI. PCR was carried out using high-fidelity DNA polymerase Pyrobest (Takara, Ohtsu, Japan). The PCR condition was as follows: denaturation at 98°C for 30 seconds, annealing at 55°C for 30 seconds, and extension at 72°C for 2 minutes for 25 cycles.
Results
Expression of an antisense DNA encoding MgcRacGAP partially blocked IL-6–induced differentiation of M1 cells
To identify key molecules that regulate cell differentiation into macrophages, we generated a cDNA library in a retrovirus vector pMXneo20 from a subclone of mouse myeloid leukemia M1 cells (termed MD1 cells) expressing a constitutively active STAT5A mutant.21 This particular mutant retained the potential to proliferate while differentiating into various stages along the monocytic differentiation pathway and giving rise to a heterogeneous population of blast, intermediate, and mature monocytic cells through the production of interleukin-6 (IL-6) (T. Kawashima et al, unpublished data). We searched for a gene for this particular phenotype by retrovirus-mediated functional screening of the MD-1–derived cDNA library; M1 cells infected with the cDNA library were screened for clones resistant to IL-6–induced terminal differentiation in the medium containing IL-6 (100 ng/mL) and G418 selection reagents (600 μg/mL) as described.16,22 After screening of 1.5 × 105 independent cDNA clones, we happened to isolate an antisense cDNA encoding the complete coding sequence of mMgcRacGAP23—from an M1 clone that became resistant to IL-6.
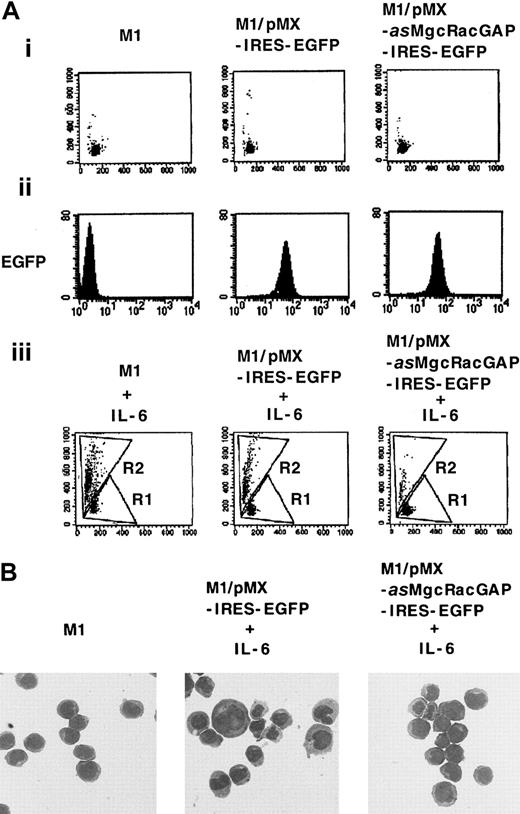
To determine if the expression of this antisense cDNA (asMgcRacGAP) indeed made M1 cells resistant to IL-6–induced macrophage differentiation, we subcloned it into a bicistronic retrovirus vector pMX-IRES-EGFP to construct pMX-asMgcRacGAP-IRES-EGFP, and we reintroduced it into M1 cells via retrovirus infection. As a control, pMX-IRES-EGFP was also introduced into M1 cells. After transduction of M1 cells with these vectors, GFP+ cells were sorted on a fluorescence-activated cell sorter (FACS) as described in “Materials and methods” and were cultured for 4 days in the presence or absence of 5-ng/mL IL-6. Flow cytometric analysis was performed to quantify morphologic changes after the culture. The increase in cell size and the granulosity of cytoplasm were evaluated based on increases in forward scatter and side scatter, respectively, in M1 cells expressing either the antisense mMgcRacGAP or the control vector after IL-6 treatment (Figure1A,C). Only 15.2% of M1 cells transduced with the antisense mMgcRacGAP showed a shift from region R1 to region R2, a hallmark of macrophage differentiation after treatment with IL-6, while 56.9% and 47.6% of the parental M1 cells and M1 cells transduced with the control vector showed similar shifts, respectively. Similarly, morphologically differentiated cells were evident in the control M1 cells but not in the antisense mMgcRacGAP-transduced M1 cells after treatment with IL-6 (Figure 1B), indicating that expression of antisense mMgcRacGAP profoundly inhibited the IL-6–induced differentiation of M1 cells. On the other hand, the growth rate of M1 cells was not affected by the expression of the antisense MgcRacGAP (data not shown).
An antisense cDNA for mMgcRacGAP inhibits IL-6–induced macrophage differentiation of M1 cells.
M1 cells were transduced with the antisense mMgcRacGAP (asMgcRacGAP) using a bicistronic retrovirus vector pMX-asMgcRacGAP-IRES-EGFP (M1/pMX-asMgcRacGAP-IRES-EGFP). As a control, M1 cells were also transduced with the blank vector pMX-IRES-EGFP (M1/pMX-IRES-EGFP). (A) Differentiation of parental M1, M1/pMX-IRES-EGFP, and M1/pMX-asMgcRacGAP-IRES-EGFP cells was evaluated by cell profiles in flow cytometry in the absence (i) or the presence (iii) of murine IL-6. The x-axis indicates forward scatter. The y-axis indicates side scatter. Undifferentiated and differentiated M1 cells were dotted in region R1 and region R2, respectively. GFP expression in parental M1, M1/pMX-IRES-EGFP, and M1/pMX-asMgcRacGAP-IRES-EGFP cells was also evaluated in flow cytometry to confirm the efficiency of expression of EGFP by bicistronic vector pMX-IRES-EGFP (ii). The x-axis indicates fluorescence intensity as a log scale ranging from 100 to 104. The y-axis indicates the number of cells. (B) Morphologic analysis of IL-6–induced differentiation in M1/pMX-IRES-EGFP and M1/pMX-asMgcRacGAP-IRES-EGFP cells. M1/pMX-IRES-EGFP and M1/pMX-asMgcRacGAP-IRES-EGFP cells were cultured in the presence of 5-ng/mL mIL-6 for 4 days, and cells were centrifuged onto glass slides and stained with May-Grunwald-Giemsa stain. Photographs were taken at 400 × magnification. The morphology of parental M1 cells is also shown as a control.
An antisense cDNA for mMgcRacGAP inhibits IL-6–induced macrophage differentiation of M1 cells.
M1 cells were transduced with the antisense mMgcRacGAP (asMgcRacGAP) using a bicistronic retrovirus vector pMX-asMgcRacGAP-IRES-EGFP (M1/pMX-asMgcRacGAP-IRES-EGFP). As a control, M1 cells were also transduced with the blank vector pMX-IRES-EGFP (M1/pMX-IRES-EGFP). (A) Differentiation of parental M1, M1/pMX-IRES-EGFP, and M1/pMX-asMgcRacGAP-IRES-EGFP cells was evaluated by cell profiles in flow cytometry in the absence (i) or the presence (iii) of murine IL-6. The x-axis indicates forward scatter. The y-axis indicates side scatter. Undifferentiated and differentiated M1 cells were dotted in region R1 and region R2, respectively. GFP expression in parental M1, M1/pMX-IRES-EGFP, and M1/pMX-asMgcRacGAP-IRES-EGFP cells was also evaluated in flow cytometry to confirm the efficiency of expression of EGFP by bicistronic vector pMX-IRES-EGFP (ii). The x-axis indicates fluorescence intensity as a log scale ranging from 100 to 104. The y-axis indicates the number of cells. (B) Morphologic analysis of IL-6–induced differentiation in M1/pMX-IRES-EGFP and M1/pMX-asMgcRacGAP-IRES-EGFP cells. M1/pMX-IRES-EGFP and M1/pMX-asMgcRacGAP-IRES-EGFP cells were cultured in the presence of 5-ng/mL mIL-6 for 4 days, and cells were centrifuged onto glass slides and stained with May-Grunwald-Giemsa stain. Photographs were taken at 400 × magnification. The morphology of parental M1 cells is also shown as a control.
Identification of the full-length form of hMgcRacGAP cDNA
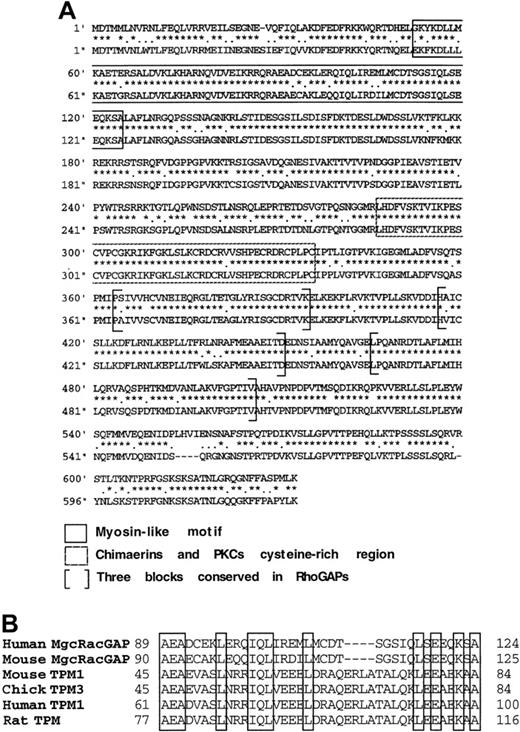
To isolate cDNA clones encoding the complete coding sequences of both murine and human MgcRacGAP, we screened libraries derived from the murine M1 cell line and the human TF-1 erythroleukemic cell line. The isolated cDNAs for 2919 base pairs of the mMgcRacGAP and 3050 base pairs of the hMgcRacGAP were then sequenced. We found that the open reading frame of the hMgcRacGAP cDNA encoded a putative protein of 632 amino acids, which is longer than the previously reported sequence of 527 amino acids by N-terminal 105 amino acids.24 The corresponding cDNA and the protein sequence of the previously reported hMgcRacGAP were not found in the FASTA/BLAST program of the National Center for Biotechnology Information or the DNA Databank of Japan/European Molecular Biology Laboratory/GenBank nucleotide sequence database. In addition, an analysis of other mRNA sources derived from Jurkat cells and phytohemagglutinin (PHA)-activated human T-cell blasts by reverse transcription PCR indicated that these cells expressed the full-length hMgcRacGAP cDNA, which was identical to the cDNA reported in the present paper (data not shown). Thus, it was shown that the hMgcRacGAP cDNA reported here is the full-length form of this RacGAP (accession No. AB030251). The surrounding sequence of the putative initiation codon ATG for the full-length form of hMgcRacGAP well matched to the Kozak consensus sequence,25 26 and a calculated molecular mass of this full-length form is 71 000 kd. Murine MgcRacGAP and the full-length form of hMgcRacGAP are 84% identical and 97% similar to each other at the amino acid level (Figure2A). Database searches revealed the similarity between the N-terminal region of the full-length hMgcRacGAP (amino acids 41-124) and myosins, mainly tropomyosins (Figure 2B). In this paper, we call the full-length form of this molecule hMgcRacGAP.
Comparison of the full-length form of hMgcRacGAP and mMgcRacGAP.
(A) The amino acid sequences of the full-length form of hMgcRacGAP (upper line) and mMgcRacGAP (lower line) were deduced from the open reading frames detected in cDNAs. Amino acid sequences that are related to the 3 consensus active site boxes of RhoGAPs are indicated by brackets. Boxed sequences in a dotted line indicate the cysteine-rich regions in which cysteine and histidine residues are conserved in similar regions of chimaerins and protein kinase C. Amino acid sequences that are related to a myosin-like motif are boxed in a straight line. (B) Alignment of the myosin-like domains of the full-length form of hMgcRacGAP, mMgcRacGAP, and tropomyosins (TPM). Homologous residues in myosin-like motif are boxed.
Comparison of the full-length form of hMgcRacGAP and mMgcRacGAP.
(A) The amino acid sequences of the full-length form of hMgcRacGAP (upper line) and mMgcRacGAP (lower line) were deduced from the open reading frames detected in cDNAs. Amino acid sequences that are related to the 3 consensus active site boxes of RhoGAPs are indicated by brackets. Boxed sequences in a dotted line indicate the cysteine-rich regions in which cysteine and histidine residues are conserved in similar regions of chimaerins and protein kinase C. Amino acid sequences that are related to a myosin-like motif are boxed in a straight line. (B) Alignment of the myosin-like domains of the full-length form of hMgcRacGAP, mMgcRacGAP, and tropomyosins (TPM). Homologous residues in myosin-like motif are boxed.
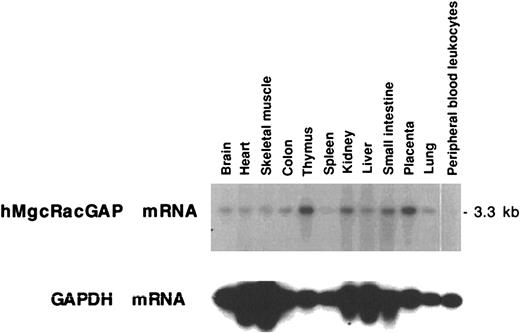
Expression of the hMgcRacGAP mRNA
Northern blot analysis revealed that a single hMgcRacGAP transcript of 3.3 kilobases (kb) was expressed in most tissues tested (Figure 3), with expression being high in thymus and placenta and low in spleen and peripheral blood leukocytes. It was noteworthy that hMgcRacGAP was highly expressed in tissues such as thymus and placenta containing immature hematopoietic cells but not in nonproliferating peripheral blood leukocytes. We also confirmed that mMgcRacGAP mRNA was expressed in murine fetal liver cells and in hematopoietic cell lines including Ba/F3 cells, CTLL-2 cells, and DA-1 cells (data not shown).
Tissue distribution of hMgcRacGAP mRNA.
Human multiple-tissue RNA blot (Clontech, Palo Alto, CA) containing poly(A)+ RNA from the indicated tissues was probed with the full-length hMgcRacGAP cDNA probe. A GAPDH cDNA was used as a control probe.
Tissue distribution of hMgcRacGAP mRNA.
Human multiple-tissue RNA blot (Clontech, Palo Alto, CA) containing poly(A)+ RNA from the indicated tissues was probed with the full-length hMgcRacGAP cDNA probe. A GAPDH cDNA was used as a control probe.
Enforced expression of hMgcRacGAP induced macrophage differentiation in the human myeloid leukemia HL-60 cells
We attempted to elucidate the role of MgcRacGAP in macrophage differentiation using murine myeloid leukemia M1 cells and human myeloid leukemia HL-60 cells. The mMgcRacGAP cDNA and the Flag-tagged hMgcRacGAP cDNA were transduced into M1 cells and HL-60 cells, respectively, using the retrovirus vector pMX-IRES-EGFP. GFP+ M1 and HL-60 cells transduced with these viruses were sorted on FACS 2 days after infection. GFP+ M1 and HL-60 cells transduced with a control pMX-IRES-EGFP vector were similarly sorted and served as negative controls.
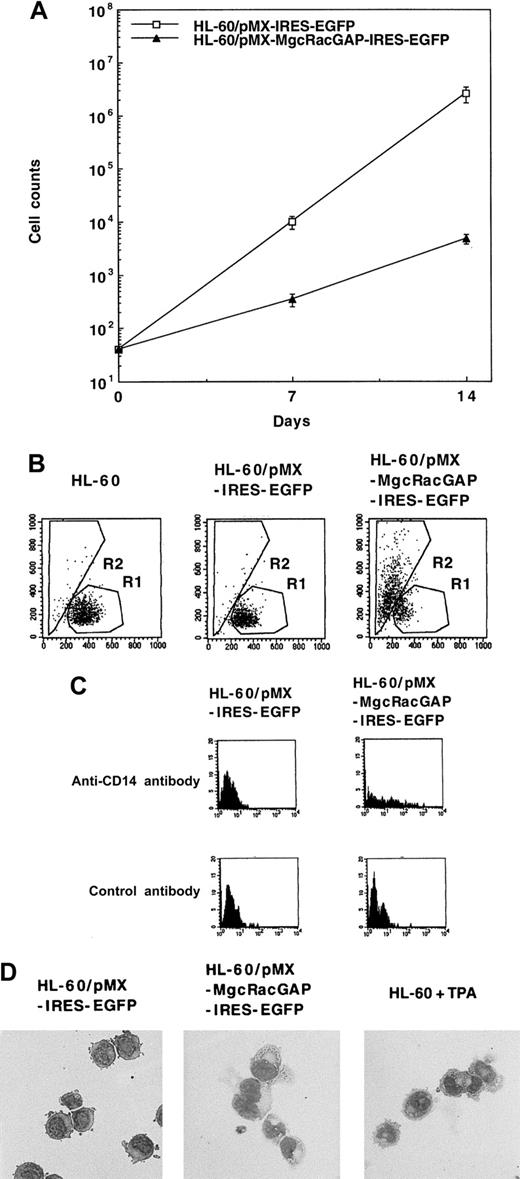
Overexpression of either mMgcRacGAP or hMgcRacGAP in M1 cells induced no significant differentiation but did moderately inhibit the growth of M1 cells (data not shown). Overexpression of mMgcRacGAP or hMgcRacGAP also retarded proliferation of a murine proB cell line Ba/F3. On the other hand, enforced expression of hMgcRacGAP in HL-60 cells profoundly suppressed growth (Figure 4A) and induced differentiation (Figure 4B-D) of the cells. Differentiation of HL-60 cells into macrophages was evaluated by FACS, expression of a monocyte marker CD14, and the morphology. First, the increase in cell size and granulosity of the cytoplasm were evaluated by increases in forward scatter and side scatter, respectively. As shown in Figure 4B, expression of hMgcRacGAP induced a shift from region R1 to region R2 in 65.8% of the transduced HL-60 cells, while expression of the control vector pMX-IRES-EGFP did not induce a significant shift. Second, expression of a myelomonocytic marker CD14 was induced in HL-60 cells expressing hMgcRacGAP but not in control HL-60 cells (Figure 4C). Morphologic changes were also noticed in HL-60 cells transduced with hMgcRacGAP; most HL-60 cells became larger with morphologic changes of monocytic differentiation and showed reduced nuclear/cytoplasmic ratios and notable hypervacuolation in the cytoplasm after transduction of hMgcRacGAP (Figure 4D). These results clearly demonstrated that overexpression of hMgcRacGAP alone induced differentiation of HL-60 cells into macrophages.
Enforced expression of hMgcRacGAP inhibits proliferation and induces macrophage differentiation of human myeloid leukemia HL-60 cells.
(A) Proliferation of HL-60 cells overexpressing hMgcRacGAP. The Flag-tagged hMgcRacGAP cDNAs were transduced into HL-60 cells using retrovirus vector pMX-IRES-EGFP. GFP+ HL-60 cells infected with these viruses were sorted on FACS 2 days after virus infection (HL-60/pMX-MgcRacGAP-IRES-EGFP cells). GFP+ HL-60 cells transduced with a blank pMX-IRES-EGFP vector were similarly sorted and used as a negative control (HL-60/pMX-IRES-EGFP cells). Both transfectants were counted at the indicated time points after the sorting. The results shown are the averages ± SD of triplicate cultures. (B) Enforced expression of the full-length form of hMgcRacGAP induces macrophage differentiation of HL-60 cells. Differentiation of parental HL-60, HL-60/pMX-IRES-EGFP, and HL-60/pMX-MgcRacGAP-IRES-EGFP cells was evaluated using flow cytometric analysis. The x-axis indicates forward scatter. The y-axis indicates side scatter. (C) CD14 expression of HL-60/pMX-IRES-EGFP cells and HL-60/pMX-MgcRacGAP-IRES-EGFP cells. The cells were stained with PE-conjugated mouse antihuman CD14 antibody or PE-conjugated isotype-matched control mouse IgG2a. (D) Morphologic changes in HL-60 cells overexpressing hMgcRacGAP. May-Grunwald-Giemsa staining profiles of HL-60/pMX-IRES-EGFP and HL-60/pMX-MgcRacGAP-IRES-EGFP cells are shown. HL-60 cells treated with TPA (16 nmol/L) for 2 days are also shown as a positive control. Cells were centrifuged onto glass slides and stained with May-Grunwald-Giemsa stain. Photographs were taken at 400 × magnification.
Enforced expression of hMgcRacGAP inhibits proliferation and induces macrophage differentiation of human myeloid leukemia HL-60 cells.
(A) Proliferation of HL-60 cells overexpressing hMgcRacGAP. The Flag-tagged hMgcRacGAP cDNAs were transduced into HL-60 cells using retrovirus vector pMX-IRES-EGFP. GFP+ HL-60 cells infected with these viruses were sorted on FACS 2 days after virus infection (HL-60/pMX-MgcRacGAP-IRES-EGFP cells). GFP+ HL-60 cells transduced with a blank pMX-IRES-EGFP vector were similarly sorted and used as a negative control (HL-60/pMX-IRES-EGFP cells). Both transfectants were counted at the indicated time points after the sorting. The results shown are the averages ± SD of triplicate cultures. (B) Enforced expression of the full-length form of hMgcRacGAP induces macrophage differentiation of HL-60 cells. Differentiation of parental HL-60, HL-60/pMX-IRES-EGFP, and HL-60/pMX-MgcRacGAP-IRES-EGFP cells was evaluated using flow cytometric analysis. The x-axis indicates forward scatter. The y-axis indicates side scatter. (C) CD14 expression of HL-60/pMX-IRES-EGFP cells and HL-60/pMX-MgcRacGAP-IRES-EGFP cells. The cells were stained with PE-conjugated mouse antihuman CD14 antibody or PE-conjugated isotype-matched control mouse IgG2a. (D) Morphologic changes in HL-60 cells overexpressing hMgcRacGAP. May-Grunwald-Giemsa staining profiles of HL-60/pMX-IRES-EGFP and HL-60/pMX-MgcRacGAP-IRES-EGFP cells are shown. HL-60 cells treated with TPA (16 nmol/L) for 2 days are also shown as a positive control. Cells were centrifuged onto glass slides and stained with May-Grunwald-Giemsa stain. Photographs were taken at 400 × magnification.
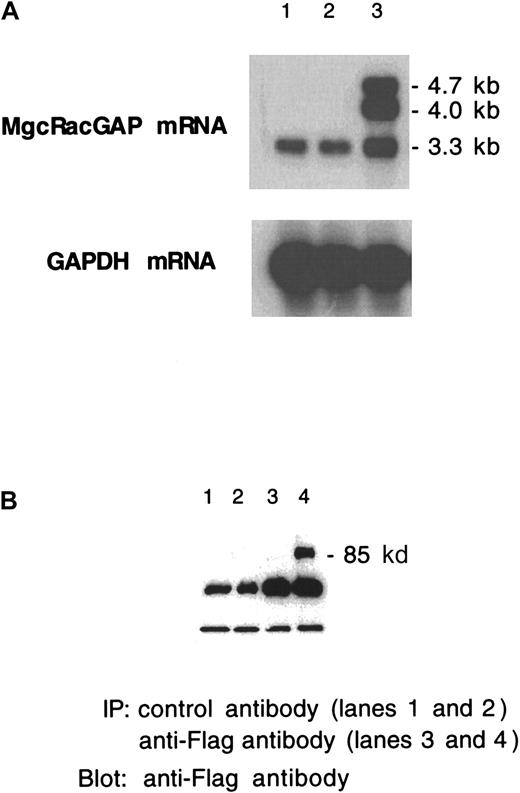
Stable expression of hMgcRacGAP transgene in HL-60 and M1 transfectants was confirmed by the Northern blot and the Western blot analysis (Figure 5A,B). In Northern blot analysis of HL-60 cells carrying the hMgcRacGAP transgene (Figure 5A, lane 3), we detected retroviral transcripts migrating at approximately 4.7 kb and 4.0 kb, which were presumably genomic and subgenomic forms of the transcripts, respectively, derived from the retrovirus vector pMX harboring splicing donor and acceptor sites as well as the endogenous hMgcRacGAP mRNA at approximately 3.3 kb. Western blot analysis demonstrated that M1 cells transduced with the Flag-tagged hMgcRacGAP transgene expressed the recombinant protein of approximately 80 kd (Figure 5B).
Stable and ectopic expression of the MgcRacGAP transgene in HL-60 and M1 transfectants.
(A) Northern blot analysis of mRNA from parental HL-60 cells, HL-60/pMX-IRES-EGFP cells, and HL-60/pMX-MgcRacGAP-IRES-EGFP cells (lanes 1, 2, and 3, respectively). (B) Western blot analysis of cell lysates from control M1 cells (lanes 1,3) and M1 cells with the introduced MgcRacGAP transgene (lanes 2,4) (3 × 107per lane).
Stable and ectopic expression of the MgcRacGAP transgene in HL-60 and M1 transfectants.
(A) Northern blot analysis of mRNA from parental HL-60 cells, HL-60/pMX-IRES-EGFP cells, and HL-60/pMX-MgcRacGAP-IRES-EGFP cells (lanes 1, 2, and 3, respectively). (B) Western blot analysis of cell lysates from control M1 cells (lanes 1,3) and M1 cells with the introduced MgcRacGAP transgene (lanes 2,4) (3 × 107per lane).
Expression of endogenous MgcRacGAP was down-regulated during IL-6– and TPA-induced macrophage differentiation of M1 and HL-60 cells, respectively
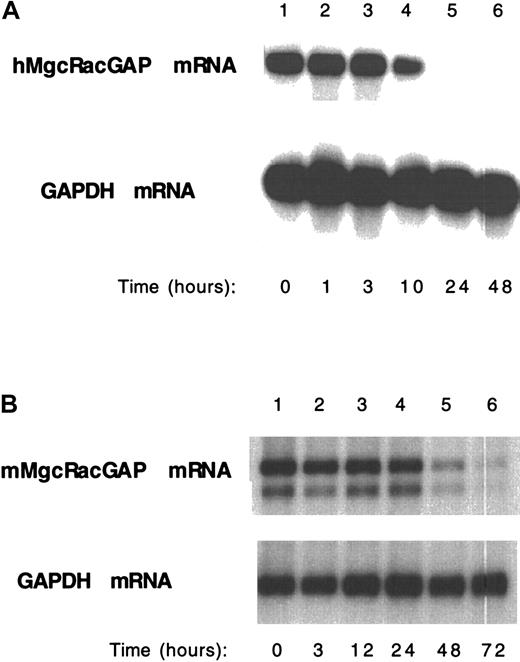
To determine whether the expression level of endogenous MgcRacGAP was altered during macrophage differentiation, M1 and HL-60 cells were stimulated with IL-6 (50 ng/ml) and 12-O-tetradecanoylphorbol-13-acetate (TPA) (16 nmol/L), respectively, and at various time points we isolated poly(A)+ RNA. Intriguingly, Northern blot analysis of these samples showed that the expression of endogenous MgcRacGAP mRNA dramatically decreased along with macrophage differentiation of both M1 and HL-60 cells (Figure6A,B).
Down-regulation of the endogenous MgcRacGAP expression along with differentiation of HL-60 cells and M1 cells into macrophages.
Poly(A)+ RNA was harvested from HL-60 (A) or M1 (B) cells before and after TPA (A) or IL-6 (B) stimulation, respectively, at the indicated time points. Poly(A)+ RNA (2 μg) from each condition was subjected to the Northern blot analysis, hybridizing with a human (A) or a mouse (B) MgcRacGAP cDNA probe. A GAPDH cDNA was used as a control probe.
Down-regulation of the endogenous MgcRacGAP expression along with differentiation of HL-60 cells and M1 cells into macrophages.
Poly(A)+ RNA was harvested from HL-60 (A) or M1 (B) cells before and after TPA (A) or IL-6 (B) stimulation, respectively, at the indicated time points. Poly(A)+ RNA (2 μg) from each condition was subjected to the Northern blot analysis, hybridizing with a human (A) or a mouse (B) MgcRacGAP cDNA probe. A GAPDH cDNA was used as a control probe.
The mutant MgcRacGAP in which the conserved arginine was replaced with an alanine exhibited no GTPase activity
A conserved arginine residue found in the first homology box of GAP domains of Rho-GAP family was also conserved in MgcRacGAP (Figure7A), and structural studies suggested that this arginine residue was a key catalytic residue.27,28 Indeed, it was reported that mutation (or deletion) of this conserved arginine residue disrupted GAP activities of n-chimaerin and Cdc42-GAP without affecting their binding activities to GTP-bound Rho GTPase.29 30 Therefore, to generate a GAP-inactive mutant of hMgcRacGAP, we replaced the conserved arginine (Arg385) of hMgcRacGAP with an alanine (R385A*MgcRacGAP). We expressed the mutant GAP domain of R385A*MgcRacGAP as GST fusion products in vitro and confirmed that the GAP activity of hMgcRacGAP toward Rac1 and Cdc42 was inactivated as a result of this mutation (Figure 7B).
A conserved arginine residue in the first homology box of GAP domains in all members of the Rho-GAP family and also GTPase-stimulating activity of the wild-type hMgcRacGAP and the mutant R385A*MgcRacGAP.
(A) Homology between members of RhoGAP family is shown. Sequence alignments were performed on the original GenBank entries for the indicated proteins using the Clustal algorithm of the DNA Star program. ABR indicates Bcr-related gene product. (B) The GTP-hydrolyzing activity of several recombinant GTPases was measured in the presence or absence of recombinant GAP domain of the wild-type hMgcRacGAP or the mutant R385A*MgcRacGAP. The amount of [γ-32P] GTP remaining bound to GTPases after 3 minutes was determined by a filter-binding assay and expressed as a percentage of the initial amount of GTP bound to each protein. The results shown are the averages ± SD of 3 independent experiments.
A conserved arginine residue in the first homology box of GAP domains in all members of the Rho-GAP family and also GTPase-stimulating activity of the wild-type hMgcRacGAP and the mutant R385A*MgcRacGAP.
(A) Homology between members of RhoGAP family is shown. Sequence alignments were performed on the original GenBank entries for the indicated proteins using the Clustal algorithm of the DNA Star program. ABR indicates Bcr-related gene product. (B) The GTP-hydrolyzing activity of several recombinant GTPases was measured in the presence or absence of recombinant GAP domain of the wild-type hMgcRacGAP or the mutant R385A*MgcRacGAP. The amount of [γ-32P] GTP remaining bound to GTPases after 3 minutes was determined by a filter-binding assay and expressed as a percentage of the initial amount of GTP bound to each protein. The results shown are the averages ± SD of 3 independent experiments.
GAP activity was dispensable, but the myosin-like domain and the cysteine-rich domain were indispensable for hMgcRacGAP-induced growth suppression and differentiation
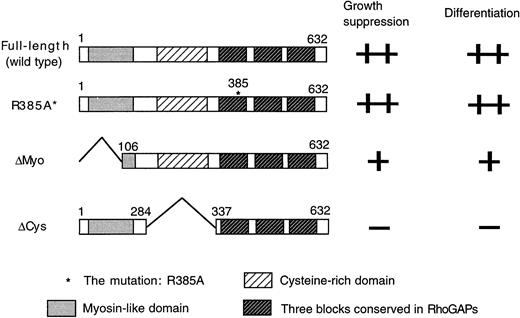
To determine if the GAP activity of hMgcRacGAP is crucial and which domain of this RacGAP is important for the growth suppression and macrophage differentiation in HL-60 cells, we used the GAP activity-negative mutant (R385A*MgcRacGAP) and 2 deletion mutants, lacking the N-terminal myosin-like domain or the cysteine-rich domain (ΔMyo-MgcRacGAP or ΔCys-MgcRacGAP, respectively) (Figure8).
Structures of the mutants of hMgcRacGAP.
Structures of the wild-type MgcRacGAP, R385A*MgcRacGAP (R385A*), and 2 deletion mutants lacking the myosin-like domain (ΔMyo) and the cysteine-rich domain (ΔCys) are shown together with the ability to induce growth suppression and differentiation of HL-60 cells.
Structures of the mutants of hMgcRacGAP.
Structures of the wild-type MgcRacGAP, R385A*MgcRacGAP (R385A*), and 2 deletion mutants lacking the myosin-like domain (ΔMyo) and the cysteine-rich domain (ΔCys) are shown together with the ability to induce growth suppression and differentiation of HL-60 cells.
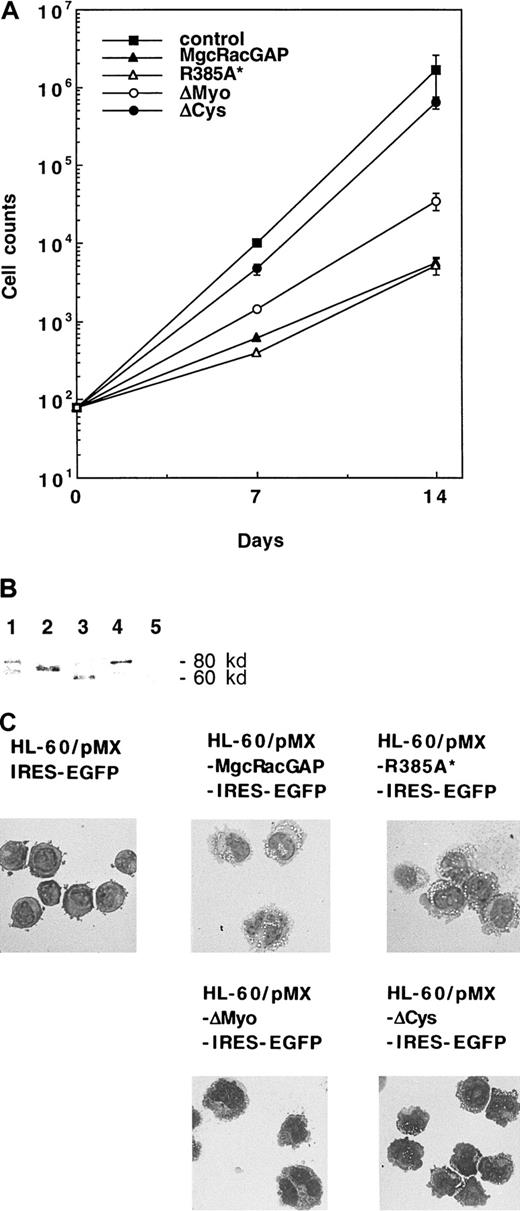
Each mutant was introduced into HL-60 cells via retrovirus infection using the retrovirus vector pMX-IRES-EGFP. GFP+cells with comparable fluorescence intensities were then sorted. As shown in Figure 9A, growth of HL-60 cells was suppressed after transduction of the GAP activity-negative mutant of hMgcRacGAP to an extent similar to that seen with HL-60 cells transduced with the wild-type hMgcRacGAP, indicating that the GAP activity of hMgcRacGAP is dispensable for the hMgcRacGAP-induced growth suppression of HL-60 cells. On the other hand, ΔMyo-MgcRacGAP showed decreased activities of growth suppression of HL-60 cells, and ΔCys-MgcRacGAP did not significantly suppress proliferation of HL-60 cells. The mutant as well as the wild-type hMgcRacGAP proteins were expressed at similar levels (Figure 9B). These results suggested that the protein kinase C–like cysteine-rich domain played a crucial role in inhibiting the proliferation of HL-60 cells.
Functional analysis of the mutants of hMgcRacGAP.
(A) Suppression of growth of HL-60 cells overexpressing the wild-type or mutated hMgcRacGAPs. Each of the wild-type, the mutant R385A*, and 2 deletion mutants was transduced into HL-60 cells using the retrovirus vector pMX-IRES-EGFP. GFP+ HL-60 cells transduced with these viruses were sorted on FACS 2 days after virus infection. GFP+ HL-60 cells, transduced with a blank pMX-IRES-EGFP vector, were sorted as a negative control. The cell number of each transfectant was plotted against time. The results shown are the averages ± SD of triplicate cultures. (B) Expression of the Flag-tagged wild-type and mutant hMgcRacGAPs in HL-60 transfectants. Cell lysates from control HL-60 cells (lane 5) and HL-60 transfectants expressing the wild-type (lane 1), ΔCys (lane 2), ΔMyo (lane 2), or R385A* (lane 4) MgcRacGAP (3 × 107 per lane) were examined by Western blotting using the anti-Flag M2 monoclonal antibody. (C) Differentiation of HL-60 cells overexpressing the wild-type or mutated hMgcRacGAPs. Morphologic differentiation of control HL-60 cells and HL-60 cells transduced with the wild-type, R385A*, or deletion mutants of hMgcRacGAP is shown. Cells were centrifuged onto glass slides and stained with May-Grunwald-Giemsa stain. Photographs were taken at 400 × magnification.
Functional analysis of the mutants of hMgcRacGAP.
(A) Suppression of growth of HL-60 cells overexpressing the wild-type or mutated hMgcRacGAPs. Each of the wild-type, the mutant R385A*, and 2 deletion mutants was transduced into HL-60 cells using the retrovirus vector pMX-IRES-EGFP. GFP+ HL-60 cells transduced with these viruses were sorted on FACS 2 days after virus infection. GFP+ HL-60 cells, transduced with a blank pMX-IRES-EGFP vector, were sorted as a negative control. The cell number of each transfectant was plotted against time. The results shown are the averages ± SD of triplicate cultures. (B) Expression of the Flag-tagged wild-type and mutant hMgcRacGAPs in HL-60 transfectants. Cell lysates from control HL-60 cells (lane 5) and HL-60 transfectants expressing the wild-type (lane 1), ΔCys (lane 2), ΔMyo (lane 2), or R385A* (lane 4) MgcRacGAP (3 × 107 per lane) were examined by Western blotting using the anti-Flag M2 monoclonal antibody. (C) Differentiation of HL-60 cells overexpressing the wild-type or mutated hMgcRacGAPs. Morphologic differentiation of control HL-60 cells and HL-60 cells transduced with the wild-type, R385A*, or deletion mutants of hMgcRacGAP is shown. Cells were centrifuged onto glass slides and stained with May-Grunwald-Giemsa stain. Photographs were taken at 400 × magnification.
To determine effects of these mutations in inducing macrophage differentiation, we performed May-Grunwald-Giemsa staining of cytospin preparations of the sorted cells (Figure 9C). Large cells, which differentiated along the monocytic differentiation pathway with distinctive hypervacuolation in the cytoplasm, were observed in HL-60 cells transduced with R385A*MgcRacGAP as well as those transduced with the wild-type hMgcRacGAP, thereby demonstrating that the GAP activity of hMgcRacGAP was not required for its activity to induce macrophage differentiation of HL-60 cells. However, well-differentiated cells were not observed in the population of HL-60 cells expressing ΔCys-MgcRacGAP. When transduced with the ΔMyo-MgcRacGAP, HL-60 cells gave rise to large cells that were frequently multinuclei but did not exhibit hypervacuolation. These results suggested that the myosin-like domain of hMgcRacGAP played some roles in cell division as well as induction of macrophage differentiation, including vacuolation.
Discussion
In this paper, we functionally cloned an antisense cDNA for mMgcRacGAP, which profoundly inhibited IL-6–induced differentiation of M1 cells. Although overexpression of sense cDNA for MgcRacGAP induced no detectable differentiation of M1 cells, it induced macrophage differentiation of human acute leukemic HL-60 cells. Moreover, overexpression of MgcRacGAP led to growth suppression of both M1 and HL-60 cells. These results indicated that MgcRacGAP was involved in control of growth and differentiation in both M1 and HL-60 cells. We also found that the GAP activity of hMgcRacGAP was dispensable for hMgcRacGAP-induced growth suppression and differentiation into macrophages.
Rac/Cdc42 small G proteins were implicated in cytoskeletal organization; membrane ruffling; production of superoxide, phagocytosis, and chemotaxis; as well as regulation of cell cycle.2 31-34 Rac/Cdc42 GAPs include Bcr, n-, β2-, α-chimaerin, IQGAP, Cdc42-GAP, Ral-BP1/RLIP1, and myosin-IXb, and they were also implicated in controlling a variety of cellular functions. However, to our knowledge, the present study is the first report demonstrating that enforced expression of a GAP protein alone is able to alter cell fate.
Interestingly, this potential of hMgcRacGAP to induce differentiation does not require its GAP activity but does require the N-terminal myosin-like domain and the cysteine-rich domain, as demonstrated by the experiments using a series of mutants. This result is reminiscent of that reported for n-chimaerin, which cooperates with Rac1 and Cdc42 in stimulating the formation of lamellipodia and filopodia, respectively; these functions ofn-chimaerin require the G protein binding capacity and the non-GAP N-terminal extension but not GAP activity.8Together, it is suggested that MgcRacGAP not only negatively regulates Rac-mediated signals through their catalytic functions, which stimulate GTP hydrolysis after binding to activated (or GTP-bound) forms of Rac GTPases, but also functions as downstream effectors of Rac proteins as a Rac-binding protein. The myosin-like domain of myosin-IXb and the cysteine-rich domain of n-chimaerin are required for interaction with actin filaments and phospholipids, respectively.11 35 Taken together, it seems likely that MgcRacGAP suppresses growth and differentiation through its multiple domains, which would interact with multiple signaling pathways but not through its GAP activity.
An N-terminus–truncated molecule of the hMgcRacGAP was previously isolated as a Rac-binding protein in a 2-hybrid experiment.24 Human MgcRacGAP was reported to be highly expressed in male germcell and was implicated in spermatogenesis. However, the reported sequence corresponded to the deletion mutant ΔMyo-MgcRacGAP in the present paper and lacked the N-terminal 105 amino acid sequence when compared with the full-length form of the hMgcRacGAP described here. We further confirmed that the previously reported hMgcRacGAP was a truncated version by detecting the full-length cDNAs in Jurkat cells and PHA-activated human T-cell blasts (unpublished results), which was identical to the hMgcRacGAP cDNA that we isolated from erythloid TF-1 cells and used in this study.
Another group has recently cloned a cDNA for mMgcRacGAP under the name of band25.36 This was cloned using the differential display techniques as a cDNA whose expression well correlated with cell growth. In addition, it was shown that expression of the band25 decreased along with terminal differentiation into myocytes of a murine myogenic cell line C2C12. Thus, the expression level of MgcRacGAP was apparently parallel to the rate of cell proliferation.
In a similar context, we found that the expression level of hMgcRacGAP was high in thymus and placenta, which contained a number of proliferating cells, but was extremely low in peripheral blood leukocytes, most of which were terminally differentiated and lost proliferative activities. In addition, expression of endogenous MgcRacGAP dramatically decreased in HL-60 and M1 cells when induced to terminally differentiate into macrophages by TPA and IL-6, respectively (Figure 6), again is correlated with cell proliferation.
Then, the important question is why overexpression of MgcRacGAP suppressed cell growth and induced differentiation of HL-60 cells. Molecular mechanisms for this phenomenon remain to be clarified, but there are 2 possibilities. The first possibility is that MgcRacGAP is a protein primarily involved in cell proliferation and that its overexpression negatively regulates or disrupts the normal control of cell growth, thereby inducing differentiation of the cells as a secondary event. In fact, our results as well as those of others indicated that MgcRacGAP was rather associated with proliferation. This hypothesis also explains why expression of endogenous MgcRacGAP became undetectable along with terminal differentiation of M1 or HL-60 cells. What, then, is the role of MgcRacGAP in cell proliferation? Of particular interest, expression of a deletion mutant ΔMyo-MgcRacGAP suppressed cell growth and induced multinucleated cells in HL-60, suggesting that MgcRacGAP played critical roles in cell division through its myosin-like domain. However, this hypothesis could not explain why expression of the antisense MgcRacGAP profoundly inhibited IL-6–induced macrophage differentiation of M1 cells while leaving cell growth unaffected, strongly indicating that MgcRacGAP was directly involved in cell differentiation in M1 cells. Therefore, we prefer an alternative possibility that MgcRacGAP plays critical roles in both cell growth and differentiation. This possibility is consistent with most results so far presented by us and others concerning MgcRacGAP. However, how MgcRacGAP is involved in both cell growth and differentiation is currently unknown. To clarify the detailed mechanism of MgcRacGAP function, we are now raising antibodies against it.
Finally, the fact that the expression of MgcRacGAP is associated with proliferation and inversely correlates with differentiation in 3 different cell types, including hematopoietic cells, preadipocytes, and myogenic cells, suggests a rather common mechanism by which MgcRacGAP controls cellular proliferation and differentiation.
Acknowledgments
We thank Dr Tatsutoshi Nakahata and Dr Tsuneo A. Takahashi for the FACS machines and Mariko Ohara and Dr Masato Nakafuku for critical reading of the manuscript.
Supported by the Chugai Pharmaceutical Company Ltd and grants from the Ministry of Education, Science, Sports, and Culture of Japan and from the Ministry of Health and Welfare of Japan.
The nucleotide sequence data for the human MgcRacGAP cDNA reported here will appear in the DNA Databank of Japan/European Molecular Biology Laboratory/GenBank nucleotide sequence database with accession No.AB030251.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Toshio Kitamura, Department of Hematopoietic Factors, The Institute of Medical Science, University of Tokyo, 4-6-1 Shirokanedai, Minato-ku, Tokyo 108-8639, Japan; e-mail:kitamura@ims.u-tokyo.ac.jp.

![Fig. 7. A conserved arginine residue in the first homology box of GAP domains in all members of the Rho-GAP family and also GTPase-stimulating activity of the wild-type hMgcRacGAP and the mutant R385A*MgcRacGAP. / (A) Homology between members of RhoGAP family is shown. Sequence alignments were performed on the original GenBank entries for the indicated proteins using the Clustal algorithm of the DNA Star program. ABR indicates Bcr-related gene product. (B) The GTP-hydrolyzing activity of several recombinant GTPases was measured in the presence or absence of recombinant GAP domain of the wild-type hMgcRacGAP or the mutant R385A*MgcRacGAP. The amount of [γ-32P] GTP remaining bound to GTPases after 3 minutes was determined by a filter-binding assay and expressed as a percentage of the initial amount of GTP bound to each protein. The results shown are the averages ± SD of 3 independent experiments.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/96/6/10.1182_blood.v96.6.2116/5/m_h81800148007.jpeg?Expires=1767818777&Signature=1kMgDS6lBxaPEexZkO0Y6oKY-F~Q81D4trJ7UsJAw~GCn8cMqxwc--aV7uBMEONHgQ8Bc5nezb-Rm82TOKF-NwCP76PRArqtZPCSo1CdAZ26NPJh~I3czQLG02-AdqCVWrrvObHwPCobs--ulutqPCnXDWiwRRK4~Cy7jpFNsZdvzpxjEOehpq0pPYHCCJGbX4n9LNfawlkYNsiRXPi-uj1bz48j5p3m31JyYdm80cMfs8RY9-mEdPrYV~gN2bY3HYwrdW~FMbhkttXoI48S-rpcGI4Znrfdfo-3LftphJGvq~yLOjkpytjRMiyv4sxR1U5OheYHiGd4WXlXm6Xk2A__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal