We used a sensitive, quantitative bisulfite PCR assay, methylation sensitive single nucleotide primer extension (Ms-SNuPE), to measure methylation of the 5′ CpG islands of c-abl andp15 in chronic myelogenous leukemia (CML) patients during progression. We found that the Pa promoter of c-abl was methylated in 81% (17/21) of the white blood cells (WBCs) of CML patients, which correlates with previous reports. In contrast, WBCs from healthy donors, acute myelogenous leukemias, acute lymphocytic leukemias, and myelodysplastic syndromes were unmethylated at thec-abl Pa promoter locus. We also observed p15hypermethylation in 24% (8/34) of CML cases. Methylation of thep15 but not c-abl Pa promoters was associated with CML progression (P = 0.047 vs 0.46), and the two events were independently acquired. We conclude that de novo methylation ofc-abl and p15 both occur in CML, and analysis of DNA methylation changes using the bisulfite-based MS-SNuPE assay allows both a sensitive and quantitative assessment of these molecular events compared to other methods currently utilized.
Several reports have shown that the c-abl Papromoter is methylated in chronic myelogenous leukemia (CML), but its significance as a prognostic indicator in the disease has been controversial.1-3 It has also become accepted that methylation of the 5′ region of the p15 tumor suppressor gene is not seen in CML,4 in contrast to other leukemia types.5 The methods used to detect DNA methylation in previous studies used the restriction endonuclease-based Southern analysis or polymerase chain reaction–based (PCR-based) assay, both of which raise issues of sensitivity and specificity, as others have already noted.3 We used sodium bisulfite–modified DNA technologies to develop a sensitive and specific quantitative PCR assay, methylation sensitive single nucleotide primer extension (Ms-SNuPE),6 to measure the methylation of both thec-abl Pa and p15 5′ regions in CML.7Our studies demonstrate that the c-abl Pa promoter is methylated in CML, but it is not associated with progression, and that the p15 5′ region can become hypermethylated in CML.
Study design
DNA was obtained from the peripheral blood and/or bone marrow of Philadelphia chromosome-positive patients with CML who were diagnosed at the Los Angeles/USC Medical Center, Los Angeles, CA, or the Medical University of Freiburg Medical Center, Freiburg, Germany. We studied 34 CML patients staged by the Southwest Oncology Group (SWOG) protocol criteria8: 17 patients were chronic, 9 patients were accelerated, and 8 patients were in blast crisis. Controls for our assay included DNA from the blood and bone marrow of patients without disease (including some CD34+ enriched samples); several leukemic cell lines (Raji, KG1a, and K562); and patients with acute myelogenous leukemia (AML), acute lymphocytic leukemia (ALL), and myelodysplastic syndrome (MDS). We extracted DNA from the samples as described previously.9 The DNA was bisulfite modified and amplified with Ms-SNuPE primers that were specific for the c-abl Paand p15 5′ regions, and the methylation at 2 and 3 sites in each 5′ region, respectively, was averaged after quantitation (PhosphorImager; Molecular Dynamics, Sunnyvale, CA) as described previously.6 7
Results and discussion
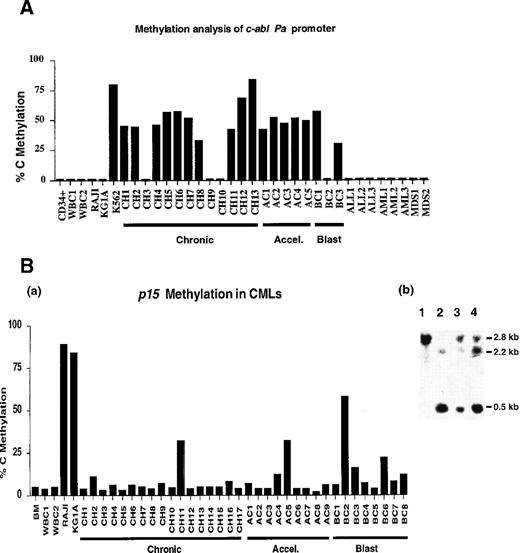
Ms-SNuPE quantitates the levels of methylation at individual CpG sites in each DNA sample analyzed. The DNA standards (generated by mixing SssI methylase–treated DNA with unmethylated DNA) gave the expected methylated results for each standard by Ms-SNuPE analysis (Figure 1). A negative control white blood cell (WBC) DNA from a healthy donor and a positive control from the Raji cell line also gave the expected p15 methylation in the Ms-SNuPE assay. The assay revealed that c-abl is methylated in CML patient samples and in the CML leukemic cell line K562, but it is unmethylated in WBCs from healthy individuals; in non-CML cell lines; and in patients with ALL, AML, and MDS (Figure2A). Methylation of c-abl was found in 10/13 (77%) patients with chronic CML, to an average extent of 41%, while 5/5 (100%) accelerated CML patients and 2/3 (67%) blast crisis CML patients were methylated to an average extent of 48% and 29%, respectively (Figure 2A). We found that c-ablwas methylated in all stages of CML, with no statistical difference between the 3 stages (P = .46, using a 2-sided 1-way analysis of variance method). In addition, equal c-abl hypermethylation was detected in all phases of CML, which suggests that it does not have prognostic significance. We also followed the methylation pattern ofc-abl during the disease progression of several CML patients, but we did not find any change in the c-abl methylation using either the PCR-based restriction enzyme1 method or the Ms-SNuPE method (data not shown).
Ms-SNuPE standard curve for the p15 5′ untranslated region (UTR) showing the quantitative results obtained from the in vitro SssI methylated DNA standards.
WBCs, which are unmethylated, are from the peripheral blood of a healthy donor, while the Raji is a leukemic cell line that is a positive methylated control. Methylation is a measure of the signal in the C lane, which represents the amount of methylated DNA molecules as a result of the [32P]dCTP incorporation. A band in the T lane is the product of [32P]dTTP incorporation and represents unmethylated cytosine molecules at a particular CpG site. NDC indicates no DNA control; 1, 2, and 3 are individual cytosines in a CpG site examined by each Ms-SNuPE primer; and T is the control for a complete bisulfite conversion, representing a C not in a CpG dinucleotide.
Ms-SNuPE standard curve for the p15 5′ untranslated region (UTR) showing the quantitative results obtained from the in vitro SssI methylated DNA standards.
WBCs, which are unmethylated, are from the peripheral blood of a healthy donor, while the Raji is a leukemic cell line that is a positive methylated control. Methylation is a measure of the signal in the C lane, which represents the amount of methylated DNA molecules as a result of the [32P]dCTP incorporation. A band in the T lane is the product of [32P]dTTP incorporation and represents unmethylated cytosine molecules at a particular CpG site. NDC indicates no DNA control; 1, 2, and 3 are individual cytosines in a CpG site examined by each Ms-SNuPE primer; and T is the control for a complete bisulfite conversion, representing a C not in a CpG dinucleotide.
Summaries of the c-abl Pa promoter and p15 5′ CpG island methylation levels.
(A) Summary of the c-abl Pa promoter methylation levels of 1 patient or healthy control. Each vertical bar represents the average of 2 CpG sites by Ms-SnuPE assay. CD34+ indicates a CD34+ enriched sample from the peripheral blood of a hematologically normal patient; WBC, white blood cell sample from a healthy individual; CH, chronic phase of CML; AC, accelerated phase of CML; BC, blast crisis phase of CML; ALL, acute lymphocytic leukemia; AML, acute myelogenous leukemia; and MDS, myelodysplastic syndrome. (B)(a) Summary of the p15 5′ CpG island methylation levels in CMLs and controls by Ms-SnuPE assay. The levels shown represent the average of 3 CpG sites. BM indicates normal bone marrow; WBC1 and WBC2, blood DNAs from healthy donors; CH, chronic CML; AC, accelerated CML; and BC, blast crisis CML. (b) Southern blotting also showed methylated p15 5′ region status in the CML patients. The following are depicted: lane 1, undigested WBC DNA; lane 2, EagI digested WBC DNA from the healthy control; lane 3, CML accelerated samples; and lane 4, CML blast samples. The samples in lanes 3 and 4 are methylated because a larger band appeared after digestion with EagI, similar to the uncut DNA in lane 1.
Summaries of the c-abl Pa promoter and p15 5′ CpG island methylation levels.
(A) Summary of the c-abl Pa promoter methylation levels of 1 patient or healthy control. Each vertical bar represents the average of 2 CpG sites by Ms-SnuPE assay. CD34+ indicates a CD34+ enriched sample from the peripheral blood of a hematologically normal patient; WBC, white blood cell sample from a healthy individual; CH, chronic phase of CML; AC, accelerated phase of CML; BC, blast crisis phase of CML; ALL, acute lymphocytic leukemia; AML, acute myelogenous leukemia; and MDS, myelodysplastic syndrome. (B)(a) Summary of the p15 5′ CpG island methylation levels in CMLs and controls by Ms-SnuPE assay. The levels shown represent the average of 3 CpG sites. BM indicates normal bone marrow; WBC1 and WBC2, blood DNAs from healthy donors; CH, chronic CML; AC, accelerated CML; and BC, blast crisis CML. (b) Southern blotting also showed methylated p15 5′ region status in the CML patients. The following are depicted: lane 1, undigested WBC DNA; lane 2, EagI digested WBC DNA from the healthy control; lane 3, CML accelerated samples; and lane 4, CML blast samples. The samples in lanes 3 and 4 are methylated because a larger band appeared after digestion with EagI, similar to the uncut DNA in lane 1.
Earlier studies looking at c-abl methylation in CML showed a correlation with progression, but hypermethylation was seen in 24%-68% of the CML patients in the chronic phase, while patients in the accelerated phase and blast crisis had hypermethylation frequencies at 73% and 80%, respectively.1 2 These studies further demonstrated a difference between methylation of c-abl in the early (less than 12 months after diagnosis) versus late chronic CML samples. Our results do not reproduce these findings, perhaps because of the more specific, sensitive, and quantitative nature of the Ms-SNuPE assay and the differences in the CML population sampled (although there was a broad range of both early and late chronic CML samples analyzed in our study).
Ms-SNuPE analysis of the p15 5′ region in CML patients also showed positive methylation, which contradicts the current view that CML has an unmethylated p15 region.4Methylation of p15 in CML was detected in all stages of disease in 8/34 (24%) patients (Figure 2Ba). The mean methylation of 3 sites in p15 was 7% in the 17 chronic patients, 9% in the 9 accelerated samples, and 16% in the 8 blast crisis cases. The small increase in mean methylation across the 3 stages of CML for p15was statistically significant (P = .047). During CML progression for more than 2 months, 1 patient also showed an increase in p15 methylation as he progressed from chronic to blast crisis (19% vs 45%) (less than 10% blasts for the chronic sample, 40% blasts in the blood, and t[7;11] for the blast crisis sample). A further increase (to 58%) was observed in a second blast crisis sample (89% blasts and t[7;11]i[17] karyotype). We confirmed the p15 methylation detected in CML by Ms-SNuPE using both Southern analysis4 and bisulfite genomic sequencing with our Ms-SNuPE primers (Figure 2Bb). Further studies show that Ms-SNuPE methylation at this region of the p15 locus is often concurrent with decreased expression of p15 at the protein level (manuscript submitted).
Our data reveal that the c-abl Pa promoter methylation is probably not a good marker for CML progression, while changes in thep15 5′ region methylation are found in CML and may also play a role in the biology of this malignancy. Although most of the samples analyzed are from an unfractionated cell population, CD34+ enriched peripheral blood from a nonleukemic control and a healthy bone marrow sample showed unmethylated c-abland p15. This suggests that methylation is a change associated with CML cells but not with hematopoietic precursors. These findings, as well as that of earlier work,2 determined that differences in methylation status of c-abl in the early chronic phase (grouped as negative, low, moderate, or high methylation) were not due to the number of blasts. All groups had similar polymorphonuclear cell (PMN) representation,2 which suggests that the c-abl andp15 methylation in CML is not due to the various differentiation stages of the cells that were examined.
Our results, obtained with a sensitive quantitative bisulfite-based assay, agree with the findings of Issa et al,2 who reported the lack of prognostic significance of c-abl methylation in CML. Our findings also suggest that to fully determine the role of p15 methylation in CML progression, future studies are needed to assess the methylation status in CMLs with respect to the type of therapy and duration. In conclusion, DNA methylation changes inc-abl and p15 are common abnormalities in CML, and our study shows that the use of a bisulfite-based Ms-SNuPE assay allows both a sensitive and specific quantitative measurement of these changes. The Ms-SNuPE assay, compared with methods that require restriction endonucleases, might be a more quantitative method to assess molecular changes associated with disease progression.
Acknowledgment
We would like to thank Dr William F. Benedict from the MD Anderson Cancer Center, Houston, TX, for graciously contributing the AML and MDS samples for our studies.
Supported by grants from the National Cancer Institutes to J.G. (RO1CA47456), N.H. (R01CA50248), and P.A.J. (1R01CA82422-01).
Reprints:Peter A. Jones, the Department of Biochemistry & Molecular Biology, USC/Norris Cancer Center, University of Southern California, Los Angeles, CA; e-mail:jones_p@ccnt.hsc.usc.edu
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.

![Fig. 1. Ms-SNuPE standard curve for the p15 5′ untranslated region (UTR) showing the quantitative results obtained from the in vitro SssI methylated DNA standards. / WBCs, which are unmethylated, are from the peripheral blood of a healthy donor, while the Raji is a leukemic cell line that is a positive methylated control. Methylation is a measure of the signal in the C lane, which represents the amount of methylated DNA molecules as a result of the [32P]dCTP incorporation. A band in the T lane is the product of [32P]dTTP incorporation and represents unmethylated cytosine molecules at a particular CpG site. NDC indicates no DNA control; 1, 2, and 3 are individual cytosines in a CpG site examined by each Ms-SNuPE primer; and T is the control for a complete bisulfite conversion, representing a C not in a CpG dinucleotide.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/95/9/10.1182_blood.v95.9.2990.009k08_2990_2992/5/m_bloo00908001w.jpeg?Expires=1769175886&Signature=kVYNZ3gXKJcL0zSmg9taXQANQt9y6tWj1qtLsqTo~1IZUYrD82C5cd83gZVq9Ti0Vyj8XjFYBhn~jHiYu25QTn50Gkz-Ycm9n66FZp6iAS6mkg3ipP3y9riyPZuRqa5W-4jKof1S0X1Mgyjva4VSEr6GkpS21NMqI0hew9R4EsxJgSgcSwEJsvKMDRQcybEpBu1sqO4GyTUaPbsVsObU546rvT6AuH4dYX71tPkMejEVx7WWHacESEvTNYSXFybkPoGMVzkZy-1c8~BlsYLc5XMkBGyMmF~oWquHHEwc6T3L3VLaHmcr1XJ7-ObqrmlXDq~8SAsuIkH1mYVDpvIS4w__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal