Most, if not all, malignancies have recurrent and disease-specific clonal chromosomal abnormalities, which play a pivotal role in tumor development. These comprise deletions or amplifications involving entire chromosomes or subchromosomal regions and, also, translocations. The latter represent the juxtaposition of fragments of DNA that are usually on different chromosomes. Most translocations are not random, involving, in a given disease, circumscribed regions of DNA within both chromosomal partners and are furthermore, reciprocal, with DNA being interchanged between both partners, implying that both chromosomal ends have to be brought into close apposition.
Some chromosomal regions and localized areas within certain genes appear to be particularly prone to chromosomal aberrations, with specific abnormalities occurring within cells of a given lineage at specific stages of differentiation.1 While some genes are involved in translocations involving only one partner gene, others are involved with multiple different partners. TheMLL gene and, in particular, an 8.3-kilobase (kb) region (the breakpoint cluster region), is recurrently involved in a large number of different translocations, principally in acute myeloid leukemias.2
The immunoglobulin (IG) loci show comparable promiscuity in their translocation partners, and recently many new translocations have been identified, using defined breaks within the IG loci to clone the translocation breakpoint. Table 1 lists the cloned translocations involving the IG loci; several other recurrent IG translocations remain to be cloned.3-5Here, we first review the IG translocations and, secondly, how the involved genes may contribute to the pathogenesis of B-cell malignancies.
Translocations involving the immunoglobulin loci
| Disease . | Frequency . | Involved genes . |
|---|---|---|
| BCP-ALL | ||
| t(5;14)(q31;q32) | <1% | IL3 |
| t(1;14)(q21;q32) | <1% | BCL9 |
| B-NHL | ||
| Burkitt's lymphoma | ||
| t(8;14)(q24;q32) | 100% | MYC |
| Follicular lymphoma | ||
| t(14;18)(q32;q21) | ∼80% | BCL2 |
| t(1;22)(q22;q11) | <1% | FCGRIIB |
| Mantle cell lymphoma | ||
| t(11;14)(q13;q32) | >95% | Cyclin D1 |
| DLCL | ||
| t(14;18)(q21;q32) | 20% | BCL2 |
| t(8;14)(q24;q32) | 10% | MYC |
| t(3;14)(q27;q32) | 5-10% | BCL6 |
| t(14;15)(q32;q11-13) | <1% | BCL8 |
| t(10;14)(q24;q32) | <1% | NFKB2 |
| Marginal zone lymphoma | ||
| Lymphoplasmacytoid lymphoma | ||
| t(9;14)(p13)(q32) | 50% | PAX5 |
| MALT | ||
| t(1;14)(p22;q32) | <5% | BCL10 |
| SLVL | ||
| t(7;14)(q21;q32), t(2;7)(p12;q21) | <5% | CDK6 |
| Others | ||
| t(11;14)(q23;q32) | <1% | PAFAH2 |
| t(11;14)(q23;q32) | <1% | RCK |
| t(12;14)(q24;q32) | <1% | BCL7A |
| t(12;22)(p13;q11) | <1% | Cyclin D2 |
| CLL | ||
| t(14;18)(q32;q21) | 1-2% | BCL2 |
| t(14;19)(q32;q13) | <1% | BCL3 |
| t(2;14)(p13;q32) | <1% | BCL11A |
| Myeloma | ||
| t(11;14)(q13;q32) | 20-25% | Cyclin D1 |
| t(4;14)(p16;q32) | 20-25% | FGFR3 |
| MMSET | ||
| t(6;14)(p25;q32) | ∼20% | IRF4 |
| t(14;16)(q32;q23) | 20-25% | C-MAF |
| t(1;14)(q21;q32) | <5% | MUM2/3 |
| Disease . | Frequency . | Involved genes . |
|---|---|---|
| BCP-ALL | ||
| t(5;14)(q31;q32) | <1% | IL3 |
| t(1;14)(q21;q32) | <1% | BCL9 |
| B-NHL | ||
| Burkitt's lymphoma | ||
| t(8;14)(q24;q32) | 100% | MYC |
| Follicular lymphoma | ||
| t(14;18)(q32;q21) | ∼80% | BCL2 |
| t(1;22)(q22;q11) | <1% | FCGRIIB |
| Mantle cell lymphoma | ||
| t(11;14)(q13;q32) | >95% | Cyclin D1 |
| DLCL | ||
| t(14;18)(q21;q32) | 20% | BCL2 |
| t(8;14)(q24;q32) | 10% | MYC |
| t(3;14)(q27;q32) | 5-10% | BCL6 |
| t(14;15)(q32;q11-13) | <1% | BCL8 |
| t(10;14)(q24;q32) | <1% | NFKB2 |
| Marginal zone lymphoma | ||
| Lymphoplasmacytoid lymphoma | ||
| t(9;14)(p13)(q32) | 50% | PAX5 |
| MALT | ||
| t(1;14)(p22;q32) | <5% | BCL10 |
| SLVL | ||
| t(7;14)(q21;q32), t(2;7)(p12;q21) | <5% | CDK6 |
| Others | ||
| t(11;14)(q23;q32) | <1% | PAFAH2 |
| t(11;14)(q23;q32) | <1% | RCK |
| t(12;14)(q24;q32) | <1% | BCL7A |
| t(12;22)(p13;q11) | <1% | Cyclin D2 |
| CLL | ||
| t(14;18)(q32;q21) | 1-2% | BCL2 |
| t(14;19)(q32;q13) | <1% | BCL3 |
| t(2;14)(p13;q32) | <1% | BCL11A |
| Myeloma | ||
| t(11;14)(q13;q32) | 20-25% | Cyclin D1 |
| t(4;14)(p16;q32) | 20-25% | FGFR3 |
| MMSET | ||
| t(6;14)(p25;q32) | ∼20% | IRF4 |
| t(14;16)(q32;q23) | 20-25% | C-MAF |
| t(1;14)(q21;q32) | <5% | MUM2/3 |
IG translocations are shown here classified by disease subtype together with their frequency of occurrence in that disease and the gene deregulated by the translocation. In some cases, variant translocations involving the IG light chain loci are also found; these have generally been omitted for clarity.
Cytogenetic abnormalities of the IG loci in B-cell malignancies
Chromosomal translocations involving the IG loci are found in some, but not all, forms of B-cell malignancy. B-cell precursor acute lymphoblastic leukemia (BCP-ALL) and B-cell chronic lymphocytic leukemia (CLL) have a low frequency of IG translocations, at least at the cytogenetic level. Some IG translocations are seen in all cases of a specific subgroup of disease. Paradigms include the involvement of MYC in all cases of Burkitt lymphoma (BL) and ofcyclin D1/BCL1 in all cases of mantle cell lymphoma.6,7 These translocations may therefore be useful as a diagnostic marker.8
In contrast, cytogenetically identical translocations may be found in several different types of disease. However, molecular analysis has shown that the same translocations in different diseases may involve different breakpoints, either within the incoming oncogene or within the IG locus.9,10 For example, sporadic BL with t(8;14)(q24;q32) shows a different spectrum of IGH breakpoints than those found in endemic (African) cases with the same translocation.9 Similarly, BCL2/IG translocations occur in both follicular lymphoma (in about 80% of cases) and, more rarely, in CLL (about 2% of cases). The former involves the 3′ portion of the BCL2 gene on chromosome 18q21 (Figure 1), whereas the latter usually involves the 5′ region of the same gene.10 These data indicate that translocations, although cytogenetically identical, may arise by different mechanisms.
t(14;18)(q32;q21) translocation.
(A) The BCL2 gene, located at chromosome 18q21.3, consists of 3 coding exons separated by an intron of about 250 kb and is involved inIG translocations in about 80% of follicular B-NHL and 1% to 2% of all cases of CLL. The transcriptional orientation of the gene is from telomere to centromere. The BCL2 open reading frame is denoted by the shaded region, and IGH switch regions are hatched. MBR denotes major breakpoint region; mcr, minor cluster region; vcr, variant cluster region; icr, intermediate cluster region. (B) Typical IGH-BCL2 translocation of follicular B-NHL involving the BCL2 MBR within the 3′ UTR of the gene and the JH segments. Note that nearly all cases of follicular B-NHL with this translocation have undergone IGH class-switch deletion. A BCL2-IGH fusion mRNA is produced. (C) A vcr rearrangement typical of CLL with t(14;18)(q32;q21). Here, the 5′ portion of the BCL2 gene, which is about 250 kb centromeric of the 3′ UTR, becomes juxtaposed with the IGHJ segments in opposite transcriptional orientation. This implies that this translocation must be associated with inversion within either chromosome 18 or 14 and suggests a different pathogenic mechanism. The precise anatomy of this type of translocation remains to be determined.
t(14;18)(q32;q21) translocation.
(A) The BCL2 gene, located at chromosome 18q21.3, consists of 3 coding exons separated by an intron of about 250 kb and is involved inIG translocations in about 80% of follicular B-NHL and 1% to 2% of all cases of CLL. The transcriptional orientation of the gene is from telomere to centromere. The BCL2 open reading frame is denoted by the shaded region, and IGH switch regions are hatched. MBR denotes major breakpoint region; mcr, minor cluster region; vcr, variant cluster region; icr, intermediate cluster region. (B) Typical IGH-BCL2 translocation of follicular B-NHL involving the BCL2 MBR within the 3′ UTR of the gene and the JH segments. Note that nearly all cases of follicular B-NHL with this translocation have undergone IGH class-switch deletion. A BCL2-IGH fusion mRNA is produced. (C) A vcr rearrangement typical of CLL with t(14;18)(q32;q21). Here, the 5′ portion of the BCL2 gene, which is about 250 kb centromeric of the 3′ UTR, becomes juxtaposed with the IGHJ segments in opposite transcriptional orientation. This implies that this translocation must be associated with inversion within either chromosome 18 or 14 and suggests a different pathogenic mechanism. The precise anatomy of this type of translocation remains to be determined.
IG translocations are usually detected by cytogenetics often supplemented with fluorescence in situ hybridization (FISH) methods using probes that span the IG loci11; the use of fiber-FISH is an elegant, although technically demanding, method to define breakpoints precisely.12 Some IGtranslocations, such as the t(4;14)(p16;q32) in myeloma, are cytogenetically cryptic due to the telomeric position of the breakpoints on the partner chromosomes and therefore need to be demonstrated by FISH.13 Cytogenetically cryptic translocations appear to be prevalent in myeloma.14 Whether similarly undetectable IG translocations are present in other diseases such as BCP-ALL remains to be determined. The use of multicolor FISH methods including spectral karyotyping has allowed the identification of previously unrecognized IG translocations in myeloma, but these have yet to be applied systematically to all diseases.15 16
IG translocations may be multiple, with different translocations involving IGH and IGL. They may occur early in the course of disease or may occur at the time of transformation from low- to high-grade disease.17 Multiple translocation events may occur at the IGHlocus.18,19 Alternatively, similar and even more complex events may occur in the absence of cytogenetically obvious translocation and may represent insertion of IGsequences.20,21 It has been shown recently that theIGH locus has been inserted into the cyclin D1/BCL1locus in a myeloma cell line, while insertion of BCL2 into the IGH locus has been observed in patients with follicular non-Hodgkin lymphoma (NHL).22 23
The origins, analysis, and consequences of IGtranslocations
Normal B cells undergo a series of double-stranded DNA breaks to produce a functional IG protein with high affinity for antigen.24 These may predispose to the targeting of translocations to the IG loci. Although the mechanisms underlying the pathogenesis of IG translocations remain unclear, it is likely that errors in each of the interrelated processes along with defects in other DNA repair genes may play a major role.
In support of this theory is the observation that most, but not all,IG translocations are targeted to sites commonly involved in normal recombination events (ie, the J segments, and, in the case ofIGH translocations, the S regions). Targeting of oncogenes to the JH regions suggests that such translocations might arise due to errors of VDJ recombination in B-cell precursors within the bone marrow. However, this is not likely: In most cases, analysis of both the productive VDJ rearrangements and both derivative chromosomes shows that translocation arises after antigen is encountered and IGH class-switching has occurred (ie, in mature B cells).12,25-27 Targeting of oncogenes to the J regions in malignancies of mature B cells may in part be explained by the reactivation of recombination activation genes (RAG) 1/2 in germinal center (GC) B cells.28
Molecular cloning of chromosome 14q32/IGH translocations has generally proceeded on the assumption that in most instances the translocation will involve either the J or the S regions. However, this is not always the case, and IG translocations may involve rearranged and unrearranged VH/VL genes, DH segments, and the JH-Cμintron, as described below and in fiber-FISH experiments on myeloma cell lines.29
The principal molecular consequences of IG translocation are deregulated expression and, in certain instances, mutation of the translocated gene. First, deregulated expression of the incoming oncogene arises due to the close physical proximity of potent B-cell transcriptional enhancers within the IG loci. This may result in abnormally high levels of protein expression or loss of normal mechanisms of control, such as the cell cycle–regulated expression ofMYC. Secondly, in B cells where the somatic hypermutation mechanism is active, mutation of the incoming gene may be observed and may contribute to the neoplastic phenotype. BCL2 mutation in cases with t(14;18)(q32;q21) has, for example, been associated with transformation of low-grade disease.30
However, other events associated with IG translocations may also occur, including, in the t(14;18)(q32;q21), deletion of control elements within the 5′ region of the translocated BCL2gene (some 250 kb telomeric of the breakpoint) and deletion within theIGH locus.12,31 The consequences of these events are not known. Similar deletions and rearrangements often hundreds of kilobases distant from the breakpoint may be observed in otherIG translocations.7 The whole of the derivative chromosome 14 may be destabilized, perhaps as a consequence of theIG translocation, with additional breaks occurring centromeric of the IGH locus, resulting in complex translocations and amplifications.32
It had been assumed that only one gene would be targeted as a result ofIG translocations and that this gene would be found on the derivative chromosome 14, close to the site of theIG-associated transcriptional enhancers. However, in translocations to S regions the intronic IG enhancer is translocated to the other partner and this, in some instances at least, may result in deregulated expression of another gene also. So far, this has been shown for the t(4;14)(p16;q32) in myeloma, where a single translocation results in deregulated expression of 2 genes (Figure2; see also “Myeloma”); whether the same phenomenon is found in other translocations to S regions remains unknown.33 34
t(4;14)(p16;q32) translocation in myeloma.
Derivative chromosomes 4 and 14 of myeloma. IGH breaks in this malignancy are usually found within the S regions. (A)FGFR3 expression is driven by the centromeric IGHtranscriptional enhancer (Eα) on the der(14), whileMMSET expression (B) is driven by the intronic enhancer (Eμ) on the der(4). Tel indicates telomere.
t(4;14)(p16;q32) translocation in myeloma.
Derivative chromosomes 4 and 14 of myeloma. IGH breaks in this malignancy are usually found within the S regions. (A)FGFR3 expression is driven by the centromeric IGHtranscriptional enhancer (Eα) on the der(14), whileMMSET expression (B) is driven by the intronic enhancer (Eμ) on the der(4). Tel indicates telomere.
IG translocations in B-cell malignancies
The various IG translocations in malignant B cells are reviewed below according to cytologic subtype of disease.8While the acute leukemias are associated with only a single chromosomal translocation, malignancies of mature B cells often exhibit enormous cytogenetic complexity, with multiple and complex translocations, deletions, and amplifications in the 1 clone. This is consistent with the slower natural history of these diseases. It also concurs with the hypothesis that several genetic abnormalities—including the concurrent activation of dominant oncogenes and inactivation of tumor suppressor genes—are necessary for the full neoplastic phenotype to emerge. Some of the additional changes have been associated with transformation from low- to high-grade disease.
Furthermore, while chromosomal translocations in ALL and acute myeloid leukemia usually result in fusion of transcription factors controlling early hemopoietic differentiation and development,IG translocations involve a plethora of different molecules that influence many different pathways. Some of these pathways are described here in detail in the context of specific chromosomal translocations.
B-cell precursor acute lymphoblastic leukemia
BCP-ALL only rarely exhibit IG translocations, which is perhaps surprising because these malignancies constitutively express RAG1/2 enzymes. However, the frequency of IG translocations may have been underestimated by cytogenetics alone. At least 10% of cases of BCP-ALL demonstrate deletions of the IGHJ segments, usually on one allele but sometimes on both.35 In most, both Cμ and Cδ are deleted with retention of Cγ3. Mapping of the breakpoints revealed that these breakpoints fell within the central portion of the intervening sequence between Cδ and Cγ3. The significance of this observation remains unknown because this portion of theIGH locus is genetically unstable and remains uncloned. Whether these unusual deletions represent internal IGH rearrangements or whether they mark cytogenetically cryptic translocations is not known.
t(5;14)(q31;q32)—interleukin-3(IL−3).
A distinct but rare subset of BCP-ALL is characterized by eosinophilia and t(5;14)(q31;q32).36 Molecular cloning demonstrated juxtaposition of JH and the IL-3 gene in a head-to-head configuration, with breakpoints occurring upstream of theIL-3 promoter, resulting in IL-3 overexpression, which is detectable in the serum. The granulocyte-macrophage colony-stimulating factor gene located only 14 kb downstream of the breakpoint—and therefore potentially within range of the IGH enhancer—is not overexpressed in this disease.
t(1;14)(q21;q32)—BCL9.
Abnormalities of chromosome 1q21-23 are common in all forms of B-cell malignancy and have been reported to be associated with poor response to therapy.37 The most common abnormality is duplication of the region: FISH analysis has shown that some of these duplications can be complex.
The t(1;14)(q21;q32) is not specific for BCP-ALL and has been reported in nearly all types of B-NHL as well as myeloma. However, the involved breakpoints on chromosome 1 appear to be specific for a given disease, and additional 1q21 breakpoints in other diseases are described below.
BCL9 was identified from the cloning of a t(1;14)(q21;q32) translocation in a pre-B ALL cell line in which this was the sole cytogenetic abnormality.38 Breaks occurred within the 3′ untranslated region (UTR) of BCL9 and within JH. BCL9 sequence analysis predicted a protein of 1394 amino acids with no known homologies. Deregulated expression ofBCL9 was seen in the cell line carrying the translocation but not in a panel of BL cell lines with other 1q21 abnormalities. Rearrangements of the locus were seen in 2 of 39 cases of B-NHL of other subtypes, including 1 case with a variant t(1;22)(q21;q11) involving the IGλ locus. At present, the function ofBCL9 and the mechanism by which this gene may mediate B-cell transformation remain unknown.
t(9;14)(p21;q32)—cyclin-dependent kinase inhibitor 2 (CDKN2).
Chromosome 9p21 harbors the CDKN2A/B tumor suppressor locus, which commonly undergoes biallelic deletion in B- and T-cell precursor ALL. In one case of BCP-ALL, the translocation t(9;14)(p21;q11) involved the T-cell receptor TCRD/A locus with this locus, resulting in inactivation of the p16/p19 ARF gene.39 This observation is of interest because it illustrates that some IGtranslocations may involve tumor suppressor genes and result in loss of gene expression. A parallel translocation t(9;14)(p21;q32) has been described in a BCP-ALL cell line, although whether this involves the same gene on chromosome 9p21 remains to be determined.40
Burkitt lymphoma/B-cell ALL Fab type L3
t(8;14)(q24;q32)—MYC.
BL is characterized by translocations involving the MYC gene at chromosome 8q24.6 In 75% of cases, a reciprocal translocation t(8;14)(q24;q32) is seen between the MYC gene and the IGH locus while, in the remainder, the reciprocal translocations t(8;22)(q24;q11) or t(2;8)(p12;q24) are seen, juxtaposing MYC to one of the IG light chain loci. These translocations are not specific for BL and are seen in many other subtypes of B-cell malignancy. They may occur as a secondary event in transformation of follicular B-NHL to high-grade disease.17 41 Variant translocations involving MYCwith a variety of other non-IG loci have also been reported.
The MYC gene consists of 2 coding exons and a noncoding first exon and, in most cases of sporadic BL, the breakpoints on 8q24.1 are located 5′ of the coding region, either in the first intron, within the first exon, or 5′ of the first exon. In contrast, in most cases of endemic BL and in cases with variant translocations, theMYC breakpoints may be considerable distances centromeric or telomeric from the MYC coding exons. In addition to the translocation, approximately 65% of BLs demonstrate MYC point mutations.42
Myc expression is normally tightly linked to the early G1phase of the cell cycle. As a result of translocation to the IGloci, control of normal Myc expression is lost, and the protein is constitutively expressed throughout the cell cycle. Despite much work, the precise functions and roles of Myc in oncogenesis remain unclear. Myc can function both as transcriptional activator and transcriptional repressor, and the identification of Myc target genes remains a major goal. Myc can function as an inducer of apoptosis and as an inducer of proliferation. These functions have been reviewed recently.43
Mantle cell lymphoma
t(11;14)(q13;q32)—BCL1/cyclin D1.
The translocation t(11;14)(q13;q32) is found in a wide variety of B-cell malignancies, including mantle cell lymphoma (MCL), splenic lymphoma with villous lymphocytes (SLVL), B-cell prolymphocytic leukemia, and multiple myeloma.44 All cases of MCL exhibit the t(11;14)(q13;q32) or, rarely, variant translocations to theIG light chains.45 The result in MCL is the juxtaposition of the0 locus originally referred to as BCL1 on chromosome 11q13 to an IGHJ segment. However, in some cases of B-NHL and in multiple myeloma, translocations occur instead toIGHS regions.46 The consequences of the translocation are the overexpression of cyclin D1, a D-type cyclin involved in control of the cell cycle. Although some of the breakpoints on 11q13 fall within the major translocation cluster, most are dispersed over a region of about 130 kb centromeric of the cyclin D1 gene.7 44
Follicular B-NHL
t(14;18)(q32;q21)—BCL2.
Approximately 80% of cases of follicular lymphoma are associated with t(14;18)(q32;q21). This results in juxtaposition ofBCL2 on chromosome 18q21.3 with IGHJ. The breakpoints within IGH all fall within JH, while the breakpoints on chromosome 18q21 occur mostly within the 3′ region of the gene (Figure 1). Unlike the breaks within the BCL1locus, BCL2 breaks are clustered. Fifty percent of the 3′BCL2 breaks falls within a major breakpoint region, a 150-breakpoint region within the 3′ UTR of the gene, and another 25% falls in the minor cluster region. Others cluster within a third, intermediate cluster region midway between the major breakpoint region and minor cluster region,47,48 while other breakpoints have been described scattered through this region. The remaining breakpoints on chromosome 18 fall within the 5′ noncoding region of the gene. 5′ BCL2 breaks are found predominantly, although not exclusively, in B-CLL and are involved predominantly in translocation to the IG light chain loci.49
The result of the translocation is deregulated expression of the 26-kd BCL2 protein, one of a family of proteins involved in the regulation of apoptosis.
t(1;22)(q22;q11)—Fc gamma receptor IIB (FcγRIIB/CD32).
A recurrent abnormality in follicular lymphoma is the t(1;22)(q22;q11) translocation, which arises at the time of transformation to high-grade disease.50 The target gene of this translocation is the IgG Fc gamma receptor IIB (FCGRIIB), a member of the low-affinity IgG Fc family of transmembrane receptors. In this case, the breakpoint was located approximately 20 kb upstream of the FCGRIIB.
Diffuse large-cell lymphoma
Unlike follicular B-NHL, diffuse large-cell lymphoma (DLCL) exhibits a plethora of different IG translocations.4 Some of these are recurrent, while others appear to be unique. The t(14;18)(q32;q21) in about 30% of DLCL is thought to represent the transformation of follicular B-NHL. The detection of this translocation in DLCL appears to have little prognostic significance, but high-level BCL2 protein expression, which correlates poorly with the presence of t(14;18), is a predictor of reduced overall and disease-free survival in DLCL.51 52IG translocations are found in about 50% of DLCL cases. Whether the DLCL cases that lack IGtranslocations constitute a distinct histologic entity with distinctive biological behavior remains to be determined.
t(3;14)(q27;q32)—BCL6.
DLCL is frequently associated with reciprocal translocations involving chromosomal band 3q27 and either one of the IG genes or another partner chromosome.53 Cloning of the t(3;14)(q27;q32) translocation allowed the identification of BCL6, a zinc-finger transcription factor.54-56 Subsequent studies have demonstrated that rearrangements of the BCL6 gene are found in 30% to 40% of DLCL, 5% to 14% of follicular lymphoma, and 20% of acquired immunodeficiency syndrome–associated DLCL, resulting in juxtaposition of the BCL6 coding region to heterologous promoters.57 The 5′ noncoding region of BCL6undergoes somatic point mutation, independent of translocation, not only in approximately 70% of DLCL and 45% of follicular lymphoma, but also in normal GC B cells.58-60
In one series, rearrangements of BCL6 were associated with extranodal presentation and a favorable clinical outcome.61However, this has not been duplicated in other studies, andBCL6 rearrangements do not seem to be associated with a particularly bad or good clinical subgroup of DLCL.62 63
BCL6 is expressed at low levels in multiple tissues, but in B cells its expression is tightly regulated. It is highly expressed only in mature B cells and within the GCs of lymph nodes but not in pre-GC cells or in differentiated plasma cells.64 However, in the mantle zone, there is expression of BCL6 messenger RNA (mRNA) but not protein.
BCL6 is a 92- to 98-kd nuclear phosphoprotein65 that contains 6 Krüppel-type zinc-finger motifs in its carboxy-terminal region and an amino-terminal POZ/BTB motif shared by various zinc-finger proteins.66,67 The amino-terminus contains a sequence-specific transcriptional repressor domain mediated by a region that includes the POZ motif.68,69 B-cell receptor (BCR)-mediated signaling via mitogen-activated protein kinase results in phosphorylation of BCL6 at Ser333 and Ser343, residues contained in PEST sites within the serine- and proline-rich central region, resulting in subsequent targeting for ubiquitination and proteosomal degradation.70 71
Gene inactivation studies have shown that BCL6 plays a key role in the activation and proliferation of B cells within the GC.72 Loss of the normal control mechanisms regulating its expression contributes to malignant transformation in GC-derived lymphomas. However, overexpression of the gene results in apoptosis and delays S-phase progression,73 and it remains unclear how BCL6 overexpression leads to transformation. No mouse transgenic models for BCL6 have been reported to date.
t(14;15)(q32;q11-13)—BCL8.
Abnormalities of 15q11-13, including the translocation t(14;15)(q32;q11-13), are seen in 3% to 4% of DLCL.74 A t(14;15)(q32;q11-q13) breakpoint has been cloned from a patient with DLCL, allowing the isolation of a new gene, BCL8, adjacent to the breakpoint. Interestingly, in this case the breakpoint fell upstream of a rearranged VH segment. The BCL8 locus was rearranged in approximately 4% of cases of DLCL by Southern blot. The gene is normally expressed only in prostate and testis, with no detectable expression in spleen or thymus. BCL8 expression was detected in all patients with 15q11-13 abnormalities analyzed and in some without such abnormalities but not in normal lymphocytes. Deregulated expression of BCL8 may therefore contribute to the pathogenesis of a subset of DLCL cases. The functions of this gene remain to be determined.
t(10;14)(q24;q32)—NFKB2.
The chromosome band 10q24 is involved in heterogeneous aberrations, including deletions and translocations to IGH in about 5% of low-grade B-NHL and less frequently in intermediate- to high-grade B-NHL.75 Molecular cloning of a t(10;14)(q24;q32) translocation breakpoint involving the Cαlocus led to the identification of NFKB2 adjacent to the breakpoint.76 The NFKB2 gene encodes the protein products p52 and p100, members of the Rel/nuclear factor (NF)-κB family.
Marginal zone lymphomas
t(9;14)(p13;q32)—paired homeobox 5 (PAX5).
Lymphoplasmacytoid lymphoma (LPL)/immunocytoma is a rare subtype of lymphoma that progresses slowly, if at all, and only rarely transforms.77 It is thought to originate from peripheral B cells that have been stimulated to undergo plasma cell differentiation.
Translocations involving chromosome 9p13 are frequently seen in LPL and involve various chromosome loci, including 1q25, 3q27, 7q11, 12q13, 12q21, 14q32, and 19p13.78
The t(9;14)(p13;q32) was initially reported in the DLCL cell line KIS-1 and is detectable in 50% of LPL.77 Molecular characterization of this translocation has shown the IGH locus to be juxtaposed in a head-to-head configuration to the PAX5locus on chromosome 9p13, resulting in deregulated expression ofPAX5 mRNA.79 80 Five of 7 cases with 9p13 translocations demonstrated rearrangements by FISH with a YAC probe spanning the PAX5 locus. The breakpoints, however, were highly heterogeneous, and whether PAX5 is the sole target gene of 9p13 translocations is unclear.
t(1;14)(p22;q32)—BCL10.
Lymphomas of the mucosa-associated lymphoid tissue (MALT) are the most common form of lymphoma arising in extranodal sites, in most cases arising in the gastric mucosa.
Low-grade MALT lymphoma is an indolent disease usually preceded by chronic inflammation, either chronic Helicobacter pyloriinfection of the stomach or autoimmune disease such as Hashimoto's thyroiditis. Low-grade gastric MALT lymphoma is often initially dependent on H pylori for growth, as some cases regress with antibiotic therapy alone.81 The molecular events leading to the development of H pylori–independent growth and high-grade transformation are not well understood. However, cytogenetic studies of aggressive low-grade cases have identified abnormalities of chromosome 1p22, particularly translocation t(1;14)(p22;q32), as uncommon but recurrent events.
Molecular analysis of the t(1;14)(p22;q32) translocation breakpoint in MALT lymphoma allowed the identification of the BCL10 gene juxtaposed in a head-to-head configuration withIGHJ.82,83 The 1p22 breakpoints in other cases of MALT lymphoma exhibiting the same translocation also map to this region. BCL10 is widely expressed in normal tissues but is highly expressed within lymph node GCs82 and in fetal liver, lung, and parts of the developing neurologic system.84
t(2;7)(p12;q21); (7;14)(q21;q32)—CDK6.
A case of CLL with the translocation t(7;14)(q21;q32) originally demonstrated the juxtaposition of IGH with a human endogenous retroviral sequence of the transposable-like human element family.85 Cloning of t(2;7)(p12;q21) in SLVL has subsequently revealed that the target gene for this translocation is the CDK6 gene,86 one of the cell cycle–associated kinases that promotes G1 to S phase transition.87 It is likely that all 7q22 translocations involving CDK6 are specific to SLVL because chromosome 7q22 is a recurrent breakpoint in this disease and not CLL.
Interestingly, the t(2;7)(p12;q21), the breakpoint on chromosome 2 involved an unrearranged Vκ gene through the 3′ heptamer nanomer recombination signal sequence (RSS).
B-cell chronic lymphocytic leukemia
t(14;19)(q32;q13)—BCL3.
The recurrent translocation t(14;19)(q32;q13) is a rare cytogenetic abnormality found in CLL but is associated with a young age of presentation and a poor prognosis. The result of the translocation is to juxtapose BCL3 and IGH in a head-to-head configuration, resulting in overexpression of BCL3 mRNA due to the 3′ IGH enhancer.88
t(2;14)(p13;q32)—BCL11A.
The translocation t(2;14)(p13;q32) is a rare but recurrent abnormality in what has been described as “childhood chronic lymphocytic leukemia.” The translocation breakpoints for 2 such cases have been cloned.89 Translocations involving the same breakpoint have been observed in a subgroup of adult patients with an aggressive atypical CLL/B-NHL (Takashi Sonoki, Tony Willis, Martin Dyer, Reiner Siebert, unpublished observations, January 2000).
t(12;22)(p13;q11)—cyclin D2 (CCND2).
Transformation of CLL to high-grade B-NHL (Richter's syndrome) occurs in about 5% of patients. A case of Richter's syndrome, which acquired a t(12;22)(p13;q11) as a secondary event during high-grade transformation, demonstrated juxtaposition of the IGλ locus and CCND2, with the additional loss of the negative regulatory elements in the upstream segment of the CCND2promoter.90
Myeloma
All cases of myeloma may exhibit IG translocations. Like DLCL, these involve numerous partner chromosomes and, like DLCL, some of these may be recurrent, while others are either unique or rare.14-16 Consistent with the mature B-cell phenotype of the disease and the expression of Ig isotypes other than IgM, most translocations occur to the IGH S regions.29 Many genes have now been shown to be involved in these translocations, but their clinical significance remains to be determined.
t(4;14)(p16;q32)—FGFR3 and MMSET/WHSC1.
The karyotypically silent t(4;14)(p16;q32) translocation is detected in approximately 20% to 25% of multiple myeloma cases and cell lines.
The effects of this translocation are unusual in that there is deregulated expression of 2 genes (Figure 2), first, on the der(14) chromosome of the fibroblast growth factor receptor 3 (FGFR3)91,92 and, secondly, on the der(4) of a novel SET domain-containing gene, MMSET (multiple myeloma SET domain), or WHSC1 (Wolf-Hirschhorn syndrome candidate 1).33 34
Cloning of the t(4;14)(p16;q32) translocation initially identifiedFGFR3 on the der(14) as the target. In each case, the breakpoint was situated 30 to 100 kb upstream of theFGFR3 gene (Figure 2A). FGFR3 is normally expressed in the developing central nervous system and in cartilage growth plates but is undetectable in lymphoid tissues; the t(4;14)(p16;q32) is associated withoverexpression of the FGFR3 transcript due to the effects of the 3′ IGH enhancer.
The FGFRs are tyrosine kinase receptors capable of binding 9 related mitogenic fibroblast growth factors (FGFs).93 Ligand binding promotes receptor homodimerization or heterodimerization, leading to complex signaling pathways regulating cell growth, survival, and differentiation. FGFR3 may promote or inhibit cell proliferation depending on cell type. FGFR3 inhibits proliferation of chondrocytes, and dominant, activating mutations in different domains of FGFR3 result in excessive inhibition of chondrocyte proliferation, resulting in dwarfism, including hypochondroplasia, achondroplasia, and the most severe form, thanatophoric dysplasia.94 However, transfection of FGFR3 into an IL3-dependent B–cell line allows IL3-independent mitogenic stimulation.95
In some cases of myeloma with t(4;14)(p16;q32), overexpression of anFGFR3 allele containing an activating mutation associated with thanatophoric dysplasia have been detected.91 One of these (Lys650Glu) has been shown by transient transfection experiments to cause ligand-independent constitutive activation of FGFR3 by autophosphorylation96 and to promote IL-6 independence in murine IL-6–dependent B cells.97 This observation may at least in part be explained by the finding that FGFR3 overexpression, like IL-6, upregulates BCL-xLexpression and inhibits apoptosis of IL-6–dependent B cells.98 Deregulated expression of unmutatedFGFR3 may allow the continuous transduction of pro-proliferative signals from bone marrow stromal cells expressing FGFs, whereas subsequent mutation of FGFR3 would allow FGF-independent growth.
Further analysis of the t(4;14)(p16;q32) translocation has demonstrated that the breakpoints on 4p16 occur telomeric to and within the 5′ introns of a novel gene, MMSET orWHSC1,33 34 which is in the opposite transcriptional orientation to the FGFR3 gene and is also deregulated as a result of the translocation (Figure 2B). TheMMSET gene is alternatively spliced, resulting in 2 different transcripts. The type I transcript contains an open reading frame coding for a 647–amino acid protein, whereas the type II transcript encodes a protein of 1365 amino acids. IGH-MMSET hybrid transcripts originating from the Iμ promoter were detected in cases and cell lines demonstrating the t(4;14) translocation but would not be expected to result in fusion proteins. The functions of MMSET, and its possible contribution to myeloma pathogenesis, are unknown.
t(6;14)(p25;q32)—IRF4.
The t(6;14)(p25;q32) is another recurrent translocation in myeloma, which cannot be easily detected because of the subtelomeric position of the 6p25 breakpoint, but it occurs in about 20% of patients.99 In the myeloma cell line SKMM1, the breakpoint on 6p25 was located just 3′ of the IRF4 (interferon regulatory factor 4) gene.100 The IRF family of transcription factors includes at least 6 molecules that share homology in their amino-terminal DNA-binding domains and are active in the regulation of gene expression in response to signaling by interferons and by other cytokines.101 IRF4 promoted transformation in vitro when overexpressed in fibroblasts100 and has been shown to control the differentiation of B cells into plasma cells and their proliferation in response to various mitogenic stimuli. IRF4-deficient mice develop progressive generalized lymphadenopathy and do not mount detectable antibody, cytotoxic, or antitumor responses, indicating a requirement for IRF4 in B- and T-cell homeostasis.102
t(14;16)(q32;q23)—c-MAF.
Approximately 25% of cases of multiple myeloma display the translocation t(14;16)(q32;q23). Cloning of translocation breakpoints from 4 myeloma cell lines with this abnormality demonstrated that these were dispersed over an approximately 500-kb region centromeric to theC-MAF proto-oncogene at 16q23.103 A further cell line with a variant t(16;22)(q23;q11) involving IGλ contained a breakpoint telomeric to C-MAF so that the breakpoints in these cell lines bracketed the C-MAF locus. Deregulated expression ofC-MAF mRNA of 2.3 and 4.4 kb was detected in these cell lines, coding for predicted proteins of 403 and 373 amino acids, respectively.
C-Maf is a member of the bZIP family of transcription factors, which includes c-Jun, c-Fos, NF-IL3, and NF-IL6. The viral homologue, v-Maf, is an oncogene produced by an avian transforming virus, and wild type c-Maf is able to transform fibroblast cells.104 Other members of the basic zipper family are involved in control of many cellular processes, including proliferation, differentiation, and responsiveness to IL6. The mechanism whereby c-Maf might contribute to myeloma pathogenesis is not known.
t(1;14)(q21;q32)—MUM2/MUM3.
Two genes within a 20-kb region spanning the 1q21 breakpoint in a myeloma cell line demonstrating t(1;14)(q21;q32) have been identified.105 Both genes (MUM2 and MUM3) are normally expressed in spleen and lymph nodes and share homologies with the FcγR family of cell surface receptors: The coding region ofMUM2 is interrupted by the translocation, yielding a structurally abnormal transcript, whereas that of MUM3 is intact. The protein homologies and their pattern of expression suggest that these genes may play a role in normal B-cell development and that aberrant signaling through these proteins may play a role in the development of a subgroup of myeloma.
Pathways to B-cell malignancy
Malignancies arising from B cells at different stages of differentiation exhibit different spectra of genetic abnormalities. Many of the genes identified through their direct involvement inIG translocations play pivotal roles not only in the development of malignancy but also in the development and activation of normal B cells, particularly within the GC. Most appear to be involved in at least 1 of 4 mechanisms of cellular regulation, including apoptosis, cell cycle progression, NF-κB activation, and signal transduction pathways from cell surface receptors.
A key experiment is the creation of mouse transgenic models in which the translocation is re-created by placing the gene of interest under the control of the IGH intronic (Eμ) enhancer. While this results in overexpression throughout the B-cell lineage rather than just in mature B cells, such models have been useful in defining the roles of these genes in B-cell neoplasia. Interestingly, most genes expressed in this context, including BCL2 and cyclin D1, do not alone result in B-cell neoplasia, again indicating the necessity for the concurrent involvement of other oncogenes and tumor suppressors.
Apoptosis
Suppression of apoptosis in the context of the indolent B-cell malignancies is a key and presumably early event, allowing the persistence of B cells that would otherwise die within the GC. When these cells acquire secondary genetic abnormalities, the full neoplastic phenotype emerges. Inappropriate high-level BCL2expression mediated either by IG translocation or by changes within the promoter plays a role in follicular B-NHL and CLL, respectively.106 However, some B-cell malignancies, including BL and a subgroup of DLCL, fail to express BCL2, and in these cases the mechanisms by which apoptosis is suppressed are not clear. Mutations of p53 play a role in some BL107 but, otherwise, p53 mutations are for the most part secondary events associated with a poor prognosis in disease of all histologic subtypes.108
The synergistic interaction of BCL2 and MYC.
BCL2 belongs to a family of proteins containing at least 15 mammalian homologues, all of which contain at least 1 of 4 conserved motifs known as BCL2 homology domains (BH1 to BH4) and which may promote or inhibit apoptosis.109,110 BCL2 prolongs cell survival by inhibiting apoptosis in response to a wide variety of stimuli and is localized to the intracellular organelles, including the cytoplasmic face of the mitochondrial outer membrane.111 Its three-dimensional structure has been determined by x-ray crystallography and nuclear magnetic resonance spectroscopy and closely resembles those of some types of bacterial toxins, such as the membrane insertion domain of the diphtheria toxin and the pore-forming colicins A and E1.112As predicted from the structure, BCL2 and BCL-xL, as well as the proapoptotic molecule BAX, can form ion channels when they are added to synthetic membranes.113-115 How interactions between these proteins control apoptosis has been reviewed.109 110
E-Bcl2 transgenic mice show an excess of mature B cells in keeping with the antiapoptotic action of Bcl2.116,117 Similarly, overexpression of Bcl2 in a variety of cell lines does not result in transformation but, rather, protection from apoptosis.109,110Eμ-Bcl2 mice do not develop malignancies until the acquisition of secondary genetic aberrations, including deregulation of Myc.118 Crossing of E-Bcl2 and Eμ-Myc mice confirms the synergistic interaction of the 2 genes, with rapid onset of lymphoid precursor malignancies.119
However, BCL2 also modulates cell cycle progression, promoting exit into G0, an effect that appears to require critical residues within the NH2-terminal domain and that is dissociable from its antiapoptotic activity.120,121 This effect may be mediated through the inhibition of p27 (KIP1) degradation, leading to reduced CDK2 activity 122 (Figure3). The secondary genetic abnormalities commonly seen in follicular B-NHL may therefore be necessary to overcome the cell cycle inhibitory effects of BCL2. Somatic mutations of the translocated BCL2 gene have been described in transformed follicular B-NHL.30 123 Many of these mutations appear to be clustered in the NH2-terminal domain, and some have been associated with a growth advantage over wild-type BCL2 in vitro, possibly as a result of the loss of the normal cell cycle inhibitory effects displayed by the wild-type protein.
Modulation of the cell cycle by IGtranslocations.
A simplified diagram of the G1- to S-phase transition. Genes that are deregulated as a result of IGtranslocation are shown in yellow. Cyclin D1/CDK6:Mitogenic signals result in transcriptional up-regulation of cyclin D1 and Akt-mediated inhibition of cyclin D1 degradation. Active cyclin D1-CDK4/6 (represented here by CDK6 for simplicity) phosphorylates Rb. The cell cycle inhibitors p27 (KIP1) and p21 (CIP1/WAF1) (represented by p27 [KIP1]) are required for assembly and activation of cyclin D-CDK4/6 complexes but mediate their inhibitory effects on the cell cycle through inactivation of cyclin E-CDK2. Overexpression of D-type cyclins or CDKs swings the equilibrium toward G1- to S-phase transition by causing sequestration of p27 (KIP1) and p21 (CIP1/WAF1), allowing activation of cyclin E-CDK2. Rb is phosphorylated further by cyclin E-CDK2, promoting its dissociation from E2F, which then drives transcription of genes required for S-phase entry, including cyclin E. A feedback loop operates whereby cyclin E-CDK2 phosphorylates and promotes proteolytic degradation of p27 (KIP1).Myc: Myc mediates its effects on the cell cycle at least partly through transcriptional up-regulation of cyclin D1/D2, which, complexed with CDKs, sequester the cell cycle inhibitors p27 (KIP1) and p21 (CIP1/WAF1). NF-κB: NF-κB also promotes transition through the cell cycle through direct up-regulation of cyclin D1 transcription.BCL2: BCL2 appears to exert its inhibitory effects on the cell cycle through modulation of the p27 (KIP1) degradation pathway. Ub indicates ubiquitin; P, phosphorylation; D1, cyclin D1; E, cyclin E.
Modulation of the cell cycle by IGtranslocations.
A simplified diagram of the G1- to S-phase transition. Genes that are deregulated as a result of IGtranslocation are shown in yellow. Cyclin D1/CDK6:Mitogenic signals result in transcriptional up-regulation of cyclin D1 and Akt-mediated inhibition of cyclin D1 degradation. Active cyclin D1-CDK4/6 (represented here by CDK6 for simplicity) phosphorylates Rb. The cell cycle inhibitors p27 (KIP1) and p21 (CIP1/WAF1) (represented by p27 [KIP1]) are required for assembly and activation of cyclin D-CDK4/6 complexes but mediate their inhibitory effects on the cell cycle through inactivation of cyclin E-CDK2. Overexpression of D-type cyclins or CDKs swings the equilibrium toward G1- to S-phase transition by causing sequestration of p27 (KIP1) and p21 (CIP1/WAF1), allowing activation of cyclin E-CDK2. Rb is phosphorylated further by cyclin E-CDK2, promoting its dissociation from E2F, which then drives transcription of genes required for S-phase entry, including cyclin E. A feedback loop operates whereby cyclin E-CDK2 phosphorylates and promotes proteolytic degradation of p27 (KIP1).Myc: Myc mediates its effects on the cell cycle at least partly through transcriptional up-regulation of cyclin D1/D2, which, complexed with CDKs, sequester the cell cycle inhibitors p27 (KIP1) and p21 (CIP1/WAF1). NF-κB: NF-κB also promotes transition through the cell cycle through direct up-regulation of cyclin D1 transcription.BCL2: BCL2 appears to exert its inhibitory effects on the cell cycle through modulation of the p27 (KIP1) degradation pathway. Ub indicates ubiquitin; P, phosphorylation; D1, cyclin D1; E, cyclin E.
Other secondary abnormalities include deregulation of Myc, usually due to IG translocation. In some instances, translocation ofBCL2 and MYC may occur at the one IGH allele. As indicated above and when seen clinically, the combination of concurrent deregulation of BCL2 and Myc results in a highly malignant phenotype that is usually resistant to combination chemotherapy.124
This is of some interest because Myc induces apoptosis when overexpressed in fibroblasts.125 An important mediator of Myc-induced apoptosis is the ARF-Mdm2-p53 pathway126(Figure 4). Myc activation in mouse embryo fibroblasts results in elevated p19 (ARF) (the alternative open reading frame of the INK4A locus) and p53 levels,127leading to apoptosis. Therefore, abrogation of Myc-induced apoptosis is necessary to allow the full transforming potential of Myc to develop. Transgenic mice expressing Myc under the control of the immunoglobulin heavy chain enhancer (Eμ-Myc) accumulate large populations of undifferentiated pre-B lymphocytes and eventually develop lymphoma.128 Eμ-Myc mice sustain spontaneous inactivation of either p53 (28%) or ARF (24%) or overexpress Mdm2 129 and develop lymphoma more rapidly in a hemizygous p53 or hemizygous ARF background than in controls.130 Lymphomas arising in these hemizygous animals have almost invariably lost the wild-type allele and are therefore either p53-null or ARF-null, demonstrating that inactivation of this pathway is an important step in Myc-mediated oncogenesis.
Synergistic interaction of BCL2 and Myc.
Genes involved in these processes, which are deregulated by IGtranslocations, are shown in yellow. BCL2: Central to the control of apoptosis is the release of cytochrome c from the mitochondrion, resulting in activation of procaspase 9. BCL2 prevents this by maintaining ADP/ATP exchange through the voltage-dependent anion channel (VDAC) within the mitochondrial membrane, inhibiting H+ accumulation in the intermembrane space and subsequent cytochrome c release. However, BCL2 also inhibits proliferation by promoting cell cycle arrest. Myc: Myc mediates both proliferation and apoptosis. Blocking of Myc-induced apoptosis by genetic events, such as BCL2 overexpression or alterations in the p19 (ARF)–Mdm2–p53 pathway, results in unrestrained proliferation.
Synergistic interaction of BCL2 and Myc.
Genes involved in these processes, which are deregulated by IGtranslocations, are shown in yellow. BCL2: Central to the control of apoptosis is the release of cytochrome c from the mitochondrion, resulting in activation of procaspase 9. BCL2 prevents this by maintaining ADP/ATP exchange through the voltage-dependent anion channel (VDAC) within the mitochondrial membrane, inhibiting H+ accumulation in the intermembrane space and subsequent cytochrome c release. However, BCL2 also inhibits proliferation by promoting cell cycle arrest. Myc: Myc mediates both proliferation and apoptosis. Blocking of Myc-induced apoptosis by genetic events, such as BCL2 overexpression or alterations in the p19 (ARF)–Mdm2–p53 pathway, results in unrestrained proliferation.
Blocking of Myc-induced apoptosis is therefore necessary for the proliferative oncogenic potential of Myc (see below) to be realized in B-cell malignancies. The mechanism involved is cell type–dependent. Loss of p19 ARF usually occurs by deletion in lymphoid malignancies and is typically seen in BCP-ALL, although it may sometimes be seen in B-NHL at transformation to high-grade disease. Mutation of p53 is commonly seen in BL and in B-cell prolymphocytic leukemia but, otherwise, these mutations are usually secondary events in B-cell malignancies.107 108 Deregulated BCL2 occurs as an antecedent event in follicular B-NHL.
Cell cycle regulation
In mammalian cells, transition through the cell cycle is controlled at different checkpoints. (Figure 3). Regulation is achieved through a family of serine/threonine protein kinases consisting of regulatory cyclin subunits that bind to and activate catalytic CDK subunits. The cyclin-CDK complexes most closely linked to the late G1phase checkpoint are the D-type cyclins (D1, D2, and D3) and their partners CDK4 and CDK6. The major targets of cyclin D/CDK4 and cyclin D/CDK6 complexes are the retinoblastoma protein (Rb) and its related family p107 and p130.131
Activation of CDKs is regulated by a variety of mechanisms, which include binding of cyclins, CDK inhibitors, and phosphorylation. The 7 mammalian CDK inhibitor genes identified thus far belong to 2 separate families and are responsible for integrating many growth-inhibitory pathways, including responses to DNA damage, senescence, and contact inhibition.132 The INK4 family includes 4 closely related ankyrin repeat-containing genes: p16 (INK4a), p15 (INK4b), p18 (INK4c), and p19 (INK4d). INK4 proteins bind to CDK4 or CDK6 and prevent D-type cyclin binding and activation. The CIP1/KIP1 family contains 3 genes: p21 (CIP1/WAF1), p27 (KIP1), and p57 (KIP2). p21/p27/p57 regulate multiple CDK enzymes, including CDK4/6-cyclin D, by forming ternary complexes with CDK and cyclin proteins. Recent evidence points to a role of p27 (KIP1) in activation of CDK4/6-cyclin D complexes and inhibition of CDK2-cyclin E.132Additionally, CDK2 and CDK4 are both regulated by phosphorylation and dephosphorylation by CAK and cdc25, respectively.
Myc.
Myc is required for progression through the cell cycle, and normally its expression correlates tightly with the proliferative state of the cell. The functions of Myc in this regard have been recently reviewed.43
Myc-null cell lines are characterized by a profound growth defect due to a lengthening of both the G1 and G2 phases of the cell cycle. The largest defect observed in these cells is a 12-fold reduction in cyclin D1-CDK4 and D1-CDK6 complexes during G1 to S phase transition. However, the activity of other cyclin-CDK complexes, including cyclin E-CDK2 and cyclin A-CDK2, is also diminished, and growth rate is not restored by overexpression of these cyclins.133 These findings are explained by the finding that Myc directly induces expression of cyclins D1 and D2 (Figure 3), which, as cyclin-CDK complexes, sequester the cell cycle inhibitors p27 (KIP1) and p21 (CIP1/WAF1),134 135 thereby releasing inhibition from cell cycle control. This is at least one of the pathways required for Myc's proliferative effects.
Cyclin D1.
Ectopic expression in G1 phase of cyclin D1 under the control of an inducible promoter causes early phosphorylation of Rb and acceleration of the cell cycle through G1.136 In lymphoma cell lines carrying the t(11;14) translocation, although cyclin D1 is still regulated in its normal cyclical manner (unlike Myc in BL), its overexpression causes acceleration through G1phase137 (Figure 3). However, although cyclin D1 overexpression is sufficient to confer transformed properties on established fibroblasts, it is insufficient to transform primary cells or to induce lymphomagenesis in Eμ-cyclin D1 transgenic mice.138 These mice demonstrated remarkably few abnormalities in their lymphoid population. Consistent with a multistep process of tumorigenesis, however, cyclin D1 collaborates strongly with Myc and modestly with p21Ras in lymphomagenesis in double transgenic animals.138 139
Cyclin D2.
CDK6.
Overexpression of CDK6 in B cells containing the translocation is likely to contribute directly to their pathogenesis by acceleration of the cell cycle through the G1- to S-phase transition in a similar way to overexpression of cyclin D1/D2 or inactivating mutations of the INK4 proteins (Figure 3). However, it is noteworthy that SLVL, the disease in which this overexpression occurs, is classically associated with a very low proliferative rate, suggesting that the overexpression of CDK6 may influence other pathways.
NF-κB activation
Mature B cells exhibit constitutive NF-κB activation.140 The genes leading to this activation are frequently targets both of chromosomal translocation and mutation.141
The Rel/NF-κB family is a family of transcription factors involved in the regulation of expression of a variety of cellular and viral genes, including those that control immune responses, acute phase reactions, and the replication of viruses.142 NF-κB transcriptional activity is mediated by dimeric complexes generated though the combination of the various members of the Rel/NF-κB family. These proteins all share a highly conserved amino-terminal Rel homology domain responsible for DNA binding, dimerization, nuclear localization functions, and cytoplasmic association with inhibitors of the IκB family.
The Rel/NF-κB family can be broadly divided into 2 groups. The first group is represented in mammalian cells by p65 (RelA), RelB, and c-Rel, which contain the amino-terminal Rel domain and distinct transactivation domains within their carboxy-terminal regions. The second group contains NF-κB1 (p105/p50) and NF-κB2 (p100/p52), which exist as 2 forms: (a) long (p105 and p100), containing the amino terminal Rel homology domain, a polyglycine region, and a carboxy-terminal ankyrin domain, and (b) short (p50 and p52), possessing only the DNA-binding Rel domain. The ankyrin domains in both long forms are homologous to the IκB family of proteins and act in a regulatory fashion.
In unstimulated cells, the ankyrin-containing IκB proteins interact with Rel/NF-κB complexes and sequester them in the cytoplasm by masking the nuclear localization signal (Figure5). On stimulation, IκB is phosphorylated, ubiquitinated, and degraded. This allows NF-κB to translocate to the nucleus and bind to specific cis-acting consensus sequences (κB sites) located in the regulatory region of inducible genes, including cytokines (eg, TNFα, IL-2, IL-6, IL-8, α-interferon), adhesion molecules (eg, intracellular adhesion molecule-1, vascular cell adhesion molecule-1), genes involved in regulation of apoptosis or response to NF-κB (eg, TRAF1/2, cIAP1 and 2, TNFR1, A20, IκBα), and those involved in cell cycle regulation and proliferation (eg, cyclin D1, MYC, p53, Rb).143Activation of NF-κB is thought to be necessary for transformation by a variety of stimuli although, in some cell types, it may also lead to apoptosis.144
Modulation of NF-κB activation by IGtranslocations.
Genes involved in these processes, which are deregulated by IGtranslocations, are shown in yellow. NF-κB activation by extracellular signals such as TNF is mediated via a signaling pathway involving the death initiator signaling complex (DISC), TRAF2, NIK, IKK, and phosphorylation and degradation of IκB. NF-κB2:Truncated NF-κB2 proteins as found in some lymphomas are constitutively active transcription factors, mediating up-regulation of NF-κB target genes, including those involved in proliferation and apoptosis, such as cyclin D1 and Bcl-xL. BCL10:BCL10 mediates both NF-κB activation, through its recruitment to the DISC, and apoptosis via mechanisms that remain unclear. Truncated BCL10 maintains its NF-κB–activating functions but loses its apoptotic activity. BCL3: BCL3 is an IκB family member, which mediates transcriptional up-regulation of NF-κB target genes through interaction with p50/p52 homodimers.
Modulation of NF-κB activation by IGtranslocations.
Genes involved in these processes, which are deregulated by IGtranslocations, are shown in yellow. NF-κB activation by extracellular signals such as TNF is mediated via a signaling pathway involving the death initiator signaling complex (DISC), TRAF2, NIK, IKK, and phosphorylation and degradation of IκB. NF-κB2:Truncated NF-κB2 proteins as found in some lymphomas are constitutively active transcription factors, mediating up-regulation of NF-κB target genes, including those involved in proliferation and apoptosis, such as cyclin D1 and Bcl-xL. BCL10:BCL10 mediates both NF-κB activation, through its recruitment to the DISC, and apoptosis via mechanisms that remain unclear. Truncated BCL10 maintains its NF-κB–activating functions but loses its apoptotic activity. BCL3: BCL3 is an IκB family member, which mediates transcriptional up-regulation of NF-κB target genes through interaction with p50/p52 homodimers.
The tumor necrosis factor (TNF) signaling pathway is the most extensively studied pathway leading to NF-κB activation, but it shares common elements with a number of other pathways, including IL-1– and CD40-mediated signaling. Binding of TNFα to TNF receptor (TNFR) results in recruitment of TNF receptor–associated factors (TRAFs), particularly TRAF2, 5, and 6.145 Recruitment of NF-κB–inducing kinase (NIK) to TRAF2 results in a kinase cascade mediated by NIK146 and IκB kinases (IKKα and β),142,147 finally resulting in phosphorylation of IκBα at Ser32 and Ser36 (Figure 4). However, this is not the only pathway to IκB phosphorylation and degradation,148 and NF-κB activation can also be mediated in the absence of IκB degradation, as seen in IgG+ B cells.140NF-κB–dependent signaling pathways play an essential role in B-cell development, as evidenced by studies on knockout mice.142
NF-κB2.
The result of the t(10;14)(q24;q32) is an NF-κB2–Cα1 fusion that retains the Rel effector and poly-G domains but is fused to an out-of-frame and prematurely truncated Cα1.76Rearrangements within the 3′ region of NFKB2 have also been reported in about 2% of other lymphoid malignancies and particularly in cutaneous lymphomas, occurring either as a result of other translocations or, more commonly, as a result of interstitial deletions of chromosome 10q24.149 The rearrangedNFKB2 genes encode either truncated NF-κB2 or fusion products to heterologous molecules: These lack some or all of the ankyrin repeats at the C-terminus but retain an intact Rel homology domain.150 Like the p52 subunit, these mutant proteins are constitutively localized to the nucleus, presumably through the loss of the IκB-like regulatory domain, where they are able to bind to κB sites and demonstrate transactivating properties 151(Figure 5).
Mice lacking the C-terminal ankyrin domain of NF-κB2 (p100−/− mice), and therefore showing abnormalities of NFKB2 similar to those seen in some patients with lymphomas, demonstrate marked gastric hyperplasia, resulting in early postnatal death.152 Both spleen and thymus are very atrophic with abnormal architecture, whereas lymph nodes are enlarged and the proliferative responses of B and T lymphocytes to several mitogens are markedly enhanced. p52-deficient mice have reduced numbers of B cells with reduced proliferative responses to antigens, reduced numbers of follicles in spleen and lymph nodes, and impaired GC formation.153 154
NF-κB2, like BCL6, is therefore a key modulator of GC formation and B-cell activation: Absence of the C-terminal domain may contribute to tumor formation by allowing abnormal levels of p52-like mutant protein to accumulate in the nucleus, resulting in enhanced NF-κB–dependent B-cell proliferation.
BCL10.
BCL10 is a 233–amino acid protein containing an amino-terminal caspase recruitment domain (CARD),155 a motif found in a number of proteins involved in the control and execution of apoptosis. This domain consists of 6 tightly packed antiparallel alpha helices156-158 and is similar in structure to the death effector domain (DED) and the death domain (DD) contained in other apoptotic proteins. CARDs mediate the binding between adapter molecules and caspases: Procaspase 2 is recruited to the death initiator signaling complex (DISC) via a CARD-CARD interaction with RAIDD/CRADD,159,160 and procaspase 9 is activated after binding to Apaf-1 via a homotypic CARD-CARD interaction.161BCL10, however, is most homologous to an equine herpes virus-2 protein E10 although this homology is limited to the CARD. The carboxy-terminus of BCL10 is rich in serine and threonine residues and is phosphorylated,162 which may allow regulation of its functions.
BCL10 activates NF-κB and induces apoptosis, both of which require oligomerization via the CARD domain. However, although CARD oligomerization is essential for both NF-κB activation and apoptosis, the CARD domain is inadequate to mediate either of these effects and appears to function as an oligomerization motif.162 No other CARD-mediated interactions with other proteins have been identified. NF-κB activation by BCL10 is mediated via activation of the NIK84 through TRAF2,163leading to phosphorylation of IKK and subsequent phosphorylation and degradation of IκB (Figure 5). BCL10 may act as a transducer in the DISC by interaction via its carboxy-terminus with the death adapter molecule TRADD.163
BCL10 may mediate its apoptotic activity through the binding of its carboxy-terminal domain to procaspase 9, resulting in autoproteolytic activation of the caspase precursor to its active form.164However, other studies have demonstrated a lack of binding to procaspase 9 and, therefore, the significance of this finding remains unclear.84,162,163 Importantly, like other proapoptotic genes, BCL10 may also act as a tumor suppressor, suppressing the transformation of primary rat embryo fibroblasts by synergistic combinations of various nuclear oncogenes.82 Whether this activity is dependent on apoptosis or on other mechanisms remains to be elucidated.
Some MALT lymphomas demonstrating the t(1;14)(p22;q32) translocation and other lymphoid tumors in which the translocation is absent contain genomic mutations of the BCL10 coding sequence.82,165 Most of these are frameshift mutations resulting in premature truncation of the protein distal to the CARD and, functionally, in loss of apoptotic activity, retention of NF-κB activation, and a “gain of function” ability to enhance transformation in rat embryo fibroblast assays.82 Other missense mutations identified reside within the carboxy-terminus and would be expected to result in loss of some of the key biological functions of BCL10.
However, not all MALT lymphomas exhibiting the t(1;14)(p22;q32) translocation exhibit mutations, and it seems likely that in these cases the apoptotic/transformation suppressor pathways are blocked by a different mechanism than loss of the carboxy-terminus. Interestingly, all cases with the t(1;14)(p22;q32) translocation, whether they contain mutated BCL10, demonstrate strong nuclear BCL10 staining different from the normal cytoplasmic pattern seen in normal B cells and in other MALT cases.166 The mechanism behind this finding is presently unknown. Overexpression of BCL10 may therefore, like some of the mutant forms of NF-κB2, result in constitutive nuclear expression. The localization of at least some isoforms of BCL10 within the nucleus suggests that this gene may have functions other than the regulation of apoptosis. Not all CARD-containing proteins are involved solely in apoptotic regulation: Some isoforms of ARC (apoptosis repressor with CARD)/Nop30 are involved in RNA processing.167 168
BCL10 is likely to play a role in the normal development of the GC, where lymphocytes proliferate in response to antigen or undergo apoptosis. Truncated BCL10 is likely to contribute to oncogenesis through the uncoupling of apoptosis from the proliferative effects of NF-κB activation.
BCL3.
BCL3 is a member of the IκB family of proteins that regulate the Rel/NF-κB family of transcription factors.169-171 The IκB family includes at least 5 different members: IκBα, IκBβ, IκBγ, IκBε, and BCL3. Each of these proteins has different specificities for interaction via the centrally located ankyrin repeat domain with the N-terminal Rel homology domains of different Rel/NF-κB homodimeric or heterodimeric complexes.142 BCL3 is unique among the IκB family in that it is a nuclear protein172 containing amino- and carboxy-terminal transactivation domains.173 It specifically associates with homodimers of p50 or p52 subunits174 175 (Figure 5). p50 and p52 subunits are similar in their primary structures. They have no defined transactivation domains and, as homodimers, may competitively inhibit the binding of transactivating NF-κB dimers to consensus DNA binding (κB) sites.
BCL3 can act as an antirepressor to remove homodimers from κB sites so that transactivating NF-κB dimers can bind,176 act as a transactivator by forming complexes with homodimers at κB sites,173,177 or enhance homodimer binding to κB sites.178
These functions are at least partly dependent on the phosphorylation status of BCL3.179 BCL3 also interacts with other nuclear cofactors, including the BRCA-1–binding protein Bard1 and the histone acetylase Tip60.180 These interactions may modulate its function.
Mice lacking Bcl3 show impaired GC formation, loss of follicular dendritic cell networks, and defects in their normal antibody responses, findings similar to those seen in NF-κB2–deficient mice.181,182 However, both NF-κB2– and Bcl3-deficient lymphocytes can form GCs and generate at least some T-cell–dependent antibody responses when adoptively transferred into RAG-1–deficient mice, suggesting that accessory cells such as follicular dendritic cells may be required for these functions. On the other hand, Eμ-Bcl3 transgenic mice are characterized by an accumulation of mature B cells in bone marrow, lymph nodes, and peritoneal cavity.183 They demonstrate a hyperresponsive humoral immunity as evidenced by an increased sensitivity to BCR cross-linking, increased levels of autoantibody to double-stranded DNA, and a threefold increase in the number of germinal cells in lymph nodes. Transgenic mice also show decreases in serum IgM and IgG3 but increases in IgG1 and IgA, consistent with the involvement of Rel/NF-κB complexes in Ig class switching. However, no lymphoid neoplasms are seen in transgenic animals.
The mechanisms underlying NF-κB–mediated transformation remain unclear. Two pathways of direct relevance to oncogenesis have emerged. First, NF-κB is able to mediate a protective role in apoptosis induced by chemotherapeutic drugs, TNFα, and by certain oncogenes, including Ras.184-186 This effect is likely to be mediated via antiapoptotic target genes, including Bcl-xL.187 Secondly, NF-κB is closely associated with the cell cycle. B cells from NF-κB–deficient mice show delays in cell cycle progression from G1 to S phase, and expression of a dominant negative mutant of IκB results in retarded phosphorylation of the Rb protein and G1/S phase transition.188,189NF-κB activates cyclin D1 transcription via NF-κB binding sites within its promoter, which induces G1/S transition.190 191 NF-κB activation may therefore have dual roles in suppression of apoptosis and in cell cycle progression. In certain situations and in certain cell types, however, NF-κB enhances apoptosis. It is likely that in B-cell malignancies this aspect will have been blocked by prior genetic events such asBCL2 overexpression.
B-cell receptor signaling
Stimulation of B cells by antigen-mediated cross-linking of the BCR induces tyrosine phosphorylation of a number of proteins, including the CD19 and CD22, the BCR signaling proteins CD79a (Igα) and CD79b (Igβ), and the kinases Syk, Lyn, PI3-K, and Btk, together with other proteins, including phospholipase Cγ (PLCγ), Shc, Grb2, and Vav192 (Figure 6). Binding of antibody to Fc receptor can result in activation, inhibition, or apoptosis. Activation responses are mediated by the immunoreceptor tyrosine activation motif (ITAM) found in FcγRI and FcγRIII. Inhibitory responses are mediated via the immunoreceptor tyrosine inhibitory motif (ITIM)193 found in FcγRIIB. B cells express only the inhibitory receptor FcγRIIB.
Modulation of antigen receptor signaling by IGtranslocations.
Genes that are deregulated as a result of IG translocation are shown in yellow. Antigen binding to the BCR results in B-cell activation via multiple effectors, including Ras, Btk, JNK, and PLCγ.Pax5: Pax5 transcriptionally up-regulates 2 key molecules in the BCR complex, CD79a and CD19, allowing enhanced B-cell proliferation. FcγRIIB: In normal B cells, FcγRIIB mediates its inhibitory signal via 1 of 2 pathways. (1) Simultaneous cross-linking of FcγRIIB with the BCR by immune complexes results in phosphorylation of the FcγRIIB ITIM and subsequent recruitment of SHIP, which, by hydrolyzing PIP3, results in dissociation from the membrane and, therefore, inactivation of Btk. (2) Alternatively, cross-linking of FcγRIIB alone results in apoptosis mediated via direct activation of Btk. In cases of follicular lymphoma demonstrating the t(1;22)(q22;q11), the t(14;18) is a prior event, resulting in overexpression of BCL2, which blocks apoptosis and may allow proliferative signals to go unhindered.
Modulation of antigen receptor signaling by IGtranslocations.
Genes that are deregulated as a result of IG translocation are shown in yellow. Antigen binding to the BCR results in B-cell activation via multiple effectors, including Ras, Btk, JNK, and PLCγ.Pax5: Pax5 transcriptionally up-regulates 2 key molecules in the BCR complex, CD79a and CD19, allowing enhanced B-cell proliferation. FcγRIIB: In normal B cells, FcγRIIB mediates its inhibitory signal via 1 of 2 pathways. (1) Simultaneous cross-linking of FcγRIIB with the BCR by immune complexes results in phosphorylation of the FcγRIIB ITIM and subsequent recruitment of SHIP, which, by hydrolyzing PIP3, results in dissociation from the membrane and, therefore, inactivation of Btk. (2) Alternatively, cross-linking of FcγRIIB alone results in apoptosis mediated via direct activation of Btk. In cases of follicular lymphoma demonstrating the t(1;22)(q22;q11), the t(14;18) is a prior event, resulting in overexpression of BCL2, which blocks apoptosis and may allow proliferative signals to go unhindered.
FcγRIIB.
The FCGRIIB gene encodes 2 isoforms, FcγRIIb1, which is the major species expressed on the surface of B cells, and IIb2, which are identical except for a 19–amino acid insertion in the cytoplasmic domain of IIb1.194 Most experimental work has been carried out on the IIb1 isoform; however, the IIb2 isoform is preferentially expressed as a result of the translocation t(1;22)(q22;q11). Whether the key biologic functions with regard to oncogenesis of these 2 isoforms are significantly different remains to be determined.
FcγRIIB mediates inhibitory signaling in normal B cells195 (Figure 6). Cross-linking of BCR and FcγRIIB results in tyrosine phosphorylation of the ITIM motif of FcγRIIB and the recruitment of the Src-homology–2 (SH2)-containing inositol polyphosphate 5-phosphatase (SHIP).196,197 SHIP, by hydrolyzing phosphatidylinositol 3,4,5-trisphosphate (PIP3), leads to dissociation of Bruton's tyrosine kinase (Btk) from the membrane198,199 and decreased activity of the survival-associated protein kinase, Akt.200,201 Btk is essential for B-cell activation and proliferation, mediating its effects through activation of PLCγ, which promotes intracellular calcium influx.202
Overexpression of an inhibitory Fc receptor might not appear to be compatible with lymphomagenesis. However, cross-linking of FcγRIIB alone results in apoptosis and activation of c-Jun N-terminal kinase (JNK), mediated via Btk, an effect that requires the transmembrane domain of FcγRIIB.203 This apoptotic response is diminished in Bcl2 transgenic mice, resulting in cell proliferation,204 presumably through uncoupling of the apoptosis effector arm from proliferative signals.
Such dissociation might provide a rationale for the acquisition of the t(1;22)(q22;q11) as a secondary abnormality in follicular lymphoma: Overexpression of BCL2 provides the necessary environment in which the apoptotic response to FcγRIIB is abrogated, allowing JNK activation or other stimulatory signals to drive proliferation.
Pax5.
Transcription of PAX5 is initiated from 2 promoters on 9p13, resulting in the splicing of 2 alternative untranslated first exons (1A or 1B) to the coding sequences (exons 2-10). PAX5 codes for the 50-kd transcription factor B-cell–specific activator protein (BSAP),205 which recognizes its target genes via the conserved paired domain contained in exons 2 and 3. Pax5 is expressed after birth only in B lymphocytes and testis. During normal B-cell lymphopoiesis, Pax5 is expressed from the earliest B-lineage committed precursor cell up to the mature B-cell stage but is subsequently down-regulated during plasma cell differentiation. Overexpression of Pax5 results in an enhanced proliferative response in splenic B cells to lipopolysaccharide (LPS) stimulation, whereas addition of antisense oligonucleotides to such cells results in an impaired LPS response,206 demonstrating that Pax5 plays a role in the regulation of activation and proliferation of mature B cells.
Pax5 plays a key role in the regulation of CD19, CD79a, and N-Myc expression: All 3 genes are down-regulated significantly in the absence of Pax5. Both CD19 and CD79a synergize with other B-cell surface receptors to transduce stimulatory signals (Figure 6), resulting in activation of a variety of signaling pathways, including the Src family phosphotyrosine kinases (Fyn, Lyn, and Lck), serine-specific protein kinases (eg, protein kinase C), p21Ras, NF-κB, and mitogen-associated protein kinase.207 CD79a, together with CD79b, serves as the signaling subunits of the BCR by establishing binding and activation sites for SH2 domain–containing molecules at phosphorylated tyrosine residues within their ITAMs. CD19 appears to function as a regulatory component central to the multiple signaling pathways activated by BCR signaling by providing docking sites for other signaling effector molecules. Overexpression of CD19 results in enhanced proliferative responses to LPS and other stimuli similar to those seen with Pax5 overexpression.
Deregulated expression of PAX5 as a result of the t(9;14)(p13;q32) in LPL may therefore contribute to malignant transformation by interfering with normal inactivation of PAX5transcription during plasma cell differentiation, resulting in enhanced proliferative signaling via the BCR.
Future prospects
MYC was first shown to be involved in the t(8;14)(q24;q32) of BL in 1982. The subsequent 18 years have seen the isolation of pathogenic genes from many other IG translocations, but raw cytogenetic data alone would suggest that more remain to be cloned. The widespread use of multicolor FISH methods, along with polymerase chain reaction methods for cloning translocation breakpoints and the resources made available by the genome sequencing project, will expedite this analysis. While these translocations may be rare, it is possible that the genes isolated by this approach will also be involved in mutations in cases lacking the translocation. Furthermore, cloning of IG translocations suggests new functions for previously cloned molecules and may define new therapeutic targets.
Many questions relating to the origin of the IG translocations remain unanswered. Study of mutant mice may allow some of the key genes involved in this process to be identified.208 However, it is unclear how the intimate associations of specific IGtranslocations with specific stages of B-cell differentiation and specific B-cell diseases might arise. The spatial arrangement and precise positioning of chromosomes and genes (8% of normal lymphocytes have MYC and IGH loci in close apposition) during the cell cycle may have a role to play.209 210
Are these translocations a feature of normal B-cell development? The t(14;18)(q32;q21) is present at low levels in many apparently normal individuals,211 and MYC translocations have been demonstrated in phenotypically normal mice.212 These data indicate that IG translocation alone is insufficient to generate disease. Whether other IG translocations are found at comparable frequency in normal B cells in human beings is not known. Application of sensitive polymerase chain reaction and FISH technologies and analysis of mouse models should allow these and other questions to be answered. If the rate at which IGtranslocations occur in normal individuals determines the propensity to develop B-cell malignancy, it may be possible to detect at-risk individuals long before the emergence of frank disease.
Note added in proof.
Two groups have recently reported the cloning of a case of extranodal DLCL with t(1;14)(q21;q32) and shown that this case directly involved the MUC1 gene on chromosome1q21.MUC1, located within a gene-rich region of chromosome 1q21, was specifically deregulated as a consequence of this translocation. MUC1 also appeared to be the target gene of some amplifications of this region. MUC1 overexpression occurs frequently in human epithelial cancers and is associated with tumor progression and poor clinical outcome, but these are the first reports of this gene involved in the pathogenesis of B-cell malignancies. How this protein, which is normally expressed only in plasma cells of the B-cell lineage, contributes to B-cell transformation is not clear.
Supported in part by grants from the Leukaemia Research Fund, the Kay Kendall Leukaemia Fund, and the Cancer Research Campaign.
Reprints:Martin J. S. Dyer, Academic Department of Haematology and Cytogenetics, Haddow Laboratories, Institute of Cancer Research, Sutton, Surrey, UK SM2 5NG; e-mail: mdyer@icr.ac.uk.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.

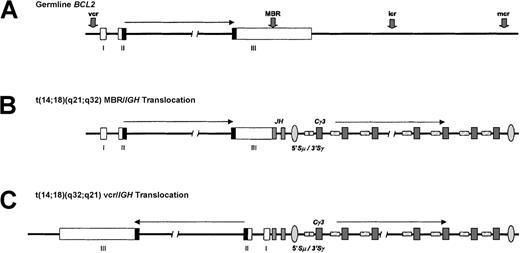

![Fig. 3. Modulation of the cell cycle by IGtranslocations. / A simplified diagram of the G1- to S-phase transition. Genes that are deregulated as a result of IGtranslocation are shown in yellow. Cyclin D1/CDK6:Mitogenic signals result in transcriptional up-regulation of cyclin D1 and Akt-mediated inhibition of cyclin D1 degradation. Active cyclin D1-CDK4/6 (represented here by CDK6 for simplicity) phosphorylates Rb. The cell cycle inhibitors p27 (KIP1) and p21 (CIP1/WAF1) (represented by p27 [KIP1]) are required for assembly and activation of cyclin D-CDK4/6 complexes but mediate their inhibitory effects on the cell cycle through inactivation of cyclin E-CDK2. Overexpression of D-type cyclins or CDKs swings the equilibrium toward G1- to S-phase transition by causing sequestration of p27 (KIP1) and p21 (CIP1/WAF1), allowing activation of cyclin E-CDK2. Rb is phosphorylated further by cyclin E-CDK2, promoting its dissociation from E2F, which then drives transcription of genes required for S-phase entry, including cyclin E. A feedback loop operates whereby cyclin E-CDK2 phosphorylates and promotes proteolytic degradation of p27 (KIP1).Myc: Myc mediates its effects on the cell cycle at least partly through transcriptional up-regulation of cyclin D1/D2, which, complexed with CDKs, sequester the cell cycle inhibitors p27 (KIP1) and p21 (CIP1/WAF1). NF-κB: NF-κB also promotes transition through the cell cycle through direct up-regulation of cyclin D1 transcription.BCL2: BCL2 appears to exert its inhibitory effects on the cell cycle through modulation of the p27 (KIP1) degradation pathway. Ub indicates ubiquitin; P, phosphorylation; D1, cyclin D1; E, cyclin E.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/96/3/10.1182_blood.v96.3.808/5/m_bloo0153003z.jpeg?Expires=1765895480&Signature=ZTenhP0Sppnzh25rOS3of6~daRtzz2FZxML18c2xhRXvUavPCX5Ydm2EdAL~Y5WPxVQhaOukzU4SsJqjsG-FJ86DRnosDPMpxeLP1EPnKaQKl3DC-XxQscs0cXnVmXiOM6gHaUP5TvcM7JyMGsLD15Oz7u8y6VnZXwtS3ypUsvX--qiovnlItb8FzV4D74Wj70z8AYkqsjJc7opCUUQ5tqj1qreHBKGwT0tYv21uuAs5IRtG1v5b3yGXcI51cGubMp0TnO63y5wRkSUreGTGUz8KcWVM7ys9jgUsRkq3Vew35~PxDzJ1DQEDB76hoVPVTnlB5gVl3IWjcG3UrP-sYg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)


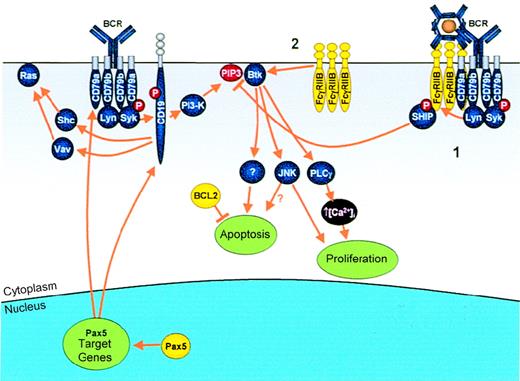
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal