Abstract
As reported previously, AML1-ETO knock-in mice were generated to investigate the role of AML1-ETO in leukemogenesis and to mimic the progression of t(8;21) leukemia. These knock-in mice died in midgestation because of hemorrhaging in the central nervous system and a block of definitive hematopoiesis during embryogenesis. Therefore, they are not a good model system for the development of acute myeloid leukemia. Therefore, mice were generated in which the expression of AML1-ETO is under the control of a tetracycline-inducible system. Multiple lines of transgenic mice have been produced with the AML1-ETO complementary DNA controlled by a tetracycline-responsive element. In the absence of the antibiotic tetracycline, AML1-ETO is strongly expressed in the bone marrow of AML1-ETO and tet-controlled transcriptional activator double-positive transgenic mice. Furthermore, the addition of tetracycline reduces AML1-ETO expression in double-positive mice to nondetectable levels. Throughout the normal murine lifespan of 24 months, mice expressing AML1-ETO have not developed leukemia. In spite of this, abnormal maturation and proliferation of progenitor cells have been observed from these animals. These results demonstrate that AML1-ETO has a very restricted capacity to transform cells. Either the introduction of additional genetic changes or the expression of AML1-ETO at a particular stage of hematopoietic cell differentiation will be necessary to develop a model for studying the pathogenesis of t(8;21).
Introduction
Acute myeloid leukemia (AML) is a major hematopoietic malignancy characterized by the proliferation of a malignant clone of myeloid progenitor cells. One of the most common targets of translocations that have been implicated in AML is theAML1 gene.1-3 The AML1 gene, also known as RUNX1, encodes a novel transcription factor that forms a complex with core binding factor β (CBFβ).4 It contains a region that is highly homologous with the Drosophila runt gene, which is required for DNA binding and heterodimerization with CBFβ.5 AML1 is involved in a number of translocations that are found in many patients with AML and childhood B-cell leukemia, including t(8;21), t(3;21), and t(12;21). In addition, CBFβ is fused to a smooth muscle myosin heavy chain in INV(16) and is associated with M4eo AML.6 The AML1 gene was isolated through a study of t(8;21) in which the runt homology domain of AML1was found to be fused to a gene termed ETO (MTG8) to form a fusion protein called AML1-ETO.7 AML1 has since been found to be crucial for normal hematopoiesis.8,9 The t(8;21) translocation is found in 40% of patients with the M2 subtype of AML.7,10 11 The frequent appearance of translocations involving AML1 and its critical function in normal hematopoiesis identifies it as an important target of chromosomal abnormalities found in human leukemias.
To study the effect of AML1-ETO on hematopoiesis, we previously generated mice in which wild-type AML1 was replaced byAML1-ETO by using a knock-in strategy.12 These mice recapitulate human t(8;21) by substituting the fusion gene for the wild-type allele. In these studies, a block in definitive hematopoiesis was seen in the heterozygous AML1+/AML1-ETO+embryos. These embryos died in midgestation because of severe hemorrhaging in the central nervous system. The resulting phenotype in these mice was similar to the phenotypes seen in AML1 andCBFβ knock-out embryos,8,9,13-15 which suggests that AML1-ETO causes the interruption of the normal function of AML1. However, yolk sac cells from the AML1+/AML1-ETO+ knock-in embryos were able to differentiate into macrophage-like cells in colony-forming unit (CFU) assays unlike the yolk sac cells from the AML1 andCBFβ knock-out embryos that form no colonies in vitro.12 In knock-in embryos generated by using a slightly different strategy, fetal liver cells formed dysplastic hematopoietic progenitors in CFU assays.16 These results suggest that AML1-ETO may have other functions, besides blocking wild-type AML1, that are critical in leukemogenesis. Because the AML1-ETO knock-in embryos exhibit an embryonic lethal phenotype, we decided to develop a murine model in which the expression of AML1-ETO is inducible. In this way, the effect of AML1-ETO on different stages of definitive hematopoiesis and leukemogenesis could be studied.
We report here the development of unique lines of transgenic mice that express AML1-ETO under the control of a tetracycline-responsive element—the tet-off system. The tet-off system has been successfully utilized to control gene expression in transgenic mice.17-20 The AML1-ETO tet-off mice show an inducible expression of AML1-ETO by both Northern blot and Western blot analyses. AML1-ETO is highly expressed in the bone marrow of mice from three different founder lines and in the peritoneal macrophages of another founder line. However, these mice have shown no disease phenotype during 24 months of observation. Although there is no onset of leukemia in these mice, we have observed a partial block of myeloid differentiation and an increased replating efficiency of progenitor cells in serial replating colony assays. These results suggest that, although AML1-ETO can block normal AML1 function during embryogenesis and regulate the expression of other hematopoietic genes, it alone may not be sufficient to cause leukemia. It is also possible that the timing and the particular target cells of AML1-ETO expression are critical for the development of a disease phenotype.
Materials and methods
Plasmid construction and generation of transgenic mice
The complementary DNA (cDNA) for AML1-ETO was obtained from S. Hiebert21 (Vanderbilt University, Nashville, TN) and was subcloned into the XbaI site of pUHD10-3, a plasmid containing a tetracycline-responsive promoter17 to form pUHD-AML1/ETO. A stop codon was added at the 3′ end of the cDNA by using the Chameleon Double Stranded, Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA).
pUHD-AML1/ETO was digested with DraI and EcoRV. A 3-kilobase (kb) band containing the AML1-ETO cDNA and the tet-responsive promoter was isolated and used for microinjection. AML1-ETO transgenic mice were produced in the transgenic facility of Beth Israel Deaconess Medical Center (Boston, MA) by using inbred FVB zygotes.
Transgenic mice containing the murine mammary tumor virus–tet-controlled transcriptional activator (MMTV-tTA) construct were obtained from Dr L. Hennighausen (National Institutes of Health, Bethesda, MD).22 These mice were bred with our pUHD-AML1/ETO mice to generate double-positive mice with inducible AML1-ETO expression.
Administration of tetracycline to transgenic mice
Slow-release tetracycline pellets (Innovative Research of America, Sarasota, FL) were implanted subcutaneously in the shoulder of female mice used for breeding, following the manufacturer's instructions. These pellets release 0.7 mg tetracycline hydrochloride per day. These females were then crossbred to produce double-positive MMTV-tTA/AML1-ETO offspring. After the pups were weaned, tetracycline was administered through their drinking water. Their water was supplemented with 1 mg/mL tetracycline and was changed every 2 days to assure the activity of the tetracycline.
Southern blot analysis
Tail DNA samples prepared as previously described23were digested with EcoRI, electrophoresed in a 0.8% agarose gel, and transferred to positively charged nylon membrane (Biotrans plus, ICN, Costa Mesa, CA). ETO cDNA (1.7-kbKpnI/XbaI DNA fragment from pKS-ETO) and the tTA-VP16 DNA (1-kb BamHI/EcoRI DNA fragment from pUHD15-1) were radiolabeled with 32P-dCTP and used as probes to hybridize with the genomic tail DNA samples to test for the presence of the 2 transgene constructs.
Isolation of RNA and Northern blot analysis
Peritoneal macrophages were harvested from the abdominal cavities of mice 48 to 72 hours after the intraperitoneal injection of thioglycollate (0.1 g/mL, 1.5 mL/mouse). Total RNA was extracted from these macrophages, various tissues from the injected mice, and cell lines by guanidine isothiocyanate extraction were followed by purification on a cesium chloride gradient. Total RNA (10 μg) for each sample was electrophoresed in a 1% agarose gel containing 0.22 mol/L formaldehyde. The RNA was transferred to positively charged nylon membrane (Biotrans plus), using capillary transfer with 20 × sodium chloride–sodium citrate (SSC). The blots were then UV cross-linked using a Stratalinker (Stratagene). The blots were subsequently hybridized with 32P-dCTP radiolabeled probes: ETO cDNA or tTA-VP16 DNA. RNA from bulk colonies was isolated by RNAzol extraction (Tel-Test Inc, Friendswood, TX). Total RNA (5 μg) for each sample was electrophoresed and blotted as described above.
Isolation of protein and Western blot analysis
One mouse from each of the 5 founder lines as well as a mouse that was positive for tTA but not AML1-ETO (negative control) was killed, and the bone marrow was flushed from the femurs with phosphate-buffered saline (PBS). The cells were centrifuged for 5 minutes at 1000 rpm in a GPKR centrifuge (Beckman, Fullerton, CA) and resuspended in a resuspension buffer (100 mmol/L NaCl, 10 mmol/L Tris pH 7.6, 1 mmol/L EDTA, 1 μg/mL aprotinin, 100 μg/mL PMSF) to which an equal volume of 2 × SDS loading buffer (4% SDS, 20% glycerol, 1.43 mol/L 2-mercaptoethanol, 125 mmol/L Tris) was then added. The samples were boiled for 10 minutes, followed by microcentrifugation for 10 minutes. Aliquots of the supernatant equivalent to 2 × 106 cells were stored at −80°C. The samples were boiled and spun as above immediately prior to loading in an 8% resolving SDS-PAGE gel (bis:acrylamide = 1:19). Nuclear extracts from wild-type Ba/F3 cells and Ba/F3 cells transfected with an AML1-ETO expression construct were also loaded as controls (35 μg each). The protein was transferred to PVDF membrane (Immobilon P, Millipore, Bedford, MA) by using a Mini Trans-Blot cell (BioRad, Hercules, CA). The blot was then blocked in 5% dry milk in TBS + 0.2% NP-40. The blot was incubated with a primary monoclonal antibody against the ETO portion of the human fusion protein (a gift from Dr P. Erickson, University of Colorado).24 The blot was then incubated with a secondary antibody conjugated to horseradish peroxidase. The blot was developed by using a chemiluminescent substrate (ECL, Amersham, Buckinghamshire, UK) and exposed on Hyperfilm (Amersham).
CFU and replating assay
Double-positive MMTV-tTA/AML1-ETO transgenic mice and wild-type control mice were killed, and the bone marrow was flushed from the femurs and the tibias. The cells were washed in PBS and resuspended in MethoCult (M3434) containing 10 ng/mL recombinant mouse (rm) interleukin (IL)-3, 10 ng/mL recombinant human (rh) IL-6, 50 ng/mL rm stem cell factor (SCF), 3 U/mL rh erythropoietin, 15% fetal bovine serum (Stem Cell Technologies, Vancouver, British Columbia, Canada), and Pen-Strep (2.5 mL/35-mm plate). The cells were then plated out at densities of 1 × 104, 2 × 104, 3 × 104, or 6 × 104 cells/plate and incubated at 37°C with 5% CO2. The colonies on the plates were counted and classified 7 to 10 days after plating. To analyze the replating efficiency of bone marrow cells from double-positive tTA/AML1-ETO mice and MMTV-tTA mice, bulk populations of colonies were harvested 7 to 10 days after plating as previously described.16 25 Cells (1 × 104) were then replated in MethoCult and 1-2 × 104 cells were cytocentrifuged onto slides for Wright-Giemsa staining. As before, colonies were counted and harvested 7 to 10 days following plating.
Results
Strong inducible expression of AML1-ETO is seen in the bone marrow of 3 of the 5 transgenic founder lines
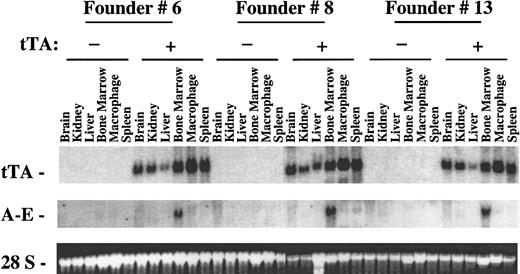
To study AML1-ETO expression in mice, we generated transgenic mice containing the AML1-ETO cDNA driven by a tetracycline-responsive promoter using pUHD-AML1/ETO. We obtained 5 unique founder lines (#2, #6, #7, #8, and #13). The mice from these 5 lines were bred with mice containing the MMTV-tTA DNA to produce double-positive mice for further study (Figure 1). As seen in Figure2, tTA is ubiquitously expressed in various tissues of the tTA-positive transgenic mice. The level of tTA expression was variable in different tissues. Founder line #2 did not show any inducible AML1-ETO expression, which could be due to the disruption or the mutation of the pUHD-AML1/ETO DNA construct during the generation of the transgenic mice (data not shown). Three (#6, #8, and #13) of the 5 founder lines had a very similar pattern of AML1-ETO expression (Figure 2). AML1-ETO was expressed in a tissue-specific manner in the bone marrow of the double-positive mice, and it was not expressed in the AML1-ETO–positive mice without the MMTV-tTA construct. A basal level of endogenous ETO expression can be seen in the brain RNA of both single- and double-positive mice (data not shown and Figure 3). AML1-ETO is also weakly expressed in the spleen, thymus, and peritoneal macrophages of the various founder lines (Figure 2 and data not shown). The level varies from founder line to founder line and is probably caused by different integration sites of the transgenic construct.
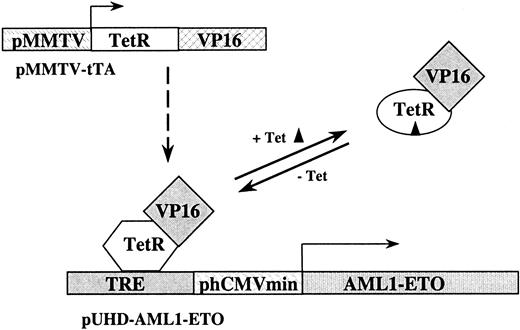
The tet-off system for AML1-ETO transcriptional control.
The tet-off system relies on 2 different DNA constructs. The first construct consists of a fusion of the wild-type tet repressor (TetR) DNA binding domain and the VP16 activation domain of herpes simplex virus driven by the promoter from the murine mammary tumor virus (pMMTV-tTA). The second construct in the system consists of the tetracycline-responsive element (TRE) just upstream from the minimal promoter of the human cytomegalovirus (CMV) driving transcription of AML1-ETO (pUHD–AML1-ETO). In the absence of tetracycline, the tet-controlled transcriptional activator (tTA), which consists of the TetR-VP16 complex, binds the TRE and activates transcription of AML1-ETO. When tetracycline is added, it binds tTA and causes a change in the conformation of the DNA-binding domain of the tet repressor. This prevents tTA from binding to the TRE and AML1-ETO transcription stops.
The tet-off system for AML1-ETO transcriptional control.
The tet-off system relies on 2 different DNA constructs. The first construct consists of a fusion of the wild-type tet repressor (TetR) DNA binding domain and the VP16 activation domain of herpes simplex virus driven by the promoter from the murine mammary tumor virus (pMMTV-tTA). The second construct in the system consists of the tetracycline-responsive element (TRE) just upstream from the minimal promoter of the human cytomegalovirus (CMV) driving transcription of AML1-ETO (pUHD–AML1-ETO). In the absence of tetracycline, the tet-controlled transcriptional activator (tTA), which consists of the TetR-VP16 complex, binds the TRE and activates transcription of AML1-ETO. When tetracycline is added, it binds tTA and causes a change in the conformation of the DNA-binding domain of the tet repressor. This prevents tTA from binding to the TRE and AML1-ETO transcription stops.
Northern blot analysis of inducible expression of AML1-ETO in transgenic mice.
Transgenic mice were generated that were positive for either pUHD–AML1-ETO or for MMTV-tTA and pUHD–AML1-ETO based on Southern blot analyses. Mice from 3 unique founder lines were killed, and total RNAs were isolated from various tissues. Total RNA (10 μg) was loaded on each lane. The RNA was then transferred to a nylon membrane and hybridized sequentially with fragments of the tTA gene and ETO cDNA. The ethidium bromide staining of the 28S ribosomal RNA is presented to show the loading of the RNA samples.
Northern blot analysis of inducible expression of AML1-ETO in transgenic mice.
Transgenic mice were generated that were positive for either pUHD–AML1-ETO or for MMTV-tTA and pUHD–AML1-ETO based on Southern blot analyses. Mice from 3 unique founder lines were killed, and total RNAs were isolated from various tissues. Total RNA (10 μg) was loaded on each lane. The RNA was then transferred to a nylon membrane and hybridized sequentially with fragments of the tTA gene and ETO cDNA. The ethidium bromide staining of the 28S ribosomal RNA is presented to show the loading of the RNA samples.
Northern blot analysis of inducible expression of AML1-ETO in founder line #7.
Transgenic mice were generated that were positive either for pUHD–AML1-ETO or for MMTV-tTA and pUHD–AML1-ETO. Mice from founder line #7 were killed, and total RNAs were isolated from various tissues. Total RNA (10 μg) was loaded on each lane. The RNA was then transferred to a nylon membrane and hybridized sequentially with tTA and ETO probes. The ethidium bromide staining of the 28S ribosomal RNA is presented to show the loading of the RNA samples.
Northern blot analysis of inducible expression of AML1-ETO in founder line #7.
Transgenic mice were generated that were positive either for pUHD–AML1-ETO or for MMTV-tTA and pUHD–AML1-ETO. Mice from founder line #7 were killed, and total RNAs were isolated from various tissues. Total RNA (10 μg) was loaded on each lane. The RNA was then transferred to a nylon membrane and hybridized sequentially with tTA and ETO probes. The ethidium bromide staining of the 28S ribosomal RNA is presented to show the loading of the RNA samples.
Strong inducible expression of AML1-ETO is seen in the bone marrow and macrophages of founder line #7
In 1 of the 5 unique founder lines that we obtained, a slightly different pattern of expression was observed (Figure 3). Founder line #7 expressed AML1-ETO RNA in peritoneal macrophages as well as in the bone marrow. The expression level in the macrophages was relatively higher than the level in the bone marrow. This pattern is possibly related to the integration site of the AML1-ETO construct in this particular line.
Functional AML1-ETO fusion protein is detectable in the inducible founder lines
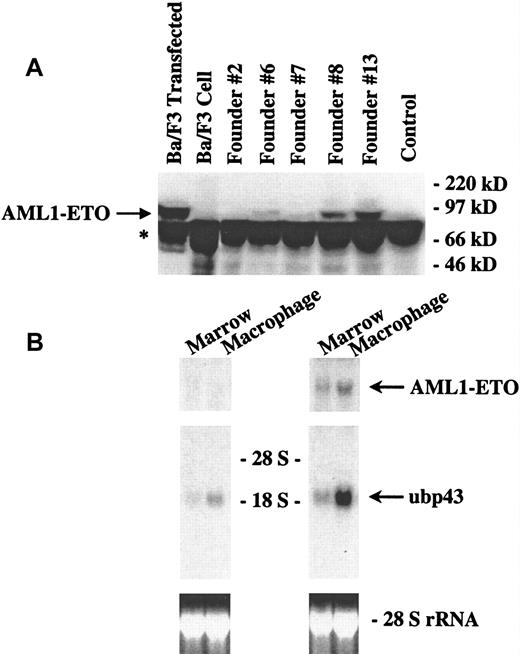
Expression of AML1-ETO was observed by Northern blot analysis of tissues from the inducible mice. To check whether there was any detectable translation of the AML1-ETO fusion protein, we performed a Western blot analysis of bone marrow samples from the double-positive transgenic mice (Figure 4A). We used protein extracts from transfected Ba/F3 cells as a control. In 4 (#6, #7, #8, and #13) of our 5 founder lines, we observed detectable levels of the AML1-ETO fusion protein. No protein expression was seen in founder line #2, the wild-type mice, or untransfected cell controls. Furthermore, the different founder lines expressed the AML1-ETO fusion protein at varying levels. This finding indicates that under tTA regulation, the transgenic construct is expressed not only at the transcriptional level but also at the protein level. To demonstrate that the expressed AML1-ETO protein is functional, we tested whether AML1-ETO expression had an effect on UBP43 gene expression. UBP43 is a novel ubiquitin-specific protease. We cloned this gene by comparing gene expression in the yolk sacs of AML1-ETO knock-in mice and wild-type mice.26 UBP43 is expressed at a relatively higher level in the knock-in mice, indicating its up-regulation by AML1-ETO. In adult mice, UBP43 is expressed at the highest level in the thymus and at a slightly lower level in the peritoneal macrophages. It is also expressed at a detectable level in the bone marrow. As shown in Figure 4B, total RNA was prepared from the bone marrow and peritoneal macrophages of either AML1-ETO transgenic mice or AML1-ETO and tTA double-positive transgenic mice from founder line #7 and subjected to Northern blot analysis. In the single-positive mice, which do not express AML1-ETO, a basal level of UBP43 expression is observed. In the mice with AML1-ETO expression, UBP43 is strongly up-regulated in the bone marrow and macrophages. This finding indicates that inducibly expressed AML1-ETO is a functional protein.
Functional AML1-ETO fusion protein is detectable in the inducible founder lines.
(A) Western blot analysis of AML1-ETO expression in the bone marrow of transgenic mice. Transgenic mice from 5 unique founder lines positive for both MMTV-tTA and pUHD-AML1/ETO as well as one negative control mouse positive for MMTV-tTA but not pUHD-AML1/ETO were killed. Bone marrow was harvested from the femurs of these mice and resuspended in SDS-PAGE loading buffer. The marrow protein samples were electrophoresed in an 8% resolving SDS-PAGE gel (bis:acrylamide = 1:19). In addition to the protein from the bone marrow of the mice, 35 μg of nuclear extracts from wild-type Ba/F3 cells (a murine pro-B cell line) and Ba/F3 cells transfected with an AML1-ETO construct were electrophoresed as controls. The protein was then transferred to PVDF membrane and hybridized with an antibody against ETO. The blot was then developed by using a chemiluminescent substrate. The arrow points to the position of AML1-ETO. * marks a nonspecific band. (B) Northern blot analysis of UBP43 expression in inducible AML1-ETO mice. Transgenic mice that were positive for either pUHD-AML1/ETO (left side panels) or for MMTV-tTA and pUHD-AML1/ETO (right side panels) from founder line #7 were killed, and total RNAs were isolated from the bone marrow and peritoneal macrophages of these mice. Total RNA (10 μg) was loaded on each lane. The RNA was then transferred to a nylon membrane and hybridized sequentially with fragments of ETO cDNA and UBP43 cDNA. The ethidium bromide staining of the 28S ribosomal RNA is presented to show the loading of the RNA samples.
Functional AML1-ETO fusion protein is detectable in the inducible founder lines.
(A) Western blot analysis of AML1-ETO expression in the bone marrow of transgenic mice. Transgenic mice from 5 unique founder lines positive for both MMTV-tTA and pUHD-AML1/ETO as well as one negative control mouse positive for MMTV-tTA but not pUHD-AML1/ETO were killed. Bone marrow was harvested from the femurs of these mice and resuspended in SDS-PAGE loading buffer. The marrow protein samples were electrophoresed in an 8% resolving SDS-PAGE gel (bis:acrylamide = 1:19). In addition to the protein from the bone marrow of the mice, 35 μg of nuclear extracts from wild-type Ba/F3 cells (a murine pro-B cell line) and Ba/F3 cells transfected with an AML1-ETO construct were electrophoresed as controls. The protein was then transferred to PVDF membrane and hybridized with an antibody against ETO. The blot was then developed by using a chemiluminescent substrate. The arrow points to the position of AML1-ETO. * marks a nonspecific band. (B) Northern blot analysis of UBP43 expression in inducible AML1-ETO mice. Transgenic mice that were positive for either pUHD-AML1/ETO (left side panels) or for MMTV-tTA and pUHD-AML1/ETO (right side panels) from founder line #7 were killed, and total RNAs were isolated from the bone marrow and peritoneal macrophages of these mice. Total RNA (10 μg) was loaded on each lane. The RNA was then transferred to a nylon membrane and hybridized sequentially with fragments of ETO cDNA and UBP43 cDNA. The ethidium bromide staining of the 28S ribosomal RNA is presented to show the loading of the RNA samples.
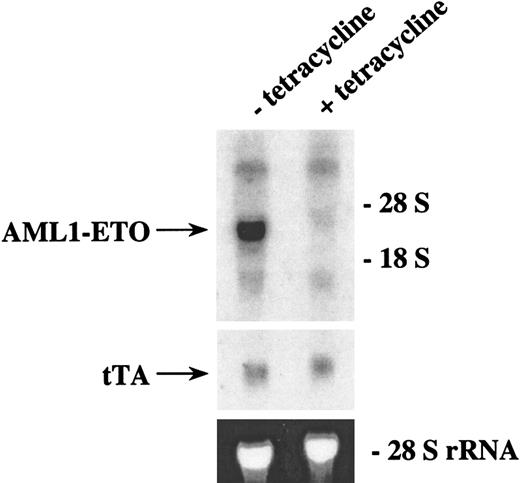
Tetracycline can control the expression of AML1-ETO in transgenic mice
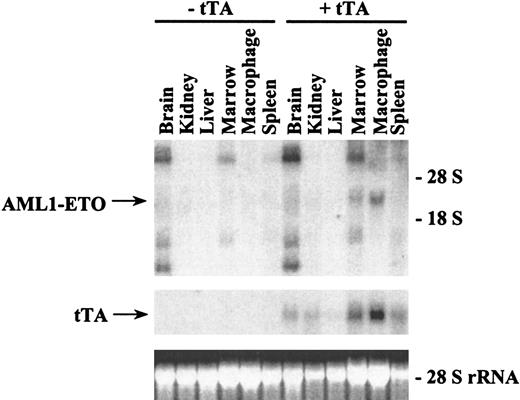
To test whether AML1-ETO expression was inducible in our double-positive transgenic mice, we bred mice in which the mother had a subcutaneous time-release tetracycline pellet to keep the AML1-ETO gene off in the developing embryos. Double-positive offspring of founder line #6 were identified by Southern blot analysis, and these mice were given tetracycline in their drinking water. When these offspring were 58 days old, the tetracycline was removed from some of these mice. After 3 weeks in the presence or absence of tetracycline, the bone marrow was harvested from these mice, and total RNA was analyzed by Northern blot analysis (Figure 5). The tet-off system of controllable gene expression was functional in these mice. The level of MMTV-tTA expression was equivalent in both the mice that continued to receive tetracycline and the mice from which tetracycline was removed. The mice from which tetracycline was removed showed a high level of AML1-ETO expression, whereas the mice that continued to receive tetracycline showed a trace level of AML1-ETO expression. This finding demonstrates that transgenic animals can be generated in which AML1-ETO expression can be controlled by administration of tetracycline.
Northern blot analysis of AML1-ETO expression in transgenic mice with or without tetracycline.
Double-positive mice from founder line #8 were generated by breeding heterozygous mice in which the females harbored a subcutaneous time-release tetracycline pellet that releases 0.7 mg tetracycline per day. F1 mice were weaned and continued to receive tetracycline (1 mg/mL) in their drinking water. Tetracycline was withdrawn from two 58-day-old mice, and the mice were maintained in the absence of tetracycline. Two other 58-day-old mice continued to receive tetracycline. Three weeks after tetracycline was removed or continued, the mice were killed and the bone marrow from the femurs was harvested. Total RNA from the marrow was isolated and 8.5 μg was loaded on each lane. The RNA was then transferred to a nylon membrane and hybridized sequentially with ETO and tTA probes. The ethidium bromide staining of the 28S ribosomal RNA is presented to show the loading of the RNA samples.
Northern blot analysis of AML1-ETO expression in transgenic mice with or without tetracycline.
Double-positive mice from founder line #8 were generated by breeding heterozygous mice in which the females harbored a subcutaneous time-release tetracycline pellet that releases 0.7 mg tetracycline per day. F1 mice were weaned and continued to receive tetracycline (1 mg/mL) in their drinking water. Tetracycline was withdrawn from two 58-day-old mice, and the mice were maintained in the absence of tetracycline. Two other 58-day-old mice continued to receive tetracycline. Three weeks after tetracycline was removed or continued, the mice were killed and the bone marrow from the femurs was harvested. Total RNA from the marrow was isolated and 8.5 μg was loaded on each lane. The RNA was then transferred to a nylon membrane and hybridized sequentially with ETO and tTA probes. The ethidium bromide staining of the 28S ribosomal RNA is presented to show the loading of the RNA samples.
Transgenic mice that express AML1-ETO have normal hematopoiesis
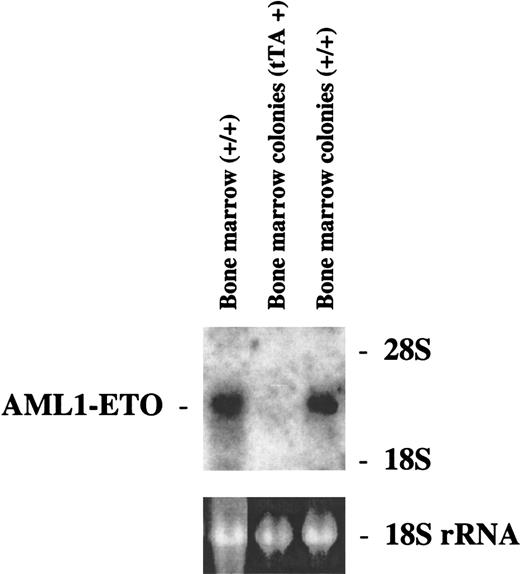
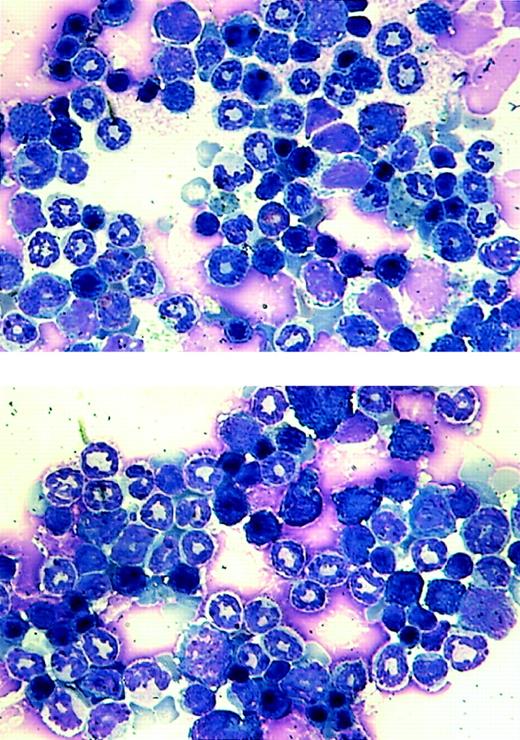
Our goal was to investigate the effect of AML1-ETO on hematopoiesis and its potential role in leukemogenesis. Because of the lethal effect of AML1-ETO on embryogenesis, we used the tet-off inducible system of gene expression. We observed adult double-positive mice that express AML1-ETO to study their hematopoiesis. These mice exhibited no outward signs of illness. Their coat appeared normal, and their level of activity was consistent with that of wild-type mice. We performed blood smears and differential counts of the blood from these mice. In all of the founder lines, the differential blood counts were normal (Table 1). We then used bone marrow cells from founder lines #7 and #8 to perform in vitro CFU assays. The numbers of different colonies observed for both double-positive and wild-type mice were approximately the same (Table1). These mice showed no abnormal hematopoiesis. CFU assays were also performed in which cells isolated from the same MMTV-tTA/AML1-ETO double-positive mice were plated in the presence or absence of tetracycline. No difference was observed in number or type of colonies generated (data not shown). Total RNA was then harvested from colonies from the CFU assay for founder line #8 and analyzed by Northern blot analysis. A similar level of AML1-ETO expression is seen in both the bone marrow of double-positive mice and colonies derived from the bone marrow of double-positive mice (Figure6). Bone marrow cells were also analyzed for their morphology (Figure 7). No significant difference can be observed between bone marrow samples from the control and AML1-ETO–expressing mice.
Percentage of colony types from CFU assays and cell types from differential blood counts of founder lines #7 and #8*
| . | Founder line #7 . | Founder line #8 . | Wild type . |
|---|---|---|---|
| Colonies from CFU assay (%) | |||
| GM | 65.5 ± 8.1 | 56.2 ± 7.1 | 57.8 ± 7.4 |
| Macrophages | 21.1 ± 9.6 | 27.8 ± 9.0 | 26.0 ± 9.0 |
| Mix | 9.4 ± 2.8 | 8.0 ± 0.8 | 10.9 ± 3.2 |
| Erythrocytes | 4.0 ± 2.1 | 7.2 ± 1.7 | 5.6 ± 2.4 |
| Differential blood counts (%) | |||
| Neutrophils | 20.7 ± 7.2 | 16.4 ± 8.8 | 18.8 ± 3.9 |
| Lymphocytes | 77.6 ± 6.8 | 81.8 ± 9.1 | 80.2 ± 3.9 |
| Monocytes | 1.7 ± 0.8 | 1.8 ± 1.1 | 1.0 ± 0.0 |
| . | Founder line #7 . | Founder line #8 . | Wild type . |
|---|---|---|---|
| Colonies from CFU assay (%) | |||
| GM | 65.5 ± 8.1 | 56.2 ± 7.1 | 57.8 ± 7.4 |
| Macrophages | 21.1 ± 9.6 | 27.8 ± 9.0 | 26.0 ± 9.0 |
| Mix | 9.4 ± 2.8 | 8.0 ± 0.8 | 10.9 ± 3.2 |
| Erythrocytes | 4.0 ± 2.1 | 7.2 ± 1.7 | 5.6 ± 2.4 |
| Differential blood counts (%) | |||
| Neutrophils | 20.7 ± 7.2 | 16.4 ± 8.8 | 18.8 ± 3.9 |
| Lymphocytes | 77.6 ± 6.8 | 81.8 ± 9.1 | 80.2 ± 3.9 |
| Monocytes | 1.7 ± 0.8 | 1.8 ± 1.1 | 1.0 ± 0.0 |
Mice positive for both the murine mammary tumor virus-tet-controlled transcriptional activator (MMTV-tTA) and pUHD-AML1/ETO constructs as well as wild-type mice were killed and the bone marrow was harvested. Colony forming unit (CFU) assays were performed and the colonies were counted. The percentage of each colony type is shown. In addition, peripheral blood was harvested from the tail and blood smears were made. The smears were stained with Wright-Giemsa and 100 to 200 cells of each slide were counted. The percentage of each cell type is shown.
Northern blot analysis of AML1-ETO expression in the bone marrow or bone marrow colonies of founder line #8.
Transgenic mice that were positive for either MMTV-tTA or for MMTV-tTA and pUHD–AML1-ETO were killed, and bone marrow was harvested from the femurs and tibias of these mice. Some of the bone marrow from these mice was used to perform CFU assays. Total RNA was harvested from the rest of the bone marrow from the double-positive mice as well as from the colonies from the CFU assays. Each RNA sample (10 μg) was electrophoresed. The RNA was then transferred to a nylon membrane and hybridized with a fragment of the ETO cDNA. The ethidium bromide staining of the 18S ribosomal RNA is presented to show the loading of the RNA samples.
Northern blot analysis of AML1-ETO expression in the bone marrow or bone marrow colonies of founder line #8.
Transgenic mice that were positive for either MMTV-tTA or for MMTV-tTA and pUHD–AML1-ETO were killed, and bone marrow was harvested from the femurs and tibias of these mice. Some of the bone marrow from these mice was used to perform CFU assays. Total RNA was harvested from the rest of the bone marrow from the double-positive mice as well as from the colonies from the CFU assays. Each RNA sample (10 μg) was electrophoresed. The RNA was then transferred to a nylon membrane and hybridized with a fragment of the ETO cDNA. The ethidium bromide staining of the 18S ribosomal RNA is presented to show the loading of the RNA samples.
Morphological analysis of bone marrow cells from the transgenic mice.
Bone marrow samples were prepared from a wild-type littermate (top panel) and an MMTV-tTA and pUHD–AML1-ETO double-positive mouse and stained with Wright-Giemsa solution.
Morphological analysis of bone marrow cells from the transgenic mice.
Bone marrow samples were prepared from a wild-type littermate (top panel) and an MMTV-tTA and pUHD–AML1-ETO double-positive mouse and stained with Wright-Giemsa solution.
Alteration of the differentiation and proliferation ability of progenitor cells expressing AML1-ETO
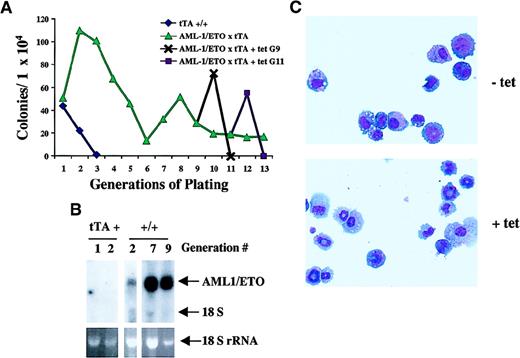
To examine the self-renewal capacity of progenitor cells expressing AML1-ETO, we performed serial replatings of bulk populations of bone marrow–derived colonies in methocellulose cultures. Seven to 10 days after plating, entire methocellulose cultures from MMTV-tTA mice and MMTV-tTA/AML1-ETO mice were harvested, and 1 × 104 cells were replated in methocellulose under conditions optimal for the differentiation of multipotential progenitors. As described earlier, primary cultures from bone marrow of MMTV-tTA/AML1-ETO mice exhibited no difference in the number or types of colonies generated as compared with MMTV-tTA mice in the presence or absence of tetracycline. However, on serial replating of the colonies, several differences became apparent. After 3 to 4 generations in methocellulose, the cultures from MMTV-tTA mice failed to form colonies (Figure 8A). Instead, large diffusely distributed cells populated the plates. On examination by cytospin and Wright-Giemsa staining, these cells were easily identifiable as fully differentiated macrophages. In contrast, colonies derived from mice expressing AML1-ETO continued to replate and to generate colonies. On these plates, we observed very few migrating, fully differentiated macrophages, but rather we saw small, compact colonies containing 50 to 100 cells. Cytospin and Wright-Giemsa staining of these colonies revealed a polymorphic population of cells, including very early progenitors as well as early myeloid cells. To be certain that these colonies were expressing AML1-ETO, we isolated RNA from colonies derived from MMTV-tTA mice and MMTV-tTA/AML1-ETO mice and performed a Northern analysis. This analysis (Figure 8B) clearly demonstrates AML1-ETO expression in the colonies from MMTV-tTA/AML1-ETO mice after culture in methocellulose.
Replating analysis of AML1-ETO–expressing progenitor cells.
(A) The effect of AML-1/ETO expression on hematopoietic progenitors. Bone marrow was harvested from femurs of mice positive for MMTV-tTA (♦) alone or MMTV-tTA/AML1-ETO (▴), and 1 × 104cells were plated in MethoCult in the absence of tetracycline. Bulk cultures were harvested after 7 to 10 days in culture, and 1 × 104 cells were replated for each sample. Each point represents the number of colonies generated per 104 cells seeded. After 9 (G9, ✖) and 11 (G11, ▪) generations, cells derived from MMTV-tTA/AML1-ETO mice were replated in MethoCult in the presence or absence of tetracycline. (B) Northern analysis of AML1-ETO expression in colonies harvested at various generations in methocellulose. RNA was harvested from bulk populations of colonies grown for 7 to 10 days in culture. Total RNA (5 μg) was loaded on each lane. The RNA was then transferred to a nylon membrane and hybridized with ETO cDNA. The ethidium bromide staining of the 18S ribosomal RNA is presented to show the loading of the RNA samples. tTA+ indicates RNA prepared from MMTV-tTA bone marrow cell culture; +/+, RNA prepared from MMTV-tTA/AML1-ETO bone marrow cell culture. (C) Morphology of cells obtained from colonies grown in the presence or absence of tetracycline. After 9 generations, cells were cultured in the presence or absence of tetracycline. Cytospins of colonies replated in the presence or absence of tetracycline after culture for 9 generations without tetracycline were stained with Wright-Giemsa solution.
Replating analysis of AML1-ETO–expressing progenitor cells.
(A) The effect of AML-1/ETO expression on hematopoietic progenitors. Bone marrow was harvested from femurs of mice positive for MMTV-tTA (♦) alone or MMTV-tTA/AML1-ETO (▴), and 1 × 104cells were plated in MethoCult in the absence of tetracycline. Bulk cultures were harvested after 7 to 10 days in culture, and 1 × 104 cells were replated for each sample. Each point represents the number of colonies generated per 104 cells seeded. After 9 (G9, ✖) and 11 (G11, ▪) generations, cells derived from MMTV-tTA/AML1-ETO mice were replated in MethoCult in the presence or absence of tetracycline. (B) Northern analysis of AML1-ETO expression in colonies harvested at various generations in methocellulose. RNA was harvested from bulk populations of colonies grown for 7 to 10 days in culture. Total RNA (5 μg) was loaded on each lane. The RNA was then transferred to a nylon membrane and hybridized with ETO cDNA. The ethidium bromide staining of the 18S ribosomal RNA is presented to show the loading of the RNA samples. tTA+ indicates RNA prepared from MMTV-tTA bone marrow cell culture; +/+, RNA prepared from MMTV-tTA/AML1-ETO bone marrow cell culture. (C) Morphology of cells obtained from colonies grown in the presence or absence of tetracycline. After 9 generations, cells were cultured in the presence or absence of tetracycline. Cytospins of colonies replated in the presence or absence of tetracycline after culture for 9 generations without tetracycline were stained with Wright-Giemsa solution.
To further examine the role played by the expression of AML1-ETO in these cells, we replated colonies expressing AML1-ETO, which had gone through 9 generations in culture without tetracycline, in the presence or absence of tetracycline. Interestingly, when AML1-ETO expression was turned off in these cells by the addition of tetracycline to the media, we saw 3-fold more colony formation as well as an increase in average colony size (150 to 300 cells). When we analyzed these colonies by cytocentrifugation and Wright-Giemsa staining, we observed an increased percentage of fully differentiated cells in both the macrophage and granulocyte lineages and a significant decrease in the percentage of blasts and immature cells (Table 2 and Figure 8C). We also observed that, following one passage in the absence of AML1-ETO expression, these cells did not replate. These data indicate that AML1-ETO expression reduces myeloid differentiation, causing the cells to pause in an immature stage. On suppression of AML1-ETO expression, these cells are fully capable of complete differentiation.
Differential counts of cells from bone marrow colonies from MMTV-tTA/AML1-ETO mice in the presence or absence of tetracycline
| . | −Tetracycline . | +Tetracycline . |
|---|---|---|
| Blasts | 14.0 ± 4.3 | 2.0 ± 1.0 |
| Immature cells | 67.7 ± 7.2 | 27.0 ± 4.7 |
| Neutrophils | 10.3 ± 2.1 | 42.7 ± 4.6 |
| Macrophages | 10.0 ± 1.7 | 21.7 ± 4.7 |
| . | −Tetracycline . | +Tetracycline . |
|---|---|---|
| Blasts | 14.0 ± 4.3 | 2.0 ± 1.0 |
| Immature cells | 67.7 ± 7.2 | 27.0 ± 4.7 |
| Neutrophils | 10.3 ± 2.1 | 42.7 ± 4.6 |
| Macrophages | 10.0 ± 1.7 | 21.7 ± 4.7 |
Murine mammary tumor virus-tet-controlled transcriptional activator (MMTV-tTA)/AML1-ETO double-positive mice were killed and the bone marrow was harvested. Colony assays were performed and replated serially through at least 13 generations in the absence of tetracycline. After 9 generations (Fig. 8A, black line), colonies were replated in the presence or absence of tetracycline. Cytocentrifugation was performed on the colonies generated and the slides were stained with Wright-Giemsa (Fig. 8C). To obtain differential counts for each sample, 100 cells were counted in triplicate (data shown here). This was repeated after 11 generations (Fig. 8A, purple line) with similar results (data not shown).
Discussion
The t(8;21) translocation is a frequent chromosomal aberration found in AML.7,10,11,27 We have previously shown that when AML1-ETO is knocked into the AML1 locus in mice, an embryonic lethal phenotype is observed.12 To further investigate the role of AML1-ETO in hematopoiesis and leukemogenesis, we generated mice that have tetracycline-inducible expression of AML1-ETO. This allowed us to control the expression of AML1-ETO by removing tetracycline to activate expression of the gene. In our mice, we have found that functional AML1-ETO is expressed in a highly inducible manner in bone marrow cells.
We generated 5 unique founder lines containing the AML1-ETO construct. We compared mice that were positive for the AML1-ETO construct with mice that were positive for both AML1-ETO and tTA. Although the expression of tTA in the double-positive mice was seen in all tissues tested, AML1-ETO was only highly expressed in the bone marrow of 4 (#6, #7, #8, and #13) of the 5 founder lines (#2, #6, #7, #8, and #13). In addition, founder line #7 showed a higher level of expression in the macrophages. The variation in the expression pattern may be associated with the different integration sites for the AML1-ETO construct in different founder lines. It is unclear why AML1-ETO is not expressed in all tissues that express tTA. One possibility is that AML1-ETO messenger RNA (mRNA) is transcribed but is not stable in some of the tissues that we have analyzed. To test this possibility, we performed nuclear run-on experiments with nuclei prepared from livers and kidneys of mice expressing AML1-ETO in the bone marrow and also from mice lacking such expression. We did not detect any AML1-ETO transcription in the livers and kidneys of mice that do express AML1-ETO in their bone marrow (data not shown). This result did not favor the RNA stability theory. Therefore, the MMTV-tTA tet-off system may be valuable for researchers interested in targeting inducible gene expression specifically to hematopoietic cells.
The purpose of this project was to create a model system in which leukemia could be brought on by the induction of AML1-ETO expression at a time after its expression would normally cause embryonic lethality. In tetracycline-inducible mice, however, we did not observe onset of leukemia. This is a clear example to support the concept proposed by Westervelt and Ley28 in their recent review that target cells are a critical component in the development of transgenic animal leukemia models. A good example of this is seen in the analysis of PML/RARA fusion protein transgenic mice. Because of the different expression patterns of cathepsin G, MRP8, and CD11b, the mice produced with the regulatory elements of these genes target different cell populations and show different leukemic phenotypes.29-32Therefore, it may be necessary for AML1-ETO to be expressed at a specific time during cell differentiation to exhibit a phenotype. Unfortunately, although the specific antibody used in this report is good for Western blot analysis, it still cannot detect AML1-ETO expression in an immunostaining assay. This difficulty makes it impossible to analyze the exact hematopoietic stage of AML1-ETO expression in t(8;21) leukemia patients or in these transgenic mice. Also, Western analysis is not practical because the number of cells necessary to produce enough protein to analyze would be difficult to acquire. Furthermore, there is no clear data correlating AML1-ETO expression during a particular stage of hematopoiesis with the development of leukemia in humans.
Although our double-positive AML1-ETO–expressing transgenic mice exhibit no disease and appear to have normal hematopoiesis as determined by analysis of differential counts of blood and bone marrow smears, we have made an important observation concerning the effect of AML1-ETO expression on the development and proliferation of a subpopulation of hematopoietic progenitors. Furthermore, we have used CFU assays to study in vitro proliferation and differentiation of bone marrow hematopoietic progenitor cells. Although we did not detect a difference between AML1-ETO–expressing bone marrow cells and nonexpressing cells in primary cultures, on serial replatings we have observed a difference in the ability of AML1-ETO–expressing cells to differentiate and proliferate. In their analysis of hematopoietic progenitors from AML1-ETO knock-in mice, Okuda et al.16also observed an increase in the self-renewal capacity of these cells. Our replating experiments with tetracycline-inducible expression of AML1-ETO indicate that hematopoietic progenitor cells from mice expressing AML1-ETO may be partially blocked from differentiation into mature myeloid cells. On addition of tetracycline to suppress the expression of AML1-ETO, these cells differentiate more efficiently into mature myeloid cells. These cells also proliferate in response to the removal of AML1-ETO expression. Taken together, these data indicate that, although differentiation is partially blocked or slowed very early in response to AML1-ETO, proliferation is not enhanced. This failure to proliferate may be the reason why we do not observe disease in these animals. To date, the AML1-ETO–expressing mice have exhibited normal hematopoiesis. Besides the concern of the time window of AML1-ETO expression as discussed above, the presence of normal hematopoiesis in these mice suggests the possibility that AML1-ETO alone is not sufficient to cause leukemia. It has been reported that the AML1-ETO transcript is detectable in t(8;21) patients with long-term remission.33-35 Therefore, it is possible to express AML1-ETO and not exhibit disease. Additional mutation or abnormal expression of another gene(s) may be necessary to promote leukemogenesis, perhaps through increased proliferation. It has been reported that retrovirus-mediated BCR/ABL or HRX-ENL expression in bone marrow cells causes leukemia.25,36,37 This demonstrates that expression of either BCR/ABL or HRX-ENL alone is sufficient to generate leukemia. When the same tetracycline-inducible system is used to direct BCR/ABL expression in the transgenic mice, MMTV-tTA/BCR/ABL double-positive mice developed leukemia.38 Conversely, when CBFβ/MYH11 is expressed in chimeric mice by a knock-in strategy, these mice do not develop leukemia in the first year of life.39 Treatment of these mice with ENU (N-ethyl-N-nitrosourea), a potent DNA alkylating mutagen, causes the mice to develop leukemia in 2 to 6 months, whereas wild-type mice treated with ENU alone do not. This finding suggests that CBFβ/MYH11 predisposes these mice to leukemia but that another mutation is required to progress to the development of leukemia. This result further supports the theory that additional event(s) may be necessary for AML1-ETO–associated leukemogenesis. Ultimately, further study is required to elucidate the contribution of AML1-ETO to the development of AML.
Acknowledgments
We wish to thank Lothar Hennighausen for the MMTV-tTA transgenic mice and Joel Lawitts, Pu Zhang, and Claudia Huettner for valuable discussion and technical assistance.
Supported by National Institutes of Health grant CA72009, and American Cancer Society grant DHP-166. D.E.Z. is a Leukemia and Lymphoma Society Scholar.
K.L.R., C.J.H., and N.H. contributed equally to this work.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Dong-Er Zhang, MEM-L51, The Scripps Research Institute, 10550 North Torrey Pines Rd, La Jolla, CA 92037; e-mail:dzhang@scripps.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal