Abstract
The anticoagulant human plasma serine protease, activated protein C (APC), inactivates blood coagulation factors Va (FVa) and VIIIa. The so-called autolysis loop of APC (residues 301-316, equivalent to chymotrypsin [CHT] residues 142-153) has been hypothesized to bind FVa. In this study, site-directed mutagenesis was used to probe the role of the charged residues in this loop in interactions between APC and FVa. Residues Arg306 (147 CHT), Glu307, Lys308, Glu309, Lys311, Arg312, and Arg314 were each individually, or in selected combinations, mutated to Ala. The purified recombinant protein C mutants were characterized using activated partial thromboplastin time (APTT) clotting assays and FVa inactivation assays. Mutants 306A, 308A, 311A, 312A, and 314A had mildly reduced anticoagulant activity. Based on FVa inactivation assays and APTT assays using purified Gln506-FVa and plasma containing Gln506-FV, it appeared that these mutants were primarily impaired for cleavage of FVa at Arg506. Studies of the quadruple APC mutant (306A, 311A, 312A, and 314A) suggested that the autolysis loop provides for up to 15-fold discrimination of the Arg506 cleavage site relative to the Arg306 cleavage site. This study shows that the loop on APC of residues 306 to 314 defines an FVa binding site and accounts for much of the difference in cleavage rates at the 2 major cleavage sites in FVa.
Protein C is a vitamin K–dependent zymogen of a plasma serine protease.1 When activated by the thrombin–thrombomodulin complex2,3 protein C acts as an anticoagulant, in concert with its cofactor protein S, on the surface of phospholipid membranes by inactivation of the blood coagulation factors Va (FVa) and VIIIa.4-6 The importance of activated protein C (APC) is illustrated by an increased risk of venous thromboembolism associated with heterozygous protein C deficiency.7 Furthermore, homozygous protein C deficiency is responsible for severe and generalized thrombotic disease.8 9
Inactivation of FVa, the primary substrate of APC, is a complex process. APC cleaves FVa at 3 locations, Arg506, Arg306, and Arg679. The significance of cleavage at Arg679 is not known, but the cleavages at Arg506 and Arg306 are primarily responsible for inactivation of FVa. Cleavage at Arg506 partially inactivates FVa, whereas cleavage at Arg306 completely inactivates FVa. In the absence of the cofactor protein S, cleavage of Arg506 is about 20-fold faster than cleavage at Arg306.10-12 But protein S accelerates cleavage of Arg306 to a rate close to that of cleavage at Arg506.13
Fisher et al14 built a molecular model of the serine protease domain of APC based on the known crystal structures of homologous serine proteases. Inspection of this model revealed a concentration of basic residues in the lower “east” quadrant of the molecule when the model was in the standard orientation for serine proteases. Subsequently, a crystal structure of a truncated version of APC confirmed the presence of this basic exosite,15 which is generally in a position similar to that of the anion binding exosite I of thrombin, an exosite that is involved in protein–protein interactions. Several basic residues in the autolysis loop of APC are part of this basic exosite. The autolysis loop of APC (residues 301-316, equivalent to chymotrypsin [CHT] residues 142-153) contains an insert of 4 residues relative to chymotrypsin and has 5 basic and 2 acidic residues. A synthetic peptide with sequence overlapping part of this autolysis loop inhibits the APC inactivation of FVa, thus implicating this region in FVa binding.16
Given the presence of so many charged residues and the inhibition of FVa inactivation by this synthetic peptide, we decided to create recombinant mutant APC molecules with individual mutations of the charged residues of the autolysis loop to investigate the role of this loop in the interaction of APC with FVa. We mutated the residues Arg306, Glu307, Lys308, Glu309, Lys311, Arg312, and Arg314 to Ala. We also made a mutant with both Arg306 and Arg312 mutated to alanines (306/312AA) and a mutant with Arg306, Lys311, Arg312, and Arg314 all mutated to alanines (306-314AAAA). These purified mutant proteins were analyzed for their anticoagulant activity and for their ability to inactivate FVa.
Materials and methods
Proteins and reagents
Factor Va and Q506-FVa were prepared as described.12,17FXa was purchased from Enzyme Research Laboratories (South Bend, IN). Phospholipid vesicles (80% phosphatidylcholine, 20% phosphatidylserine) were prepared as described.18 The chromogenic substratel-pyroglutamyl-l-prolyl-l-argininyl-p-nitroanilide hydrochloride (S-2366) was purchased from Chromogenix (Franklin, OH). The chromogenic substrate H-D-lysyl (g-Cbo)-prolyl-argininyl-p-nitroanilide (Spectrozyme PCa) was purchased from American Diagnostica (Greenwich, CT). The chromogenic substrate CBS 34-47 was purchased from American Bioproducts (Parsippany, NJ). Normal human citrate-anticoagulated plasma was purchased from Precision Biologicals (Dartmouth, Novia Scotia). Factor V–deficient plasma was purchased from George King Bio-Medical, Inc (Overland Park, KS). Biotinylated goat antirabbit-IgG, streptavidin alkaline phosphatase conjugate, nitroblue tetrazolium chloride (NBT), and 5-bromo-4-chloro-3′-indolylphosphate-p-toluidine salt (BCIP) were purchased from Pierce (Rockford, IL). Polyvinylidene fluoride membrane was purchased from Millipore (Bedford, MA).
Expression and purification of recombinant protein C
Mutant protein C expression vectors were constructed and purified recombinant protein C was prepared as described.19,20Briefly, crude recombinant protein C was loaded on a fast-flow Q-Sepharose column in 50 mmol/L Tris, 150 mmol/L NaCl, pH 7.4. Then protein C was eluted with 50 mmol/L Tris, 120 mmol/L NaCl, and 15 mmol/L CaCl2. This protocol selects for properly γ-carboxylated protein C because undercarboxylated protein C is not eluted by CaCl2.21 Some preparations of recombinant protein C were purified further by chromatography on a calcium-dependent sheep polyclonal antiprotein C antibody-Sepharose column.22 The protein C was loaded onto the column in 50 mmol/L Tris, pH 7.4, 10 mmol/L CaCl2, 1 mol/L NaCl, 0.02% Tween 20, and 0.05% NaN3. After washing the column with this buffer, protein C was eluted with 50 mmol/L Tris, pH 7.4, 10 mmol/L EDTA, 0.1 mol/L NaCl, 0.02% Tween 20, and 0.05% NaN3. The protein C–containing fractions were pooled and dialyzed into HBS (50 mmol/L HEPES, pH 7.4, 150 mmol/L NaCl) for storage at −80°C. Final concentration of protein C was determined using the Asserachrom Protein C ELISA from American Bioproducts.
Functional assays
Protein C was activated by thrombin. Protein C in HBS plus 2 mmol/L EDTA and 0.5% bovine serum albumin (BSA), at a concentration of 50 μg/mL, was incubated for 2.5 hours with 5 μg/mL thrombin at 37°C followed by the addition of 1.1 U hirudin per unit of thrombin to inactivate the thrombin. Controls were done in amidolytic assays, activated partial thromboplastin time (APTT) clotting assays, and FVa inactivation assays to verify that the thrombin and hirudin used had no effect on these assays. Wild-type APC and the mutants 306/312AA and 306-314AAAA were quantitated using an active site titration adapted from Chase and Shaw23 using APC at approximately 8 μmol/L in HBS and p-nitrophenol-guanidino benzoate at 0.1 mmol/L with an extinction coefficient for p-nitrophenol of 9890 M-1cm-1 calculated for the observed pH. Amidolytic activity of each APC was measured using the chromogenic substrate S-2366 (0.83 mmol/L) in TBS, 0.5% BSA, 0.02% NaN3, pH 8.2 and an EL312 Microplate Bio-Kinetics reader (Bio-Tek Instruments, Winooski, VT).18 Alternatively the substrate Spectrozyme PCa was used in HBS, 0.5% BSA, 5 mmol/L CaCl2, 0.1 mmol/L MnCl2, pH 7.4. Concentrations of single site APC mutants were calibrated relative to wild-type APC based on amidolytic activity for the substrate S2366. By varying chromogenic substrate concentration from 1.43 to 0.0446 mmol/L we derived Km andkcat values using Eadie-Hofstee plots.
For APTT clotting assays, 50 μL of plasma was mixed with 50 μL of APTT reagent (Platelin LS; Organon Technika Corp, Durham, NC) and preincubated at 37°C for 3 minutes. Then 2 μL of APC was added followed by 50 μL of HBS, 0.5% BSA, and 25 mmol/L CaCl2. The clotting time was recorded using an ST4 coagulometer (Diagnostica Stago, Asnieres, France).
Inactivation of FVa was measured as follows. A mixture of 1 nmol/L FVa with 25 μmol/L phospholipid vesicles was made in 50 mmol/L HEPES, pH 7.4, 100 mmol/L NaCl, 0.5% BSA, 5 mmol/L CaCl2, 0.1 mmol/L MnCl2. Inactivation was initiated by the addition of APC. Aliquots of 1 μL were removed at time points and added to 40 μL of 1.25 nmol/L factor Xa with 25 μmol/L phospholipid vesicles, followed by 10 μL of 3 μmol/L prothrombin (final concentrations: 1 nmol/L FXa, 20 pmol/L FVa, 25 μmol/L phospholipid vesicles, and 0.6 μmol/L prothrombin). After 2.5 minutes a 15-μL aliquot of this mixture was quenched by addition to 55 μL HBS containing 10 mmol/L EDTA, 0.5% BSA, pH 8.2. Chromogenic substrate CBS 34-47 was added and the rate of thrombin formation was assessed by measuring the change in absorbance at 405 nm as described earlier.
Curve fitting of these pseudo–first-order time courses of FVa inactivation was done according to Nicolaes and colleagues11 using the following equation:
Vat is the cofactor activity determined at time t. Vao is the cofactor activity determined before APC is added. B is the relative cofactor activity of factor Vaint, the partially inactivated Va cleaved only at Arg506 (expressed as a fraction of the cofactor activity of intact FVa).k506 is the apparent second-order rate constant for cleavage at Arg506 in native FVa.k306 is the apparent second-order rate constant for cleavage at Arg306 in FVaint andk′306 is the apparent second-order rate constant for cleavage at Arg306 in intact FVa.k′306 was determined from the inactivation time courses of Q506-FVa fit to a single exponential and was taken as a constant in the above equation. Data were fit to the equations using nonlinear least-squares regression analysis.
These apparent second-order rate constants in units of M-1s-1 are equivalent tokcat/Km, so the change in transition-state stabilization energy (ΔΔGT‡) can be calculated using the equation:
in which R is the gas constant and T is the absolute temperature.24
Plasma inactivation of APC and the inactivation of APC by the serpin inhibitors, protein C inhibitor (PCI), and α1-antitrypsin were measured essentially according to Heeb and coworkers.25 For Western blots of FVa inactivated by APC, 20 nmol/L FVa was incubated with the indicated concentrations of wild-type APC or 306-314AAAA-APC in the buffer described above. Aliquots were removed at the indicated time points and boiled in sodium dodecyl sulfate (SDS) sample buffer containing 10 mmol/L EDTA, then electrophoresed in a 4% to 12% Bis-Tris gradient gel (Novex, San Diego, CA) and transferred to PVDF membrane. FVa and FVa cleavage products were monitored by Western blotting with a polyclonal antibody against the human FVa heavy chain26 and a monoclonal antibody against the human FVa heavy chain (Enzyme Research Laboratory).12
Results
Production and characterization of protein C mutants
Wild-type and mutant recombinant protein C was recovered at levels varying from 0.1 to 1.5 mg/mL of conditioned media. After purification the concentrations of the recombinant protein C were determined by absorbance at 280 nm and by enzyme-linked immunosorbent assay (ELISA). From ELISA and silver-stained SDS–polyacrylamide gel electrophoresis (SDS-PAGE) we estimated purity to range from about 75% to 95% (data not shown). These silver-stained gels also showed no changes in apparent molecular weight for the mutants, indicating that the mutations had no apparent effect on N-linked glycosylation. The lowest purity levels were observed for the proteins that were recovered at the lowest levels (307A, 308A, and 309A protein C mutants). All the protein C was activated with thrombin as described. All the mutants except 306-314AAAA had Km values for S2366 that were indistinguishable from that of wild-type and theKm for 306-314AAAA was increased by about 10% (data not shown).
Anticoagulant activity of single-point mutants
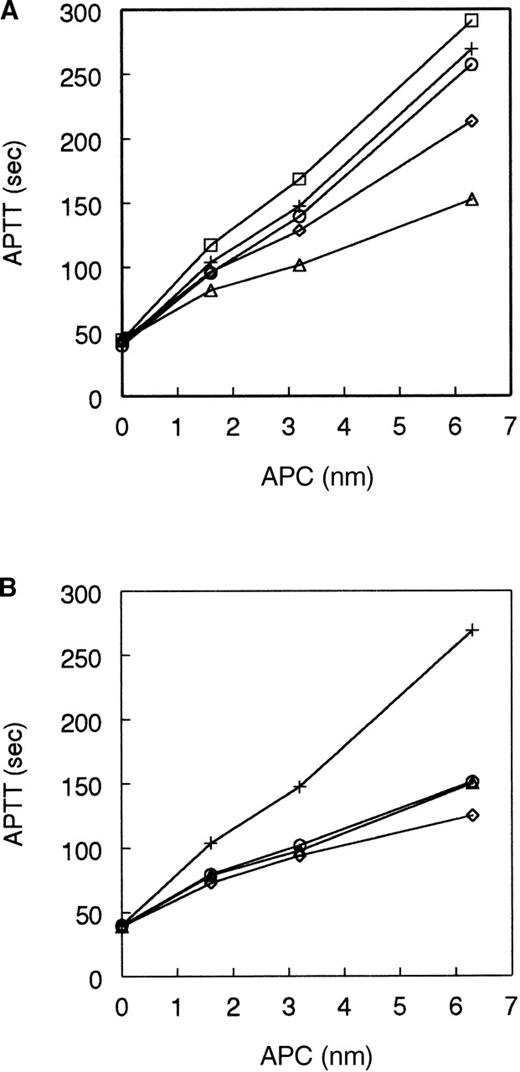
The anticoagulant activity of the single-point mutant APCs was measured in an APTT assay with normal human plasma (Figure1). In this clotting assay the mutant 306A-APC had significantly reduced activity. Mutant 307A-APC had near wild-type activity; 308A-APC had a moderate reduction in activity; and 309A-APC had somewhat increased anticoagulant activity (Figure 1A). Figure 1B shows that mutants 311A-APC, 312A-APC, and 314A-APC all had a significantly reduced level of anticoagulant activity.
Anticoagulant activity of APC single-point mutants.
The anticoagulant activity of the autolysis loop single mutants was tested in an APTT assay as described in “Materials and methods” over a range of APC concentration from 1.6 to 6.3 nmol/L. (A) APC: wild-type (+), 306A (▵), 307A (○), 308A (◊), 309A (□). (B) APC: wild-type (+), 311A (▵), 312A (○), 314A (◊). All curves are averaged from 4 to 8 individual experiments.
Anticoagulant activity of APC single-point mutants.
The anticoagulant activity of the autolysis loop single mutants was tested in an APTT assay as described in “Materials and methods” over a range of APC concentration from 1.6 to 6.3 nmol/L. (A) APC: wild-type (+), 306A (▵), 307A (○), 308A (◊), 309A (□). (B) APC: wild-type (+), 311A (▵), 312A (○), 314A (◊). All curves are averaged from 4 to 8 individual experiments.
Inactivation of FVa by single-point mutants
Inactivation of FVa was tested in a purified system over a time course as described in “Materials and methods.” Results are presented in 2 panels with inactivation curves for wild-type APC, 306A-APC, 307A-APC, 308A-APC, and 309A-APC in Figure2A and inactivation curves for wild-type APC, 311A-APC, 312A-APC, and 314A-APC in Figure 2B. The inactivation curves were biphasic on a log scale. Investigators at several laboratories have postulated that this is due to rapid cleavage at Arg506 resulting in a FVa reaction intermediate that retains partial cofactor activity in prothrombin activation and that this partially active form is then fully inactivated by a slower cleavage at Arg306.10,11,26,27 These inactivation time courses are pseudo–first-order time courses because they were performed under first-order conditions, that is, when the inactivation rate was directly proportional to the residual concentration of FVa because the FVa concentration was below the Km for the inactivation cleavages. Therefore, rate constants for cleavage at Arg506 and for cleavage at Arg306 in the partially inactive form were determined by fitting the data to the biphasic exponential equation described in “Materials and methods” according to Nicolaes and colleagues.11 This analysis yielded second-order rate constants in units of M-1 s-1 that are equivalent tokcat/Kmfor the reaction of APC with the respective cleavage site. In the equation that was used, the value of k306represents the second-order rate of cleavage at Arg306 in the intermediate form of FVa that is already partially inactivated by Arg506 cleavage. Cleavage at Arg506 is represented by the second-order rate constant k506. A value for the fraction of remaining FVa activity following cleavage at Arg506 was first determined, using wild-type APC, to be 0.66 and was then fixed for all further line fits. A value for the apparent second-order rate of cleavage at Arg306 in the absence of Arg506 cleavage was determined in the time course of inactivation of Q506-FVa, which was fit to a single exponential equation. This value (k′306) was determined for each mutant and the wild-type APC using purified Q506-FVa (Table1) and was fixed in the equation used to fit the inactivation time courses of wild-type FVa.
Inactivation of Factor Va by APC single-point mutants.
FVa was incubated at a concentration of 1 nmol/L with 50 pmol/L wild-type or mutant APC and inactivation was followed over time as described in “Materials and methods.” (A) APC: wild-type (+), 306A (▵), 307A (○), 308A (◊), 309A (□). (B) APC: wild-type (+), 311A (▵), 312A (○), 314A (◊). Data points shown are averages of between 3 to 6 experiments for each APC. Standard deviations for all the data points shown averaged ± 3.4%. Error bars are shown for wild-type APC in panel B for illustrative purposes. Other error bars are left out because many of the curves are so close to each other. Curves were fit to the equation in “Materials and methods” according to Nicolaes and colleagues.l11 Coefficient of determination (r2) values were 0.995 or greater for all the curves except for that of 314A-APC (0.977).
Inactivation of Factor Va by APC single-point mutants.
FVa was incubated at a concentration of 1 nmol/L with 50 pmol/L wild-type or mutant APC and inactivation was followed over time as described in “Materials and methods.” (A) APC: wild-type (+), 306A (▵), 307A (○), 308A (◊), 309A (□). (B) APC: wild-type (+), 311A (▵), 312A (○), 314A (◊). Data points shown are averages of between 3 to 6 experiments for each APC. Standard deviations for all the data points shown averaged ± 3.4%. Error bars are shown for wild-type APC in panel B for illustrative purposes. Other error bars are left out because many of the curves are so close to each other. Curves were fit to the equation in “Materials and methods” according to Nicolaes and colleagues.l11 Coefficient of determination (r2) values were 0.995 or greater for all the curves except for that of 314A-APC (0.977).
Rate constants for APC cleavages of FVa and Q506 FVa
| Mutant . | APC sequence 306-314 . | FVa k506 M−1 s−1 . | k306 M−1 s−1 . | Q506-FVa k′306 M−1 s−1 . |
|---|---|---|---|---|
| Wild-type | REKEAKRNR | 2.5 × 108 | 7.7 × 106 | 4.6 × 106 |
| 306A | AEKEAKRNR | 8.8 × 107 | 6.7 × 106 | 3.1 × 106 |
| 307A | RAKEAKRNR | 1.5 × 108 | 8.7 × 106 | 4.5 × 106 |
| 308A | REAEAKRNR | 2.3 × 108 | 9.3 × 106 | 4.6 × 106 |
| 309A | REKAAKRNR | 2.2 × 108 | 7.0 × 106 | 4.7 × 106 |
| 311A | REKEAARNR | 6.2 × 107 | 3.8 × 106 | 2.9 × 106 |
| 312A | REKEAKANR | 6.0 × 107 | 3.8 × 106 | 3.1 × 106 |
| 314A | REKEAKRNA | 9.0 × 107 | 4.3 × 106 | 1.9 × 106 |
| 306/312AA | AEKEAKANR | 2.2 × 107 | 3.9 × 106 | 2.5 × 106 |
| 306-314AAAA | AEKEAAANA | 3.5 × 106 | 1.2 × 106 | 9.7 × 105 |
| Mutant . | APC sequence 306-314 . | FVa k506 M−1 s−1 . | k306 M−1 s−1 . | Q506-FVa k′306 M−1 s−1 . |
|---|---|---|---|---|
| Wild-type | REKEAKRNR | 2.5 × 108 | 7.7 × 106 | 4.6 × 106 |
| 306A | AEKEAKRNR | 8.8 × 107 | 6.7 × 106 | 3.1 × 106 |
| 307A | RAKEAKRNR | 1.5 × 108 | 8.7 × 106 | 4.5 × 106 |
| 308A | REAEAKRNR | 2.3 × 108 | 9.3 × 106 | 4.6 × 106 |
| 309A | REKAAKRNR | 2.2 × 108 | 7.0 × 106 | 4.7 × 106 |
| 311A | REKEAARNR | 6.2 × 107 | 3.8 × 106 | 2.9 × 106 |
| 312A | REKEAKANR | 6.0 × 107 | 3.8 × 106 | 3.1 × 106 |
| 314A | REKEAKRNA | 9.0 × 107 | 4.3 × 106 | 1.9 × 106 |
| 306/312AA | AEKEAKANR | 2.2 × 107 | 3.9 × 106 | 2.5 × 106 |
| 306-314AAAA | AEKEAAANA | 3.5 × 106 | 1.2 × 106 | 9.7 × 105 |
In Figure 2A and Table 1, 307A-APC, 308A-APC, and 309A-APC appeared very similar to wild-type in their cleavage rates for both Arg506 and Arg306. In this assay 307A-APC had slightly decreased activity toward Arg506 but a slightly increased activity toward Arg306 (Table 1). The mutants 308A-APC and 309A-APC had values very close to that of wild-type, but 308A-APC had somewhat higher activity toward Arg306 than wild-type. This is in contrast to the results from APTT assays in which 308A-APC had somewhat decreased activity and 309A-APC had activity greater than wild-type. In Figure 2A, 306A-APC was distinct from the other mutants because it had significantly decreased activity toward Arg506 with a cleavage rate of 8.8 × 107M-1 s-1. However, 306A-APC was only slightly decreased in activity toward Arg306 relative to wild-type with a cleavage rate of 6.7 × 106 M-1s-1 (Table 1). In the case of 306A-APC the final FVa activity at the end of the time course was only slightly greater than that of the FVa inactivated by wild-type APC. This illustrated that in this prothrombinase assay, final FVa activity (below 60%) was largely dictated by cleavage at Arg306. In contrast, anticoagulant activity in APTT assays correlated better with the rate of cleavage at Arg506.
The FVa inactivation time courses for mutants 311A-APC, 312A-APC, and 314A-APC are shown in Figure 3B. All 3 had similar inactivation time courses with reduced cleavage rates for both Arg506 and Arg306 in FVa as shown in Table 1. Table 1 also shows cleavage rates for the mutant APCs for Arg306 of purified Q506-FVa. All of the APCs cleaved Arg306 in Q506-FVa with a second-order rate constant that was about 20% to 50% less than the rate of cleavage of Arg306 following Arg506 cleavage in wild-type FVa. The mutants 307A-APC, 308A-APC, and 309A-APC had essentially wild-type activity toward the Arg306 cleavage site in Q506-FVa. The mutants 306A-APC, 311A-APC, and 312A-APC had 65% to 70% of the activity of wild-type APC toward the Arg306 cleavage site in Q506-FVa. Finally, the mutant 314A-APC had about 40% of the activity of wild-type APC toward the Arg306 cleavage site in Q506-FVa. Table 1 illustrates that cleavage at Arg506 was more sensitive to mutation in the autolysis loop than cleavage at Arg306. For example, mutation of Arg306 in APC to Ala reduced k506 by 2.8-fold but only reducedk306 by 1.1-fold andk′306 by 1.5-fold.
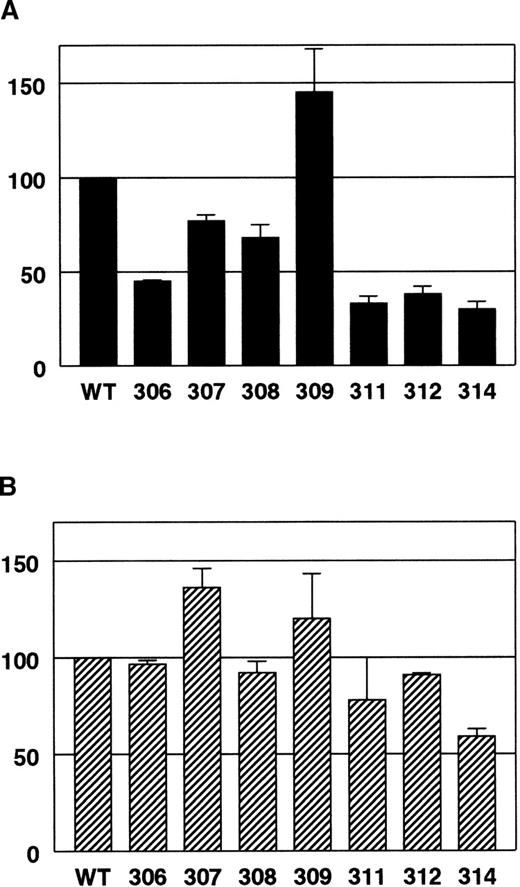
Anticoagulant activity of APC single-point mutants relative to wild-type APC in normal human plasma versus homozygous Q506-FV plasma.
(A) Normal human plasma. (B) Q506-FV plasma. APTT assays were done using plasma mixtures containing 10% normal plasma or 10% Q506-FV plasma and 90% FV-deficient plasma. The percentage of wild-type activity for each APC mutant was calculated as follows. The wild-type APC dose-response data were fit to a line and then that equation was used to calculate a relative amount of wild-type APC for the respective clotting time values for each mutant. For each experiment, values from 2 to 3 concentration points for each mutant were averaged together to get a percent of wild-type APC activity for that experiment. The results of 2 to 3 experiments were averaged together and the SDs are shown as error bars.
Anticoagulant activity of APC single-point mutants relative to wild-type APC in normal human plasma versus homozygous Q506-FV plasma.
(A) Normal human plasma. (B) Q506-FV plasma. APTT assays were done using plasma mixtures containing 10% normal plasma or 10% Q506-FV plasma and 90% FV-deficient plasma. The percentage of wild-type activity for each APC mutant was calculated as follows. The wild-type APC dose-response data were fit to a line and then that equation was used to calculate a relative amount of wild-type APC for the respective clotting time values for each mutant. For each experiment, values from 2 to 3 concentration points for each mutant were averaged together to get a percent of wild-type APC activity for that experiment. The results of 2 to 3 experiments were averaged together and the SDs are shown as error bars.
Clotting assays with Q506-FV plasma
We assessed cleavage at Arg306 in FVa based on APTT clotting assays using homozygous Q506-FV human plasma. Figure 3 illustrates the reduction in activity of the mutant APCs relative to wild-type in both normal plasma (Figure 3A) and Q506-FV plasma (Figure 3B). These APTT assays were done using plasma mixtures that were 90% FV-deficient plasma and 10% either normal or Q506-FV plasma. This equalized the quantities of all the coagulation factors other than FV coming from the normal and Q506-FV plasmas. Furthermore, because the resulting plasma mixtures had only 10% of the usual FV levels, the sensitivity of the assay toward APC was increased. In Figure 3 it is clear that mutagenesis in the autolysis loop caused less reduction of the anticoagulant activity of APC toward the Arg306 cleavage site (Q506-FV plasma) compared with cleavage at both sites (normal plasma). Most notably, 306A-APC had 45% of normal anticoagulant activity in normal human plasma but 97% of normal anticoagulant activity in Q506-FV plasma (Figure 3).
Double and quadruple mutants
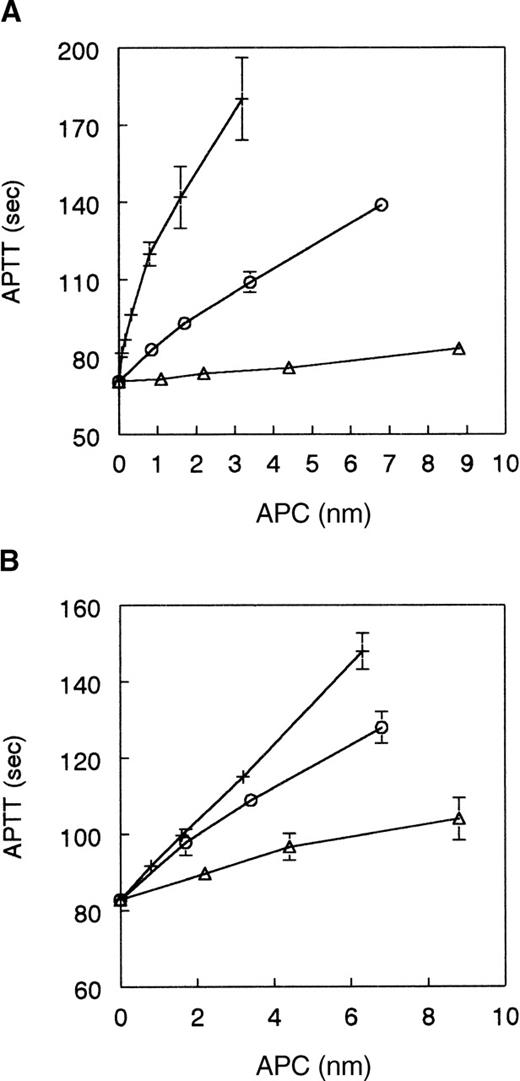
Given the apparent reduction in cleavage site specificity for the single site mutants, 306A-APC, 311A-APC, 312A-APC, and 314A-APC, we decided to make selected combined mutants to determine the cumulative effects of these mutations. The assumption was that the effects of the individual mutants might be additive. The APC mutants constructed were a double mutant of R306A and R312A (306/312AA) and a quadruple mutant of R306A, K311A, R312A, and R314A (306-314AAAA). Figure4 shows results of APTT assays using normal and Q506-FV plasma. Both of the mutants had significantly reduced anticoagulant activity relative to recombinant wild-type APC in normal plasma where 306/312AA-APC had 17% activity and 306-314AAAA-APC had 1.6% of wild-type activity (Figure 4A). In contrast both mutants had relatively high activity in Q506-FV plasma where 306/312AA-APC had 75% activity and 306-314AAAA-APC had 28% activity (Figure 4B). This is a 4.4-fold difference in activity for 306/312AA-APC and an 18-fold difference in activity for 306-314AAAA-APC.
Anticoagulant activity of mutants 306/312AA-APC and 306-314AAAA-APC versus wild-type APC.
(A) Normal human plasma: wild-type APC (+), 306A/312AA-APC (○), 306A-314AAAA-APC (▵). (B) Q506-FV human plasma: wild-type APC (+), 306A/312AA-APC (○), 306A-314AAAA-APC (▵). APTT assays were performed using plasma mixtures containing 10% normal human plasma or 10% Q506-FV human plasma as described above. Data shown are the average of 4 experiments for each line and SDs are shown as error bars.
Anticoagulant activity of mutants 306/312AA-APC and 306-314AAAA-APC versus wild-type APC.
(A) Normal human plasma: wild-type APC (+), 306A/312AA-APC (○), 306A-314AAAA-APC (▵). (B) Q506-FV human plasma: wild-type APC (+), 306A/312AA-APC (○), 306A-314AAAA-APC (▵). APTT assays were performed using plasma mixtures containing 10% normal human plasma or 10% Q506-FV human plasma as described above. Data shown are the average of 4 experiments for each line and SDs are shown as error bars.
To confirm that loss of anticoagulant activity in APTT assays was not caused by an increased neutralization of these mutants by plasma serpins, we followed the rate of inactivation of wild-type APC and the least active mutant, 306-314AAAA-APC, in plasma. The half-life of the mutant 306-314AAAA-APC in plasma was not shorter than the normal half-life (20 minutes) of wild-type APC (data not shown). Thus, the more than 98% reduction of anticoagulant activity of 306-314AAAA-APC in plasma APTT assays was not due to more rapid neutralization by plasma serpins.
The activity of these mutants was also tested in the FVa inactivation assay with purified components. The rate of cleavage of Arg306 in the absence of any possible Arg506 cleavage was determined using purified Q506-FVa. Rates of cleavage at Arg506 and cleavage at Arg306 following Arg506 cleavage were determined using normal FVa. Apparent second-order rate constants for these cleavages show remarkable differences in effects of the mutations on k506 versusk306. Mutant 306-314AAAA-APC cleaved Arg506 at a rate that was reduced 71-fold relative to wild-type APC (Table 1). However, 306-314AAAA-APC cleaved Arg306 at a rate that was reduced 6.4-fold relative to wild-type APC following Arg506 cleavage (k306) and 4.7-fold in Q506-FVa (k′306). From these apparent second-order rate constants we were able to calculate the change in transition-state stabilization energy (ΔΔGT‡) for each of the mutants relative to wild-type (Table 2). Changes in free energy for both 306/312AA-APC and 306-314AAAA-APC were nearly additive for both cleavage sites. As implied by the APTT results, mutation of 4 residues in the autolysis loop had a much larger effect on Arg506 cleavage than on Arg306 cleavage, such that these 4 residues appear to provide for 11- to 17-fold of the discrimination of the Arg506 cleavage site over the Arg306 cleavage site.
Change in transition-state stabilization energy (▵▵GT‡) for cleavages of FVa
| . | FVa Arg506 kcal/mol . | Arg306 kcal/mol . | Q506-FVa Arg306 kcal/mol . |
|---|---|---|---|
| 306A | +0.62 | +0.08 | +0.23 |
| 307A | +0.30 | −0.07 | +0.01 |
| 308A | +0.05 | −0.11 | 0.00 |
| 309A | +0.08 | +0.06 | −0.01 |
| 311A | +0.83 | +0.42 | +0.27 |
| 312A | +0.84 | +0.42 | +0.23 |
| 314A | +0.60 | +0.34 | +0.52 |
| 306/312AA | +1.44 | +0.40 | +0.36 |
| 306-314AAAA | +2.53 | +1.10 | +0.92 |
| . | FVa Arg506 kcal/mol . | Arg306 kcal/mol . | Q506-FVa Arg306 kcal/mol . |
|---|---|---|---|
| 306A | +0.62 | +0.08 | +0.23 |
| 307A | +0.30 | −0.07 | +0.01 |
| 308A | +0.05 | −0.11 | 0.00 |
| 309A | +0.08 | +0.06 | −0.01 |
| 311A | +0.83 | +0.42 | +0.27 |
| 312A | +0.84 | +0.42 | +0.23 |
| 314A | +0.60 | +0.34 | +0.52 |
| 306/312AA | +1.44 | +0.40 | +0.36 |
| 306-314AAAA | +2.53 | +1.10 | +0.92 |
To rule out any possibility that reduction in activity of the APC mutants was merely due to a defect in phospholipid binding, we studied FVa inactivation using wild-type APC and the least active mutant, 306-314AAAA-APC, in the absence of phospholipid vesicles. Within 6 minutes wild-type APC inactivated FVa to about 60% activity, at which point the activity leveled off because cleavage at Arg306 requires phospholipid.28 FVa inactivation by the 306-314AAAA-APC mutant was greatly reduced relative to wild-type and was so slow that the activity of FVa was not appreciably decreased during the course of the assay (30 minutes, data not shown). Hence, the 306-314AAAA-APC mutant had the same reduction in activity involving cleavage at Arg506 in both the presence and absence of phospholipid vesicles, thus verifying that reduction in activity for the APC mutant was not a result of defective phospholipid binding.
To confirm that the decreases in anticoagulant activity of the APC mutants were not due to extensive disruption of the active site, theKm and kcat for the chromogenic substrate Spectrozyme PCa were determined for wild-type APC and the least active mutant, 306-314AAAA-APC, in the same conditions in which the FVa inactivation assays were performed. The catalytic efficiency (kcat/Km) of wild-type APC toward Spectrozyme PCa was 1.3 × 105 M-1 s-1and the catalytic efficiency of 306-314AAAA was 0.98 × 105 M-1 s-1(Table 3). Thus, compared with wild-type APC, 306-314AAAA-APC retained 75% activity toward the chromogenic substrate Spectrozyme PCa, whereas it exhibited only 1.6% anticoagulant activity. A similar modest reduction in catalytic efficiency was seen with the substrate S2366 (data not shown). We also compared the rates of reaction of purified plasma serpins, PCI, and α1-antitrypsin, with wild-type and 306-314AAAA-APC.25 For reaction of PCI with wild-type APC and 306-314AAAA-APC the second-order rate constants were 480 M-1 s-1 and 350 M-1s-1, respectively, and for α1-antitrypsin they were 0.84 M-1 s-1 and 0.47 M-1s-1, respectively. The reductions for 306-314AAAA-APC in serpin reactivity were comparable to the reductions seen in chromogenic substrate reactivity and are minor relative to the effect on cleavage of Arg506 in FVa. Thus, we can make the assumption that mutation-induced changes in APC activity toward FVa are mainly due to altered interactions specific for the APC-FVa complex, rather than due to alterations in the conformation of the immediate active site that would have nonspecific effects on any substrate.
Hydrolysis of spectrozyme PCa by wild-type and 306-314AAAA-APC
| . | Km mmol/L . | kcat s−1 . | kcat/Km M−1 s−1 . |
|---|---|---|---|
| Wild-type | 0.44 ± 0.04 | 57 ± 4 | 1.3 × 105 |
| 306-314AAAA | 0.50 ± 0.08 | 49 ± 3 | 0.98 × 105 |
| . | Km mmol/L . | kcat s−1 . | kcat/Km M−1 s−1 . |
|---|---|---|---|
| Wild-type | 0.44 ± 0.04 | 57 ± 4 | 1.3 × 105 |
| 306-314AAAA | 0.50 ± 0.08 | 49 ± 3 | 0.98 × 105 |
Western blots of FVa inactivation
To verify that these cleavage rates calculated from FVa inactivation curves do correspond to cleavages at Arg506 and Arg306 in the FV heavy chain, we performed SDS-PAGE analysis of aliquots of FVa inactivated by wild-type APC and 306-314AAAA-APC. Figure5A shows the time course of inactivation of FVa by wild-type APC. The gel was blotted with anti-FV heavy chain antibodies. The observed pattern of cleavage is typical of published results.10,11 26 A band at about 75 kd and a triplet of bands at 26 to 28 kd, which appear early in the time course, were the result of cleavage at Arg506. Over a longer period of time the 75 kd band disappeared, as a band at about 45 kd appeared that was the fragment (residues 1-306) that results from cleavage at Arg306.
Western blot of FVa inactivation by wild-type APC and 306-314AAAA-APC.
(A) Wild-type APC, 200 pmol/L. (B) 306-314AAAA-APC, 2 nmol/L. FVa at a concentration of 20 nmol/L was incubated in 50 mmol/L HEPES, pH 7.4, 100 mmol/L NaCl, 5 mmol/L CaCl2, 0.1 mmol/L MnCl2, 0.1% BSA with 25 μmol/L phospholipid vesicles. APC was added and aliquots were removed at 1 through 40 minutes. The Western blot was developed as described in “Materials and methods.” Molecular weight standards are indicated on the left in kilodalton units. On the right fragments of the FVa heavy chain are labeled. HC = intact FVa heavy chain.
Western blot of FVa inactivation by wild-type APC and 306-314AAAA-APC.
(A) Wild-type APC, 200 pmol/L. (B) 306-314AAAA-APC, 2 nmol/L. FVa at a concentration of 20 nmol/L was incubated in 50 mmol/L HEPES, pH 7.4, 100 mmol/L NaCl, 5 mmol/L CaCl2, 0.1 mmol/L MnCl2, 0.1% BSA with 25 μmol/L phospholipid vesicles. APC was added and aliquots were removed at 1 through 40 minutes. The Western blot was developed as described in “Materials and methods.” Molecular weight standards are indicated on the left in kilodalton units. On the right fragments of the FVa heavy chain are labeled. HC = intact FVa heavy chain.
In contrast to the wild-type APC cleavage pattern, a blot of a time course of FVa cleavage by the mutant 306-314AAAA-APC shows a very different pattern (Figure 5B). This FVa inactivation reaction was done with the mutant APC at a 10 times higher level than the wild-type APC reaction to obtain similar levels of remaining intact FVa. In this blot, one can see that the band at 45 kd due to Arg306 cleavage appeared early in the reaction time course and at higher levels than the 75-kd band due to Arg506 cleavage alone. In Figure 5B the faint 75-kd band disappeared soon afterward and never accumulated to the level that it did in the reaction with wild-type. This was presumably due to subsequent, relatively fast cleavage at Arg306 in the 75-kd fragment comprising residues 1 to 506. However, the 26- to 28-kd triplet band that was from the fragment 506 to 709 was still present and levels of the band remained fairly constant over the time course. This pattern of inactivation was very similar to that seen when protein S is added to a FVa inactivation assay, which is known to increase the cleavage rate at Arg306 about 18-fold.13
Thus, it appears that significant amounts of intact FVa heavy chain are cleaved first at Arg306 by the quadruple mutant APC directly resulting in production of the 1 to 306 fragment and the 307 to 709 fragment. The FVa heavy chain that is cleaved first at Arg506 by the 306-314AAAA-APC is rapidly cleaved further at Arg306 so that little of the 1 to 506 fragment ever accumulates.
Discussion
Systematic mutational analysis of a protein–protein interaction is an established method for characterizing the molecular details of that interaction. However, it is important that the effects of the substitution made can be limited to the interaction under analysis. The ideal substitution is a nondisruptive deletion of a side chain that removes the chemical group involved in a specific interaction but has no other effects on the structure of the protein.29 The technique of charged-to-alanine scanning mutagenesis has proven effective in isolating specific interactions.30 Given the exposed and flexible nature of the autolysis loop in the 3-dimensional structure of APC,14,15 we reasoned that mutation of all charged residues to alanine would be unlikely to have large global structural effects on protein C. Analysis of the kinetics of cleavage of 2 different amidolytic tripeptide substrates and analysis of inactivation by 2 serpin inhibitors showed minor effects of these mutations in APC on these interactions of various small molecules and macromolecules with the active site of APC. The effect of the most severely affected APC mutation (306-314AAAA) on reactivity with amidolytic tripeptide substrates is a reduction in activity to about 70% to 75% of wild-type. Similarly the effect of the mutation 306-314AAAA on APC reactivity with the serpins PCI and α1-antitrypsin is a reduction in the second-order rate constants of inactivation to 56% to 73% of wild-type. In contrast, the cleavage rate for Arg506 in FVa by the mutant 306-314AAAA-APC is reduced to 1.4% of the wild-type APC; that is, wild-type APC is 70-fold more active than the tetra-Ala mutant against FVa but only about 1.7-fold more active with serpins. Hence, the effects on FVa inactivation of these mutations are mainly the result of specific interactions of FVa with the autolysis loop rather than the result of major structural perturbations of the APC active site. In a somewhat parallel finding, the thrombin derivative γ-thrombin, which is cleaved in its homologous autolysis loop, has only slightly reduced activity toward chromogenic substrates but a dramatic decrease (greater than 90%) in its ability to clot fibrinogen. 31
If the sites of 2 mutations in a protein that interacts with another protein are thermodynamically independent of each another, then the free energy changes observed for the 2 individual mutants should be additive in a double mutant of those 2 sites.24,32 33 We analyzed the double mutant 306/312AA-APC and the quadruple mutant 306-314AAAA-APC and found that for both the Arg506 and the Arg306 cleavages the ΔΔGT‡ values were very close to being additive of the ΔΔGT‡values for the individual mutants. In both cases though, the ΔΔGT‡ values for the combined mutants were slightly less than the added values of the individual mutants. This is not surprising because all the mutants are adjacent to each other on the autolysis loop. The result of this additivity of free energy is that the relative effect of the combined mutations on apparent second-order cleavage rate constants for Arg506 versus Arg306 is multiplied. For the 4 single-point mutants that had reduced activity, the changes in free energy for the 2 different cleavage sites in FVa, Arg506 and Arg306, were not the same. The ΔΔGT‡ values for the 2 cleavages were uniformly smaller for the Arg306 cleavage than for the Arg506 cleavage. Therefore, k506 is 33-fold greater thank306 and 54-fold greater thank′306 for wild-type APC but only 2.9-fold and 3.6-fold greater than these 2 rate constants, respectively, for 306-314AAAA-APC (Table 1). This is essentially an 11- to 15-fold reduction in discrimination between the Arg506 cleavage site and the Arg306 cleavage site.
This reduction in preference for the Arg506 cleavage relative to the Arg306 cleavage is closely reflected in the APTT clotting assays (Figures 3 and 4). Clearly the complexity of the many reactions in plasma does not allow us to monitor unambiguously FVa inactivation by APC in a plasma-based clotting assay. However, the close correlation of results between APTT assays and FVa inactivation assays in purified mixtures suggests that APTT assays are primarily measuring relative FVa inactivation rates. In normal human plasma, APC anticoagulant activity determined by prolongation of the APTT seems to be primarily sensitive to cleavage at Arg506.12 34 Therefore, the effect of these mutations on APC prolongation of clotting times in normal human plasma is probably primarily reflecting a reduction in the cleavage rate at Arg506. Thus, the effects in APTT assays with normal human plasma can be compared to the calculated second-order rate constants for cleavage at Arg506 (k506). The effects in APTT assays with Q506-FV plasma can be compared to the second-order rate constants for cleavage at Arg306 in the absence of Arg506 cleavage (k′306). For example, in the case of 306-314AAAA-APC, the correlations in activity are striking. In the normal human plasma APTT assay, wild-type APC has 63 times the activity of the mutant 306-314AAAA-APC, whereas k506for FVa inactivation is correspondingly 71-fold greater for wild-type APC than for 306-314AAAA-APC. In the Q506-FV plasma APTT assay, wild-type APC has only 3.6-fold greater anticoagulant activity than the mutant 306-314AAAA-APC, whereas k′306for Q506-FVa inactivation is correspondingly 4.7-fold greater for wild-type APC than for 306-314AAAA-APC. Thus, when the quadruple mutant is compared with wild-type APC, the APTT assay implies a 17-fold reduction in discrimination between Arg506 and Arg306 and the FVa inactivation assay shows a 15-fold reduction in discrimination between Arg506 and Arg306. Therefore, the APTT assays for APC activity also support the conclusion that the autolysis loop contributes to the discrimination of the 2 cleavage sites on FVa by APC.
Kinetic analyses of the cleavages of normal FVa and Q506-FVa by normal APC performed by Nicolaes et al11 demonstrated that the primary difference between cleavage at Arg506 and Arg306 was on theKm for the interaction of APC with the respective cleavage sites (wild-type FVa,Km = 20 nmol/L; Q506-FVa,Km = 196 nmol/L). Therefore, it is likely that the main effect of mutations in the autolysis loop of APC is on the Km values for the 2 substrates. Although our experimentally determined second-order rate constants should be equivalent tokcat/Kmfor the reactions of APC with the 2 cleavage sites, we did not independently determine kcat andKm so this was not conclusively determined in our study.
The serine protease domain of APC contains a cluster of positively charged residues made up of residues in the autolysis loop as well as the loop from 225 to 235 (Ca++ ion binding loop) and the loop of residues from 191 to 193. If individual basic residues Arg222 and Arg352 are also included, at least 13 Arg and Lys residues may be included in this basic exosite on the surface of the APC protease domain.14,15 Thrombin contains a similar basic exosite, anion binding exosite I, in this region on its surface. Among the blood coagulation serine proteases, thrombin is most closely related to protein C.35 In thrombin this positive exosite is involved in binding to fibrinogen, hirudin, and thrombomodulin.36 In APC this basic exosite was proposed to be involved in heparin binding14 and has also been implicated in FVa binding by studies involving inhibition of FVa inactivation by a synthetic peptide with the sequence of residues 311 to 325 of APC,16 which partially represents the autolysis loop. Our results directly demonstrate that this basic exosite in APC is important for FVa binding.
An insertion in the autolysis loop of 4 residues is present in human, monkey, mouse, and rat protein C but is absent in dog, cat, goat, horse, and bovine protein C. However, in the autolysis loop of the known protein Cs, the sequence KRNR is absolutely conserved and an Arg or Lys residue homologous to Arg306 in human protein C is conserved.37 Thrombin is the only other serine protease that has an insertion in this autolysis loop, but there is little sequence homology between thrombin and protein C in this loop. It is likely that the interactions of the autolysis loop of protein C with FVa are not with residues directly adjacent to the FVa cleavage sites on the primed side of the FVa cleavage sites, that is, with residues 507 to 511 or 307 to 311 of FVa. Rather, the APC autolysis loop likely interacts with surface exosites of FVa that complement the basic exosite of APC. Fisher and coworkers14 modeled the Arg506 cleavage site loop of FVa into a model of APC and proposed contacts with APC as distant as the P4′ residue of FVa (Arg510) but did not propose any specific interactions of FVa with the APC autolysis loop. The similar anion binding exosite I of thrombin binds to regions of fibrinogen that are distant from the fibrinopeptide cleavage site.38
It is not necessary that each of the basic residues in the autolysis loop be involved in a specific salt bridge with a corresponding acidic residue in FVa. The crystal structure of thrombin in complex with hirudin showed that, although the C-terminal tail of hirudin contains 5 negatively charged residues, only 2 of them, Asp55 and Glu58, make specific salt bridge contacts with thrombin.39,40 However, all 5 of the negatively charged residues in this C-terminal region of hirudin made similar contributions to binding energy. This can be explained by the interaction of these charges with the general positive electrostatic potential of thrombin resulting in stabilization of the complex and positive effects on the kinetics of complex formation due to “electrostatic steering.”41 In support of this concept a 3-dimensional model of a complex of FVa and APC bound to the Arg506 cleavage site shows a significant number of negatively charged residues on the surface of FVa in close proximity to the positive exosite of APC (Pellequer et al, in preparation). Therefore, we speculate that differences in the negative electrostatic surface potential surrounding the 2 FVa cleavage sites at Arg506 and Arg306 may result in differing complementary acidic exosites on FVa that are responsible for the discrimination of the basic exosite on APC toward the 2 FVa cleavage sites via an electrostatic steering effect. Future experimentation will further characterize the contributions of other residues in the basic exosite on APC as well as the complementary contributions of specific residues in FVa near the 2 main cleavage sites, Arg506 and Arg306.
Supported in part by NIH grants R37HL52246 and R01HL21544. A.J.G. is a fellow of the Leukemia & Lymphoma Society. M.J.H. is partly supported by a grant from the American Heart Association.
Reprints:John H. Griffin, Department of Molecular and Experimental Medicine, MEM-180, The Scripps Research Institute, 10550 N. Torrey Pines Road, La Jolla, CA 92037; e-mail:jgriffin@scripps.edu.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal