Abstract
In acute myeloid leukemia (AML) patients, a variety of clinical and biologic parameters, including phenotype, have been examined for potential value in predicting treatment response and survival. The European Group for the Immunological Classification of Leukaemias (EGIL) has proposed that AML be defined immunologically by the expression of 2 or more of the following myeloid markers: myeloperoxidase, CD13, CD33, CDw65, and CD117. With regard to this classification, the prognostic significance of 21 antigens taken separately and with immunophenotypic subgroups were evaluated and compared with other clinical and biological variables in 177 adult AML patients. None of the antigens tested were associated with treatment outcome. In contrast, patients with blasts disclosing a full expression of panmyeloid phenotype (defined by the expression of all 5 myeloid markers) had a higher complete remission rate (P < .0001) and differed significantly in disease-free survival (P = .02) and overall survival (P = .008) than patients whose cells expressed fewer than 5 of these markers. In multivariate analysis, only age, panmyeloid phenotype, performance status, and permeability glycoprotein activity influence treatment outcome. Cytogenetics was significant in univariate analysis but not in multivariate analysis, most likely because of the redundancy with panmyeloid phenotype and a higher sensitivity of immunophenotyping. Patients whose cells exhibit the panmyeloid phenotype appear to define a relatively homogeneous biological subset of AML. The 4 independent prognostic factors were used to create a prognostic score, defined by the number of factors present. This score permitted a stratification of patients with AML, thereby allowing for the consideration of innovative therapies to improve outcome in the poorer outcome groups.
Immunophenotyping is a widely used method to diagnose and classify acute leukemias, thereby complementing morphology and cytochemistry.1-7 A variety of clinical and biologic parameters, including immunophenotype, have been examined for potential value in predicting treatment response and survival.8Several reports suggested a relationship between some antigens (eg, CD7, CD9, CD11b, CD13, CD14, CD15, CD33, CD34, CD56, and Tdt [deoxynucleotidal transferase terminal]) and acute myeloid leukemia (AML) patient prognosis.9-19 But subsequent studies have produced conflicting results.9-19
Leukemic myeloblasts express a variety of leukocyte differentiation antigens, which reflect commitment to the myeloid lineage as well as a level of maturation. The European Group for the Immunological Classification of Leukaemias (EGIL) has proposed an immunological classification of acute leukemias.6,7 In this classification, AMLs are defined immunologically by the expression of 2 or more of the following myeloid markers: myeloperoxidase (MPO), CD13, CD33, CDw65, and CD117.6,7 With regard to this classification, we attempted to evaluate the prognostic significance of different immunophenotypic subgroups. We also compared the results of other prognostic features within the context of a large clinical trial of adult AML patients treated with chemotherapy. Using multivariate analysis, few studies compared the prognostic value of immunophenotype with the other well-known prognostic factors.12-14 19 Blast cells from 177 AML patients were analyzed with a uniform panel of monoclonal antibodies (mAbs).
Patients, materials, and methods
Patients
From January 1994 through December 1998, 177 consecutively admitted, untreated AML patients (without acute promyelocytic leukemia [APL]) who were diagnosed in a single center were enrolled in this study. These 177 patients were analyzed for the myeloid phenotype as defined immunologically by the expression of 2 or more of the following panmyeloid markers: MPO, CD13, CD33, CDw65, and CD117. APL patients were not analyzed in this study because they had received retinoic acid treatment. For each patient in the study, we analyzed several clinical and biological characteristics: age, white blood cell (WBC) count at diagnosis, sex, performance status, French-American-British (FAB) morphology, lactate hydrogenase (LDH) level, platelet level, hemoglobin level, karyotype, and permeability glycoprotein (Pgp) expression and function.
Patients with t(9;22) were excluded from the study. Cytogenetic risk groups were defined as previously reported.20 Bone marrow for cytogenetic analysis was cultured according to standard methods: 20 or more cells were fully analyzed to exclude clonal abnormalities, which were defined in accordance with the International System for Human Cytogenetic Nomenclature (ISCN) guidelines.21 A successful analysis was available for 167 patients, representing 94% of the AML cases in the trial. A diagnostic result was not available in 10 cases because cytogenetic studies were not performed (n = 3) or the studies failed (n = 7). Clinical and biologic features are summarized in Table1.
Patient population
| . | All patients, no. (N = 177) . | Patients expressing all 5 myeloid markers, no. (n = 50) . | Patients expressing 1-4 myeloid markers, no. (n = 127) . | P . |
|---|---|---|---|---|
| Age, y, mean/range | 57/18-82 | 51 ± 15 | 60 ± 16 | .05* |
| WHO PS < 2, n | 133 | 44 | 89 | NS* |
| WBC at diagnosis, ×109 cells per L, mean/range | 50/0.3-525 | 68 ± 60 | 39 ± 58 | NS* |
| LDH, U/L, mean/range | 1803/257-15 000 | 1740 ± 1362 | 1853 ± 1990 | NS* |
| FAB morphology, n (% of total) | .03† | |||
| M0 | 4 (2) | 0 | 4 | |
| M1 | 35 (20) | 12 | 22 | |
| M2 | 63 (36) | 16 | 47 | |
| M4 | 35 (20) | 15 | 20 | |
| M4E | 7 (4) | 4 | 3 | |
| M5 | 25 (14) | 3 | 22 | |
| M6 | 7 (4) | 0 | 7 | |
| M7 | 1 (<1) | 0 | 1 | |
| Karyotype, n (% of total) | .003† | |||
| Good prognosis, n | 22 (13) | 10 | 12 | |
| Inv(16) | 7 | 4 | 3 | |
| t(8;21) | 15 | 6 | 9 | |
| Intermediate prognosis | 113 (63) | 35 | 78 | |
| No abnormality | 78 | 25 | 53 | |
| +8 | 15 | 5 | 10 | |
| 11q23 | 6 | 1 | 5 | |
| Other abnormalities | 14 | 4 | 10 | |
| Unfavorable prognosis | 32 (22) | 2 | 30 | |
| Complex | 10 | 0 | 10 | |
| −7 | 9 | 0 | 9 | |
| Del(5q) or −5 | 8 | 2 | 6 | |
| Abn(3q) | 5 | 0 | 5 | |
| Failure or not done | 10 (6) | 1 | 9 | |
| Pgp expression, mean | 0.15 ± 0.09 | 0.11 ± 0.07 | 0.19 ± 0.12 | NS* |
| Pgp activity, mean (n = 150) | 0.42 ± 0.19 | 0.37 ± 0.19 | 0.52 ± 0.21 | .05* |
| In vitro sensitivity to DNR, μmol/L (n = 90)‡ | 0.53 ± 0.41 | 0.27 ± 0.27 | 0.78 ± 0.72 | .04* |
| In vitro sensitivity to AraC, μmol/L (n = 90)‡ | 13.4 ± 8.5 | 8.5 ± 5.7 | 17.9 ± 16.4 | .04* |
| . | All patients, no. (N = 177) . | Patients expressing all 5 myeloid markers, no. (n = 50) . | Patients expressing 1-4 myeloid markers, no. (n = 127) . | P . |
|---|---|---|---|---|
| Age, y, mean/range | 57/18-82 | 51 ± 15 | 60 ± 16 | .05* |
| WHO PS < 2, n | 133 | 44 | 89 | NS* |
| WBC at diagnosis, ×109 cells per L, mean/range | 50/0.3-525 | 68 ± 60 | 39 ± 58 | NS* |
| LDH, U/L, mean/range | 1803/257-15 000 | 1740 ± 1362 | 1853 ± 1990 | NS* |
| FAB morphology, n (% of total) | .03† | |||
| M0 | 4 (2) | 0 | 4 | |
| M1 | 35 (20) | 12 | 22 | |
| M2 | 63 (36) | 16 | 47 | |
| M4 | 35 (20) | 15 | 20 | |
| M4E | 7 (4) | 4 | 3 | |
| M5 | 25 (14) | 3 | 22 | |
| M6 | 7 (4) | 0 | 7 | |
| M7 | 1 (<1) | 0 | 1 | |
| Karyotype, n (% of total) | .003† | |||
| Good prognosis, n | 22 (13) | 10 | 12 | |
| Inv(16) | 7 | 4 | 3 | |
| t(8;21) | 15 | 6 | 9 | |
| Intermediate prognosis | 113 (63) | 35 | 78 | |
| No abnormality | 78 | 25 | 53 | |
| +8 | 15 | 5 | 10 | |
| 11q23 | 6 | 1 | 5 | |
| Other abnormalities | 14 | 4 | 10 | |
| Unfavorable prognosis | 32 (22) | 2 | 30 | |
| Complex | 10 | 0 | 10 | |
| −7 | 9 | 0 | 9 | |
| Del(5q) or −5 | 8 | 2 | 6 | |
| Abn(3q) | 5 | 0 | 5 | |
| Failure or not done | 10 (6) | 1 | 9 | |
| Pgp expression, mean | 0.15 ± 0.09 | 0.11 ± 0.07 | 0.19 ± 0.12 | NS* |
| Pgp activity, mean (n = 150) | 0.42 ± 0.19 | 0.37 ± 0.19 | 0.52 ± 0.21 | .05* |
| In vitro sensitivity to DNR, μmol/L (n = 90)‡ | 0.53 ± 0.41 | 0.27 ± 0.27 | 0.78 ± 0.72 | .04* |
| In vitro sensitivity to AraC, μmol/L (n = 90)‡ | 13.4 ± 8.5 | 8.5 ± 5.7 | 17.9 ± 16.4 | .04* |
Values are expressed as the mean plus or minus SE. The Pvalue indicates the comparison of a patient group with the presence of all 5 myeloid markers and a group with the absence of one or several myeloid markers. The normal value of LDH is less than 618 U/L. NS indicates not significant. Del indicates chromosome deletion, and Abn indicates abnormal chromosome. DNR indicates daunorubicin and AraC, cytosine arabinoside.
Indicates that the Mann-Whitney U test was used.
Indicates that the Fisher exact test was used.
Indicates that the MTT assay was used.
Treatment
None of the patients had a history of prior therapies with anticancer drugs or a diagnosis of myelodysplastic syndrome. All patients of this study were given a combination of cytosine arabinoside (AraC) and anthracyclin with or without etoposide. Antileukemic treatments were differentiated according to age, but the treatments were similar.
Patients aged 60 years or younger received 100 mg/m2 AraC per day for 10 days and either 45 mg/m2 daunorubicin (DNR), 10 mg/m2 idarubicin, or 7 mg/m2 mitoxantrone per day for 3 days with or without 100 mg/m2 etoposide per day for 5 days. Those patients who achieved complete remission (CR) after 1 or 2 cycles of therapy received 1 cycle of consolidation therapy (with the same induction of anthracyclin). Patients achieving CR were subsequently scheduled to proceed to allogeneic bone marrow transplantation if a matched sibling donor was available (20 patients); patients greater than 45 years of age or lacking a suitable donor received an autograft transplantation.
Patients 61 years of age and older were given a combination of 100 mg/m2 AraC per day for 10 days and either 7 mg/m2 mitoxantrone per day for 3 days or 45 mg/m2 DNR per day for 3 days with or without 100 mg/m2 etoposide per day for 3 days. Those patients who achieved CR after 1 or 2 cycles of therapy received 1 cycle of consolidation therapy (with the same induction of anthracyclin). Patients less than 70 years old who achieved CR were subsequently scheduled to proceed to autograft transplantation; patients greater than 70 years of age received a second course of consolidation therapy.
Differing induction regimens did not appear to make a difference in clinical response (ie, achievement of CR, DFS, or OS). In addition, the distribution of variables analyzed as the panmyeloid phenotype (defined by the expression of all 5 myeloid markers), WHO performance status, cytogenetics, FAB subtypes, WBC count at diagnosis, LDH level, and Pgp expression and activity was not different between different treatment regimens. In the “intent to treat” analysis, there was no difference in clinical response (DFS or OS) between the different consolidation regimens in our department. In addition, there were no significant differences in the treatment received according to antigen expression.
Immunological phenotyping
The immunophenotype was performed by multiparameter flow cytometry (FACSORT flow cytometer, Becton Dickinson, Le Pont de Claix, France). Flow cytometry was performed on blast cells gated on their abnormal light scatter characteristics using mAbs (Becton Dickinson and Immunotech, Marseille, France) for the following 21 antigens: CD2 (Leu5b), CD3 (Leu4), CD4 (Leu3a), CD5 (Leu1), CD7 (Leu9), CD8 (Leu2), CD10 (J5), CD13 (LeuM7), CD14 (LeuM3), CD19 (SJ25C1), CD20 (Leu16), CD22 (Leu14), CD33 (LeuM9), CD34 (HPCA2), CD41 (SZ22), CD56 (Leu19), CDw65 (88H7), and CD117 (95C3). In addition, the cells were labeled with antibodies to TdT, HLA-DR, and MPO. Several panels containing 3 conjugated antibodies each were used for immunophenotyping analyses: HLA-DR-CD33-CD34; CD79a-CD34-CD20; CD10-CD19-CD34; MPO-cCD3-CD34; CD4-CD8-mCD3; CD10-CD22-CD19; CDw65-CD117-CD34; CD7-CD4-CD34; CD2-CD5-CD34; TdT-cCD22-CD34; CD34-CD14-CD13; CD56-CD33-CD34; and CD41-CD33-CD34. A membrane marker was considered positive when more than 20% of the blast cells expressed it. A cytoplasmic marker was considered positive when more than 20% of the blast cells expressed it. These values were selected by reference to previous published immunophenotyping AML studies and to the recent proposals of the EGIL group.6 7
Level of Pgp expression
Pgp expression was measured by labeling fresh viable cells with the UIC2 mAb (Immunotech) and phycoerythrin (PE)-labeled second antibody as described previously.22 The expression of Pgp was established with blast cells selected by the CD34 mAb (2-color assay) or other markers (eg, CD33/CD7, CD33/CD2, CD33/CD19, or CD33/CD22 by 3-color assays) whenever possible. Physical characteristics were used to establish expression of Pgp only if blast cells did not express characteristic markers. Fluorescence was analyzed on a FACSORT flow cytometer. The D value generated for Pgp expression compared gated leukemic blasts stained with UIC2 versus isotype control by means of the Kolmogorov-Smirnov (KS) test. This statistic, denoted D, measures the difference between 2 distribution functions and generates a value ranging from −1.0 to 1.0.22Correlations with clinical outcome were largely performed using the D value as a continuous variable, in accordance with consensual recommendations.23,24 Staining with UIC2 was considered positive with a D value of at least 0.15. This D value cutoff point was derived based on observations of previous works.22
Functional analysis of Pgp using calcein-am
Pgp function was measured as described previously.22Briefly, cells exposed to the nonfluorescent calcein-am(Sigma, St Quentin-Fallarrer, France) become fluorescent following the intracytoplasmic cleavage of calcein-am by cellular esterases, which produced the fluorescent derivate calcein. Pgp actively extruded the calcein-am. When we measured the calcein-am uptake by flow cytometry, we assessed the amount of fluorescent calcein that had been converted from the nonfluorescent calcein-am. When the Pgp protein was active, less calcein-am was retained and less was converted to fluorescent calcein. Therefore, calcein-am uptake (with specific modulators of Pgp) could be used to assess whether Pgp was functional. In our previous studies, calcein-am uptake with or without cyclosporin A (CsA) provided a functional test for AML cells as specific and sensitive as Rh123 ± CsA, the most specific and sensitive Pgp functional test.22
We performed this functional test in 150 adult AML patients among the 177 total AML patients. The cells were incubated with 0.1 μmol/L calcein-am for 15 minutes at 37°C in Roswell Park Memorial Institute medium (RPMI 1640) without or with 2 μmol/L CsA. The cells were washed twice in cold phosphate-buffered saline (PBS), and the samples were analyzed with a FACSORT flow cytometer. All samples were analyzed without fixation. As for Pgp expression, the D value measures the difference between 2 distribution functions (with and without a modulator). The function of Pgp was established with blast cells selected as above. Correlations with clinical outcome were performed using the D value as a continuous variable, in accordance with several consensual recommendations.23,24 Positive uptake of calcein-am was defined as a D value of at least 0.3. This D value cutoff point was derived based on observations of previous works.22
Methyl-thiazol-tetrazolium cytotoxicity test
In vitro cytotoxicity was measured as described previously.25 Previous studies, using the methyl-thiazol-tetrazolium (MTT) assay for the prediction of chemoresistance in adult AML, suggested that the assay may be helpful for risk group stratification in adult AML.25,26,27Stratification also has a strong value in the prediction of clinical response in childhood leukemias.28,29 Therefore we used the MTT assay to assess the in vitro resistance to drugs. In our previous study, patients who exhibited high lethal concentration of 50% (LC50) DNR and/or high LC50 of AraC had a poorer prognosis than the other patients.25
In vitro sensitivity of cells to DNR, Ara-C, and etoposide was determined by planting 2 × 105 cells in a 200 μL growth medium, without any specific growth factor, containing several dilutions of the drug in 96-well microtitre plates. Each concentration of drugs was repeated in 6 wells. After incubation at 37°C for 3 days with 5% carbon dioxide (CO2), cell viability was determined using the MTT assay as described by Kaspers et al.30 Briefly, 20 μL MTT (5 mg/mL in PBS) was added to each well and incubated for 6 hours. The medium and MTT were then removed from the wells by centrifugation, and formazan crystals were dissolved in 200 μL dimethyl sulfoxide (DMSO). The absorbance was recorded in a microplate reader (Model MR5000; Dynatex Laboratories, Issy-les-Moulineaux, France) at the wavelength of 550 nm. The effect of drugs on growth inhibition could be assessed as: % of growth inhibition = 1 − [(absorbance of drug-treated cells/absorbance of untreated cells) × 100]. LC50 was determined as the drug concentration that resulted in a 50% growth inhibition. The samples were considered evaluable if the drug-free control wells contained more than 80% of leukemic cells before culture and more than 70% of leukemic cells after 3 days of culture. The MTT assay gave reliable results under these conditions.30 The percentage of blast cells was determined by May-Grünwald-Giemsa staining and by immunophenotyping, which was performed by flow cytometry. We performed this functional test on 90 adult AML patients among the 177 total AML patients.
Statistical analysis
The association between variables was analyzed by the Fisher exact test for categorical variables and by the Mann-Whitney U test for continuous variables. Clinical and biological factors were investigated for their influence on remission rate by the Fisher exact test for binary variables and by the Mann-Whitney U or Kruskal-Wallis tests for continuous variables. DFS was measured from the establishment of CR until relapse or death from any cause, with observation censored for patients last known alive without report of relapse. OS was measured from diagnosis until death from any cause, with observation censored for patients last known alive. DFS and OS were estimated by the Kaplan-Meier method31 and compared by the log-rank test. Analyses of prognostic factors for treatment outcomes were based on proportional hazards regression models for DFS and OS.32Significance was defined as P ≤ 0.5, as determined by the 2-tailed test. The Cox proportional model was used for the multivariate analyses on DFS and OS.32 The median follow-up time for censored patients was 716 days. We included the 177 patients in this study from January 1994 through December 1998: 16 patients in 1994, 37 in 1995, 41 in 1996, 42 in 1997, and 41 in 1998. The time-point used for the proportion of DFS and OS was August 31, 1999.
Results
Immunophenotype
The expression of 21 antigens for the group of 177 assessable adult AML patients is presented in Table 2. Among different markers, the most positive markers were the following (percentage of positivity noted in parentheses): the myeloid lineage antigens CD13 (95%), CD33 (91%), and MPO (73%) and the hematopoietic progenitor cell markers HLA-DR (87%), CD117 (73%), and CD34 (68%). Another myeloid lineage marker, CDw65, was positive in only 40% of the patients. CD7, a stem cell marker, was positive in 37% of the patients. We detected the T-cell markers CD2 in 18% of patients, CD5 in 4%, cCd3 in 2%, and CD8 in 0%, whereas we detected the B-cell markers CD19 in 16%, CD10 in 10%, cCd22 in 2%, and CD20 in 0%. CD4 and CD14 were positive in 63% and 25% of the patients, respectively.
Correlation of antigen expression with clinical and biological variables
| . | HLA-DR . | CD34 . | MPO . | CD33 . | CD13 . | CDw65 . | CD117 . | CD14 . | CD2 . | CD4 . | CD5 . | CD7 . | CD10 . | CD19 . | TdT . | CD41 . | CD56 . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Patients, N | 177 | 177 | 177 | 177 | 177 | 177 | 177 | 177 | 142 | 177 | 130 | 177 | 177 | 177 | 177 | 99 | 42 |
| Positive patients, n (%) | 154 (87) | 120 (68) | 129 (73) | 161 (91) | 168 (95) | 71 (40) | 129 (73) | 44 (25) | 26 (18) | 111 (63) | 5 (4) | 65 (37) | 18 (10) | 28 (16) | 32 (18) | 6 (6) | 10 (24) |
| Age, y, Mean +/ mean − | 54/62 | 54/56 | 53/58 | 54/60 | 55/43 | 53/58 | 58/61 | 53/54 | 58/54 | 55/54 | ND | 49/58 | 59/53 | 49/55* | 62/53* | 48/53 | 50/57 |
| WBC at diagnosis, Mean +/ mean − | 54/79 | 45/75* | 59/57 | 61/8* | 55/85 | 59/58 | 42/48 | 88/46* | 53/54 | 58/48 | 90/54 | 76/42* | 75/54 | 45/58 | 39/58 | 100/59 | 40/43 |
| WHO PS, n (%) | |||||||||||||||||
| <2 | 116 (87) | 85 (64) | 100 (75) | 122 (92) | 126 (95) | 54 (41) | 104 (78)† | 31 (23) | 20 (18) | 82 (62) | 4 (4) | 50 (38) | 13 (10) | 20 (15) | 22 (16) | 5 (7) | 7 (22) |
| ≥2 | 38 (86) | 35 (79) | 29 (66) | 39 (89) | 42 (95) | 17 (39) | 25 (57)† | 13 (29) | 6 (19) | 29 (66) | 1 (3) | 15 (34) | 5 (11) | 8 (18) | 10 (23) | 1 (4) | 3 (27) |
| LDH, Mean +/ mean − | 1818/2395 | 1818/2395 | 1801/2599 | 2077/1652 | 1959/3346 | 2839/1727 | 1573/2484 | 2972/1746* | 1930/1949 | 2071/1609 | 1972/2103 | 2363/1824 | 2532/2020 | 1411/2180 | 1923/2116 | 2167/2236 | 2639/1357 |
| Karyotype, n = positive patients (%) | |||||||||||||||||
| Good | 22 (100) | 21 (95)† | 22 (100)† | 21 (95) | 22 (100) | 10 (45) | 15 (68) | 5 (23) | 3 (16) | 9 (41)† | 0 (0)† | 5 (23) | 1 (5) | 13 (59)† | 1 (5) | 0 (0) | 0 (0)† |
| Intermediate‡ | 93 (82) | 64 (57)† | 87 (77)† | 104 (92) | 105 (93) | 49 (43) | 85 (75) | 26 (23) | 20 (22) | 79 (70)† | 1 (1)† | 47 (42) | 12 (11) | 7 (6)† | 23 (20) | 4 (7) | 5 (17)† |
| Poor | 28 (87) | 26 (81)† | 14 (44)† | 26 (81) | 31 (97) | 8 (25) | 25 (78) | 5 (17) | 2 (8) | 15 (47)† | 4 (19)† | 12 (38) | 3 (9) | 6 (19)† | 5 (16) | 2 (13) | 4 (66)† |
| Failure or not done | 10 (100) | 9 (90) | 6 (60) | 10 (100) | 10 (100) | 4 (40) | 4 (40) | 8 (80) | 1 (12) | 8 (80) | 0 (0) | 1 (10) | 2 (20) | 2 (20) | 3 (30) | 0 (0) | 1 (50) |
| FAB morphology, n = positive patients (%) | |||||||||||||||||
| M0 | 4 (100)† | 4 (100)† | 2 (50)† | 4 (100) | 4 (100) | 0 (0)† | 3 (75)† | 0 (0)† | 0 (0)† | 3 (75)† | 1 (25) | 1 (25)† | 0 (0)† | 2 (50)† | 2 (50)† | 0 (0)† | 0 (0) |
| M1 | 23 (66)† | 25 (71)† | 26 (74)† | 30 (86) | 32 (91) | 13 (37)† | 24 (69)† | 1 (3)† | 3 (12)† | 15 (43)† | 1 (4) | 9 (26)† | 0 (0)† | 3 (9)† | 4 (11)† | 0 (0)† | 0 (0) |
| M2 | 54 (86)† | 52 (82)† | 52 (82)† | 58 (92) | 62 (98) | 15 (24)† | 56 (89)† | 1 (2)† | 3 (5)† | 34 (54)† | 1 (2) | 19 (30)† | 0 (0)† | 20 (32)† | 7 (11)† | 0 (0)† | 5 (38) |
| M4 | 33 (94)† | 20 (57)† | 31 (88)† | 34 (97) | 34 (97) | 20 (57)† | 28 (80)† | 18 (51)† | 9 (36)† | 30 (86)† | 2 (8) | 17 (49)† | 7 (20)† | 2 (6)† | 15 (43)† | 2 (11)† | 3 (50) |
| M4E | 7 (100)† | 6 (86)† | 7 (100)† | 7 (100) | 7 (100) | 4 (57)† | 7 (100)† | 4 (57)† | 4 (66)† | 6 (86)† | 0 (0) | 2 (29)† | 1 (14)† | 0 (0)† | 0 (0)† | 1 (50)† | 0 (0) |
| M5 | 25 (100)† | 5 (20)† | 9 (36)† | 20 (80) | 21 (84) | 18 (72)† | 4 (16)† | 20 (80)† | 7 (35)† | 23 (92)† | 0 (0) | 17 (68)† | 10 (40)† | 1 (4)† | 7 (28)† | 2 (20)† | 2 (29) |
| M6 | 7 (100)† | 7 (100)† | 2 (29)† | 7 (100) | 7 (100) | 1 (14)† | 6 (86)† | 0 (0)† | 0 (0)† | 0 (0)† | 0 (0) | 0 (0)† | 0 (0)† | 0 (0)† | 0 (0)† | 0 (0)† | 0 (0) |
| M7 | 1 (100)† | 1 (100)† | 0 (0)† | 1 (100) | 1 (100) | 0 (0)† | 1 (100)† | 0 (0)† | ND | 0 (0)† | ND | 0 (0)† | 0 (0)† | 0 (0)† | 0 (0)† | 1 (100)† | ND |
| Pgp expression, Mean +/ mean − | .27/.29 | .27/.30 | .26/.30 | .27/.38* | .28/.31 | .31/.26 | .31/.30 | .29/.27 | .25/.29 | .26/.28 | .3/.27 | .32/.25 | .30/.28 | .27/.27 | .15/.27 | .20/.24 | |
| Pgp activity, Mean +/ mean − | .47/.45 | .55/.3* | .49/.42 | .45/.64* | .48/.31 | .36/.54* | .51/.36 | .32/.5* | .33/.5 | .41/.51 | .3/.46 | .4/.5 | .25/.49* | .55/.46 | .36/.5* | .4/.5 | .37/.47 |
| . | HLA-DR . | CD34 . | MPO . | CD33 . | CD13 . | CDw65 . | CD117 . | CD14 . | CD2 . | CD4 . | CD5 . | CD7 . | CD10 . | CD19 . | TdT . | CD41 . | CD56 . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Patients, N | 177 | 177 | 177 | 177 | 177 | 177 | 177 | 177 | 142 | 177 | 130 | 177 | 177 | 177 | 177 | 99 | 42 |
| Positive patients, n (%) | 154 (87) | 120 (68) | 129 (73) | 161 (91) | 168 (95) | 71 (40) | 129 (73) | 44 (25) | 26 (18) | 111 (63) | 5 (4) | 65 (37) | 18 (10) | 28 (16) | 32 (18) | 6 (6) | 10 (24) |
| Age, y, Mean +/ mean − | 54/62 | 54/56 | 53/58 | 54/60 | 55/43 | 53/58 | 58/61 | 53/54 | 58/54 | 55/54 | ND | 49/58 | 59/53 | 49/55* | 62/53* | 48/53 | 50/57 |
| WBC at diagnosis, Mean +/ mean − | 54/79 | 45/75* | 59/57 | 61/8* | 55/85 | 59/58 | 42/48 | 88/46* | 53/54 | 58/48 | 90/54 | 76/42* | 75/54 | 45/58 | 39/58 | 100/59 | 40/43 |
| WHO PS, n (%) | |||||||||||||||||
| <2 | 116 (87) | 85 (64) | 100 (75) | 122 (92) | 126 (95) | 54 (41) | 104 (78)† | 31 (23) | 20 (18) | 82 (62) | 4 (4) | 50 (38) | 13 (10) | 20 (15) | 22 (16) | 5 (7) | 7 (22) |
| ≥2 | 38 (86) | 35 (79) | 29 (66) | 39 (89) | 42 (95) | 17 (39) | 25 (57)† | 13 (29) | 6 (19) | 29 (66) | 1 (3) | 15 (34) | 5 (11) | 8 (18) | 10 (23) | 1 (4) | 3 (27) |
| LDH, Mean +/ mean − | 1818/2395 | 1818/2395 | 1801/2599 | 2077/1652 | 1959/3346 | 2839/1727 | 1573/2484 | 2972/1746* | 1930/1949 | 2071/1609 | 1972/2103 | 2363/1824 | 2532/2020 | 1411/2180 | 1923/2116 | 2167/2236 | 2639/1357 |
| Karyotype, n = positive patients (%) | |||||||||||||||||
| Good | 22 (100) | 21 (95)† | 22 (100)† | 21 (95) | 22 (100) | 10 (45) | 15 (68) | 5 (23) | 3 (16) | 9 (41)† | 0 (0)† | 5 (23) | 1 (5) | 13 (59)† | 1 (5) | 0 (0) | 0 (0)† |
| Intermediate‡ | 93 (82) | 64 (57)† | 87 (77)† | 104 (92) | 105 (93) | 49 (43) | 85 (75) | 26 (23) | 20 (22) | 79 (70)† | 1 (1)† | 47 (42) | 12 (11) | 7 (6)† | 23 (20) | 4 (7) | 5 (17)† |
| Poor | 28 (87) | 26 (81)† | 14 (44)† | 26 (81) | 31 (97) | 8 (25) | 25 (78) | 5 (17) | 2 (8) | 15 (47)† | 4 (19)† | 12 (38) | 3 (9) | 6 (19)† | 5 (16) | 2 (13) | 4 (66)† |
| Failure or not done | 10 (100) | 9 (90) | 6 (60) | 10 (100) | 10 (100) | 4 (40) | 4 (40) | 8 (80) | 1 (12) | 8 (80) | 0 (0) | 1 (10) | 2 (20) | 2 (20) | 3 (30) | 0 (0) | 1 (50) |
| FAB morphology, n = positive patients (%) | |||||||||||||||||
| M0 | 4 (100)† | 4 (100)† | 2 (50)† | 4 (100) | 4 (100) | 0 (0)† | 3 (75)† | 0 (0)† | 0 (0)† | 3 (75)† | 1 (25) | 1 (25)† | 0 (0)† | 2 (50)† | 2 (50)† | 0 (0)† | 0 (0) |
| M1 | 23 (66)† | 25 (71)† | 26 (74)† | 30 (86) | 32 (91) | 13 (37)† | 24 (69)† | 1 (3)† | 3 (12)† | 15 (43)† | 1 (4) | 9 (26)† | 0 (0)† | 3 (9)† | 4 (11)† | 0 (0)† | 0 (0) |
| M2 | 54 (86)† | 52 (82)† | 52 (82)† | 58 (92) | 62 (98) | 15 (24)† | 56 (89)† | 1 (2)† | 3 (5)† | 34 (54)† | 1 (2) | 19 (30)† | 0 (0)† | 20 (32)† | 7 (11)† | 0 (0)† | 5 (38) |
| M4 | 33 (94)† | 20 (57)† | 31 (88)† | 34 (97) | 34 (97) | 20 (57)† | 28 (80)† | 18 (51)† | 9 (36)† | 30 (86)† | 2 (8) | 17 (49)† | 7 (20)† | 2 (6)† | 15 (43)† | 2 (11)† | 3 (50) |
| M4E | 7 (100)† | 6 (86)† | 7 (100)† | 7 (100) | 7 (100) | 4 (57)† | 7 (100)† | 4 (57)† | 4 (66)† | 6 (86)† | 0 (0) | 2 (29)† | 1 (14)† | 0 (0)† | 0 (0)† | 1 (50)† | 0 (0) |
| M5 | 25 (100)† | 5 (20)† | 9 (36)† | 20 (80) | 21 (84) | 18 (72)† | 4 (16)† | 20 (80)† | 7 (35)† | 23 (92)† | 0 (0) | 17 (68)† | 10 (40)† | 1 (4)† | 7 (28)† | 2 (20)† | 2 (29) |
| M6 | 7 (100)† | 7 (100)† | 2 (29)† | 7 (100) | 7 (100) | 1 (14)† | 6 (86)† | 0 (0)† | 0 (0)† | 0 (0)† | 0 (0) | 0 (0)† | 0 (0)† | 0 (0)† | 0 (0)† | 0 (0)† | 0 (0) |
| M7 | 1 (100)† | 1 (100)† | 0 (0)† | 1 (100) | 1 (100) | 0 (0)† | 1 (100)† | 0 (0)† | ND | 0 (0)† | ND | 0 (0)† | 0 (0)† | 0 (0)† | 0 (0)† | 1 (100)† | ND |
| Pgp expression, Mean +/ mean − | .27/.29 | .27/.30 | .26/.30 | .27/.38* | .28/.31 | .31/.26 | .31/.30 | .29/.27 | .25/.29 | .26/.28 | .3/.27 | .32/.25 | .30/.28 | .27/.27 | .15/.27 | .20/.24 | |
| Pgp activity, Mean +/ mean − | .47/.45 | .55/.3* | .49/.42 | .45/.64* | .48/.31 | .36/.54* | .51/.36 | .32/.5* | .33/.5 | .41/.51 | .3/.46 | .4/.5 | .25/.49* | .55/.46 | .36/.5* | .4/.5 | .37/.47 |
Other markers tested included (number of samples tested/% of positive cases): CD3 (30/0%), cCD3 (175/2%), CD8 (20/0%), CD20 (51/0%), CD22 (31/3%), and cCD22 (175/0%). ND indicates not determined. Shaded areas indicate P values no more than .05.
Indicates the Mann-Whitney U test was used; P ≤ .05.
Indicates the Fisher exact test was used; P ≤ .05.
This includes a normal karyotype and another intermediate prognostic karyotype.
The expression of the 5 myeloid antigens (ie, MPO, CD13, CD33, CDw65, and CD117) defining the panmyeloid phenotype was analyzed in the 177 patients. Leukemic blasts from these 177 patients expressed 2 of these antigens in 13 patients (7%), 3 antigens in 49 patients (27%), 4 antigens in 65 patients (36%), and 5 antigens in 50 patients (28%).
Correlations of antigen expression and other clinical and biological variables
The correlations of antigen expression and other clinical and biological variables are shown in Table 2. Age was significantly correlated with CD19 and Tdt; WBC count with CD34, CD33, CD14, and CD7; WHO performance status with CD117; LDH level with CD14; cytogenetics with CD34, MPO, CD4, CD5, and CD19; FAB morphology with all markers except CD33, CD13, CD5, and CD56; Pgp expression with CD33; and Pgp activity with CD34, CD33, CDw65, CD14, CD10, and Tdt. Interestingly CD34 and CD19 were seen in good and poor cytogenetic groups. In contrast, CD4 expression was lower in good and poor cytogenetic groups than in the intermediate cytogenetic group. No significant correlations were found between platelet level, hemoglobin level, and markers.
Expression of panmyeloid antigens and other clinical and biological variables
Table 1 shows significant associations between the presence of all 5 myeloid markers (panmyeloid phenotype) and cytogenetics (P = .003), FAB subtypes (P = .03), young age (P = .05), and low Pgp activity (P = .05). Associations were not noted with WBC count, LDH level, WHO performance status, and Pgp expression.
Relationship between immunophenotype and treatment outcome
Out of 177 patients, 101 (57%) achieved CR. None of the antigens tested were associated with a higher or lower CR rate. The analysis of DFS and OS curves likewise failed to show any prognostic significance for the antigens tested. In contrast, patients with leukemic blasts disclosing the expression of the panmyeloid phenotype had a higher CR rate compared with those patients who did not disclose expression (80% vs 48%, respectively; P < .0001) (Table3). DFS and OS of patients expressing the panmyeloid phenotype also differed significantly from patients whose leukemic cells expressed only 1-4 antigens (Figure1); for DFS, 52% vs 16%, with an unattained median vs 360 days, respectively (P = .02), and for OS, 48% vs 17%, with a median of 780 days vs 190 days, respectively (P = .008) (Table 3). An examination of the possible prognostic relevance of the other combination of antigens for the CR rate, DFS, and OS yielded no further information (data not shown).
Relationship between clinical and biological variables and treatment outcome
| . | Patients, no. . | CR, % . | P3-150,3-151 . | P3-152 . | 3-y DFS, %/median d . | P3-150,3-153 . | P3-152 . | 3-y OS, %/median d . | P3-150,3-153 . | P3-152 . |
|---|---|---|---|---|---|---|---|---|---|---|
| Panmyeloid antigens | <.0001 | .02 | .02 | .04 | .008 | .04 | ||||
| + | 50 | 80 | 52/NR | 48/680 | ||||||
| − | 127 | 48 | 16/390 | 17/231 | ||||||
| Age | <.0001 | .001 | .01 | .02 | <.0001 | .002 | ||||
| <60 | 80 | 35 | 14/290 | 19/164 | ||||||
| ≥60 | 97 | 75 | 41/580 | 43/784 | ||||||
| WHO performance status | .01 | .01 | .01 | .03 | .005 | .02 | ||||
| <2 | 133 | 62 | 40/439 | 37/484 | ||||||
| ≥2 | 44 | 41 | 14/392 | 20/180 | ||||||
| Cytogenetic | .005 | NS | .01 | NS | .01 | NS | ||||
| Good | 22 | 82 | 59/NR | 53 | ||||||
| Intermediate | 113 | 59 | 33/518 | 31/345 | ||||||
| Poor | 32 | 34 | 12/189 | 19/201 | ||||||
| Failure or not done | 10 | 50 | ||||||||
| FAB subtypes | NS | NS | NS | NS | NS | NS | ||||
| M0 | 4 | 75 | 0/170 | 20/170 | ||||||
| M1 | 35 | 54 | 0/130 | 0/185 | ||||||
| M2 | 63 | 59 | 34/579 | 34/531 | ||||||
| M4 | 35 | 54 | 60/NR | 51/NR | ||||||
| M4E | 7 | 86 | 40/380 | 50/NR | ||||||
| M5 | 25 | 52 | 22/170 | 21/190 | ||||||
| M6 | 7 | 57 | 0/227 | 40/200 | ||||||
| WBC count, ×109/L | NS | NS | NS | NS | NS | NS | ||||
| ≥30 | 59 | 47 | 29/430 | 23/295 | ||||||
| <30 | 118 | 62 | 38/588 | 36/300 | ||||||
| LDH level, U/L | NS | NS | NS | NS | NS | NS | ||||
| ≥2000 | 48 | 46 | 32/350 | 32/277 | ||||||
| <2000 | 129 | 61 | 33/537 | 34/599 | ||||||
| Pgp expression3-155 | NS | NS | NS | NS | NS | NS | ||||
| + | 87 | 50 | 14/370 | 16/286 | ||||||
| − | 90 | 63 | 37/352 | 24/245 | ||||||
| Pgp activity (n = 150)3-154 | .04 | .05 | .04 | .05 | .04 | .05 | ||||
| + | 48 | 50 | 0/321 | 8/231 | ||||||
| − | 102 | 68 | 14/360 | 33/348 |
| . | Patients, no. . | CR, % . | P3-150,3-151 . | P3-152 . | 3-y DFS, %/median d . | P3-150,3-153 . | P3-152 . | 3-y OS, %/median d . | P3-150,3-153 . | P3-152 . |
|---|---|---|---|---|---|---|---|---|---|---|
| Panmyeloid antigens | <.0001 | .02 | .02 | .04 | .008 | .04 | ||||
| + | 50 | 80 | 52/NR | 48/680 | ||||||
| − | 127 | 48 | 16/390 | 17/231 | ||||||
| Age | <.0001 | .001 | .01 | .02 | <.0001 | .002 | ||||
| <60 | 80 | 35 | 14/290 | 19/164 | ||||||
| ≥60 | 97 | 75 | 41/580 | 43/784 | ||||||
| WHO performance status | .01 | .01 | .01 | .03 | .005 | .02 | ||||
| <2 | 133 | 62 | 40/439 | 37/484 | ||||||
| ≥2 | 44 | 41 | 14/392 | 20/180 | ||||||
| Cytogenetic | .005 | NS | .01 | NS | .01 | NS | ||||
| Good | 22 | 82 | 59/NR | 53 | ||||||
| Intermediate | 113 | 59 | 33/518 | 31/345 | ||||||
| Poor | 32 | 34 | 12/189 | 19/201 | ||||||
| Failure or not done | 10 | 50 | ||||||||
| FAB subtypes | NS | NS | NS | NS | NS | NS | ||||
| M0 | 4 | 75 | 0/170 | 20/170 | ||||||
| M1 | 35 | 54 | 0/130 | 0/185 | ||||||
| M2 | 63 | 59 | 34/579 | 34/531 | ||||||
| M4 | 35 | 54 | 60/NR | 51/NR | ||||||
| M4E | 7 | 86 | 40/380 | 50/NR | ||||||
| M5 | 25 | 52 | 22/170 | 21/190 | ||||||
| M6 | 7 | 57 | 0/227 | 40/200 | ||||||
| WBC count, ×109/L | NS | NS | NS | NS | NS | NS | ||||
| ≥30 | 59 | 47 | 29/430 | 23/295 | ||||||
| <30 | 118 | 62 | 38/588 | 36/300 | ||||||
| LDH level, U/L | NS | NS | NS | NS | NS | NS | ||||
| ≥2000 | 48 | 46 | 32/350 | 32/277 | ||||||
| <2000 | 129 | 61 | 33/537 | 34/599 | ||||||
| Pgp expression3-155 | NS | NS | NS | NS | NS | NS | ||||
| + | 87 | 50 | 14/370 | 16/286 | ||||||
| − | 90 | 63 | 37/352 | 24/245 | ||||||
| Pgp activity (n = 150)3-154 | .04 | .05 | .04 | .05 | .04 | .05 | ||||
| + | 48 | 50 | 0/321 | 8/231 | ||||||
| − | 102 | 68 | 14/360 | 33/348 |
NR indicates not reached; NS indicates not significant.
Indicates univariate analysis.
Indicates that the Fisher exact test was used.
Indicates multivariate analysis.
Indicates that the log-rank test was used.
Staining with UIC2 was considered positive when D ≥ 0.15. This D value cutoff point was derived based on observations of previous works.21
Uptake of calcein-am was considered positive when D ≥ 0.3. This D value cutoff point was derived based on observations of previous works.21

DFS and OS of patients with or without all myeloid markers.
DSF is shown in panel A and OS in panel B.
In most cases, except for a few markers, these results were confirmed for both groups of patients: less than 60 years and 60 years or older (data not shown). For younger patients (less than 60 years), CDw65 was associated with a higher CR rate than older patients (90% vs 60%, respectively; P = .01). Similarly, for older patients (at least 60 years), CD34+ (P = .05), CD10 (P = .06), and TdT (P = .06) negatively influenced 3-year OS.
Other clinical and biological parameters influencing outcome of treatment
The factors influencing achievement of CR are summarized in Table 3. CR rate significantly decreased with increasing age (P < .0001), increasing WHO performance status (P = .01), cytogenetics (P = .005), and increasing Pgp activity (P = .04). However, CR rate was not associated with the other variables.
In univariate analysis (Table 3), the estimated probability of a 3-year DFS and OS, respectively, was significantly poorer for patients with unfavorable cytogenetics (P = .01 and P = .01). The probability decreased significantly with increasing age (P = .01 and P < .0001), increasing performance status (P = .01 and P = .005), and increasing Pgp activity (P = .04 and P = .04).
Multivariate analysis
In multivariate analysis, only age (P = .001), panmyeloid antigens (P = .02), WHO performance status (P = .01), and Pgp activity (P = .05) influenced achievement of CR (Table 3). A Cox multivariate analysis of DFS and OS was performed. The following patient characteristics (predictive, in univariate analysis, for an unfavorable outcome) were included in the model: age, WHO performance status, presence of all 5 myeloid markers (panmyeloid phenotype), cytogenetics, and Pgp activity. DFS and OS were influenced, respectively, by age (P = .02 andP = .002), the subgroup of panmyeloid phenotype (P = .04 and P = .04), WHO performance status (P = .03 and P = .02), and Pgp activity (P = .05 and P = .05).
Prognostic score
We have included in this prognostic score all independent prognostic factors: age (less than 60 years vs at least 60 years), WHO performance status (less than 2 vs at least 2), Pgp activity (D < 0.15 vs D ≥ 0.15), and the subgroup of panmyeloid phenotype (panmyeloid markers, less than 5 vs 5). We have pooled patients in accordance with the number of independent prognostic factors: good prognostic (no poor prognostic factors), intermediate prognostic (1 poor prognostic factor), and poor prognostic (2-4 poor prognostic factors). The estimated probability of 3-year DFS and OS for each group is shown in Figure 2A and 2B, respectively.
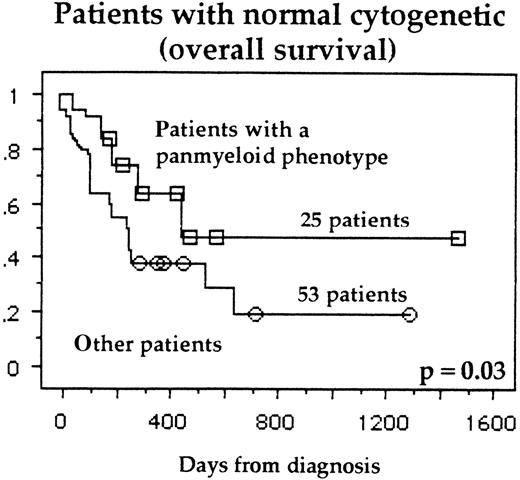
OS of patients with normal cytogenetics, expressing all 5 myeloid markers, or lacking 1 or more of these myeloid markers.
Expression of panmyeloid phenotype and correlation with in vitro resistance variables
We compared patients with leukemic blasts disclosing expression of all 5 myeloid markers with those who did not. The results showed, respectively: a lower activity of Pgp (0.37 ± 0.26 vs 0.52 ± 0.25, P = .05); a higher sensitivity to DNR (0.27 ± 0.27 μmol/L vs 0.78 ± 0.72 μmol/L,P = .04); and a higher sensitivity to AraC (8.5 ± 5.7 μmol/L vs 17.9 ± 16.4 μmol/L, P = .04) (Table1).
Normal cytogenetics and panmyeloid markers
In patients with normal cytogenetics, a favorable subgroup consisted of patients who had the following: expression of all 5 myeloid markers (Figure 3), low WHO score, young age, and low Pgp activity in univariate analysis. In this subgroup of normal cytogenetic patients, a multivariate analysis revealed that there were the same 4 good independent prognostic indicators of survival: young age (P = .03), WHO score (P = .03), Pgp activity (P = .04), and panmyeloid phenotype (P = .05). Using our prognostic score, the estimated probability of a 3-year DFS and OS in each group is shown in Figure 2C and 2D, respectively. The other subgroups of cytogenetics were too small to analyze the prognostic significance of the panmyeloid phenotype.
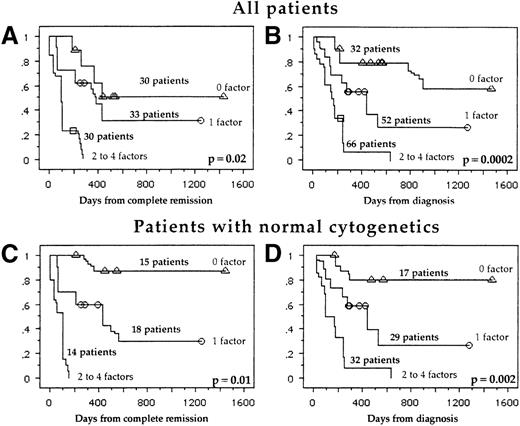
Score system stratifying AML patients.
DFS (A) and OS (B) of all patients and DFS (C) and OS (D) of patients with normal cytogenetics. The factors of poor prognosis include a WHO performance status greater than 2, a positive Pgp activity, expression of only 1-4 myeloid markers, and older age (at least 60 years).

Score system stratifying AML patients.
DFS (A) and OS (B) of all patients and DFS (C) and OS (D) of patients with normal cytogenetics. The factors of poor prognosis include a WHO performance status greater than 2, a positive Pgp activity, expression of only 1-4 myeloid markers, and older age (at least 60 years).
Discussion
The results concerning the prognostic implications of surface antigen expression in AML have been controversial.8-19 But the comparability of the results can be hampered by methodologic differences in the detection of antigen expression as well as by differences in patient populations studied and treatment regimens administered.8-19 Our study involved immunophenotyping examinations in addition to several clinical and biological parameters in a large number of adults with newly diagnosed AML.
Our results do not indicate that the expression of 1 antigen can be applied for risk stratification in adult AML at diagnosis. In fact, the majority of markers are associated with both poor and good prognostic factors. For example, several studies reported a poor response to induction chemotherapy in patients with CD34 and/or CD7 AML.10,15 In our study, CD34+ patients were associated with both good prognostic factors (cytogenetics of t(8;21) and Inv(16) and low WBC count at diagnosis) as well as poor prognostic factors (cytogenetics of poor prognosis and high Pgp activity). AML patients expressing 1 antigen, such as CD34 or CD7, do not comprise a sole biological entity of AML but correspond to a heterogeneous group. Using the EGIL classification,6,7 we have shown that patients expressing all the 5 myeloid markers had a better prognosis than other patients. Perhaps this subgroup of patients, expressing a combination of several markers, recognized a more homogeneous biological entity of AML. These patients have a younger age, a lower Pgp activity, and a lower in vitro resistance to DNR and AraC than other patients. Similarly, they are associated with particular FAB subtypes as well as chromosomal abnormalities. In the same way, normal cytogenetics (an intermediate prognostic factor)20 regroups patients with heterogeneous disease.33-36 In this subgroup of patients, those expressing all the 5 myeloid markers have a better prognosis than others. Therefore patients expressing the panmyeloid phenotype (28% of patients) defined a subgroup of AML with a particularly good prognosis.
Besides the panmyeloid phenotype, other independent prognostic factors identified in the current series included older age, higher Pgp activity, and higher performance status. Even if the treatment of younger and older patients was very similar, treating patients differently based on age may confound conclusions about the significance of age in determining treatment outcome. Cytogenetics was not selected by this multivariate analysis because of a probable redundancy with panmyeloid phenotype and a higher sensitivity of immunophenotyping. These features are key components of our prognostic model for adult AML. This model classified patients by 3 risk groups. This score stratifies patients well; innovative therapies may be used to improve outcome in the poorer outcome groups, while for patients with a better prognosis, diminished toxicity with standard effective therapy can be proposed.
The correlation between Pgp activity and CD34 in AML was first emphasized by Te Boekhorst et al37 and is now established.38,39 In accordance with other publications, we also found a strong correlation between CD34 expression and Pgp activity.39 However, the relationship of Pgp expression or activity to other markers largely remains unclear and a debated question.40,41 In this study, Pgp activity was positively correlated with CD34 expression and negatively correlated with CD10, CD14, CD33, CDw65, and TdT expression. In a recent study by Wuchter et al,42 CD14 and CD65 were also negatively correlated with Pgp activity. Interestingly, more of these markers (CD10, CDw65, and TdT) influenced treatment outcome in subgroups of patients (CDw65 for younger patients, CD10 and TdT for older patients) in our study. This also emphasized the importance of the patient population studied. In contrast to other markers, the positive value of the presence of all 5 myeloid markers was found in the whole population including older and younger patients. This displays the value of panmyeloid phenotype as prognostic factor.
In conclusion, the expression of the 5 myeloid markers is an independent prognostic factor for outcome in patients with adult AML. Patients who express all 5 myeloid markers probably define a relatively homogeneous biological entity of AML. The results from this study will be used to help develop treatment strategies that are based on the risk factors of the individual patient.
Supported in part by grant 9637 from Association de la Recherche sur le Cancer, Villejuif, France.
Reprints:Jean-Pierre Marie, Hôpital Hôtel-Dieu, 1 Place du Parvis Notre Dame, Service d'Hématologie, 75181 Paris Cedex 04, France; e-mail:jean-pierre.marie@htd.ap-hop-paris.fr.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal