Abstract
The ubiquitin–proteasome pathway is responsible for selective degradation of short-lived cellular proteins and is critical for the regulation of many cellular processes. We previously showed that ubiquitin (Ub) secreted from hairy cell leukemia cells had inhibitory effects on clonogenic growth of normal hematopoietic progenitor cells. In this study, we examined the effects of exogenous Ub on the growth and survival of a series of human hematopoietic cells, including myeloid cell lines (HL-60 and U937), a B-cell line (Daudi), and T-cell lines (KT-3, MT-4, YTC-3, and MOLT-4). Exogenous Ub inhibited the growth of various hematopoietic cell lines tested, especially of KT-3 and HL-60 cells. The growth-suppressive effects of Ub on KT-3 and HL-60 cells were almost completely abrogated by the proteasome inhibitor PSI or MG132, suggesting the involvement of the proteasome pathway in this process. Furthermore, exogenous Ub evoked severe apoptosis of KT-3 and HL-60 cells through the activation of caspase-3. In interleukin-6 (IL-6)-dependent KT-3 cells, STAT3 was found to be conjugated by exogenous biotinylated Ub and to be degraded in a proteasome-dependent manner, whereas expression levels of STAT1, STAT5, or mitogen-activated protein kinase were not affected. Moreover, IL-6-induced the up-regulation of Bcl-2 and c-myc, and JunB was impaired in Ub-treated KT-3 cells, suggesting that the anti-apoptotic and mitogenic effects of IL-6 were disrupted by Ub. These results suggest that extracellular Ub was incorporated into hematopoietic cells and mediated their growth suppression and apoptosis through proteasome-dependent degradation of selective cellular proteins such as STAT3.
Hematopoiesis is regulated by a wide variety of external stimuli, including those from hematopoietic growth factors. On binding to cell-surface receptors, the growth factors activate multiple signaling molecules, resulting in the induction of specific target genes required for physiologic processes such as cell growth and survival (reviewed in Ihle1 and Darnell2). Recently, several transcription factors such as STATs (signal transducers and activators of transcription) and AP-1 have been reported to be implicated as important mediators of growth signals from membrane to nucleus, where they contribute to cell-cycle progression through transcriptional regulation of cell-cycle regulatory genes, including cyclins, cyclin-dependent kinases (CDKs), and CDK inhibitors. Hematopoietic growth factors also exert anti-apoptotic effects through the activation of Ras/mitogen-activated protein kinase (MAPK) pathways, phosphatidylinositol 3′-kinase (PI3-K)/Akt pathways, or STATs (reviewed in Miyajima et al3). In addition, Bcl-2 family proteins behave as cell-death antagonists (eg, Bcl-2, Bcl-XL, and Mcl-1) or agonists (eg, Bax, Bad, and Bak) (reviewed in Reed4 and Tsuimoto5 ) and play crucial roles in anti-apoptotic effects of hematopoietic growth factors. It was reported that expression levels of Bcl-XL proteins were tightly regulated by hematopoietic growth factors, and the overexpression of Bcl-XL effectively protected IL-3–dependent myeloid cells from factor-deprived apoptosis.6 Furthermore, IL-3–activated Akt was shown to phosphorylate serine residues of BAD in a PI-3K–dependent manner, thereby protecting the cells from intrinsic death machinery.7 8
The expression levels of cell-cycle regulatory molecules and signal transduction molecules are regulated at transcriptional and posttranscriptional levels, and the ubiquitin (Ub)-proteasome system plays the most important role in the latter step (reviewed in Hochstrasser9 and Vershavsky10). Ub is a highly conserved 76-amino acid polypeptide, and the Ub-proteasome–mediated proteolysis is executed in a series of enzymatic reactions. First, Ub is charged with adenosine triphosphate by Ub-activating enzyme (E1) and then transferred to the substrate by a Ub-conjugating enzyme (E2). An element within the target protein termed a degron is recognized by E2 either alone or in combination with a ubiquitin ligase (E3). After recognition of the degron, the Ub-target protein conjugates are formed through an isopeptide bond between the carboxyl-terminal glysine of Ub and a lysine residue on the target protein. Repeated rounds of ubiquitylation result in highly ubiquitylated target protein, which is recognized and destroyed rapidly by the 26S proteasome. At present, a number of molecules are known to be the targets of the Ub-proteasome system as follows: cell-cycle regulatory molecules, E2F, cyclin D1, cyclin E, cyclin A, cyclin B, CDC25B, p21WAF,1p27Kip,1 and p53; signal transduction molecules, c-jun, c-myc, STAT1, STAT3, SOCS1, NFκB, IκB, β-catenin, HIF-1, and HSF2 (reviewed in Hochstrasser9 and Vershavsky10). Thus, the Ub-proteasome system is required for the normal regulation of cell growth, differentiation, and survival, and abnormalities in ubiquitin-mediated processes in hematopoietic system are suggested to cause pathologic conditions, including malignant transformation and uncontrolled hematopoiesis.
Hairy cell leukemia (HCL) is a hematologic malignancy characterized by a unique morphology of leukemic cells bearing hairy cytoplasmic projections.11 A major clinical manifestation in patients with HCL is marked pancytopenia. We previously hypothesized that HCL cells might secrete a factor capable of inhibiting normal hematopoiesis. Supporting this hypothesis, the conditioned media of hairy cells were found to contain an approximately 8-kd protein that inhibits clonogenic growth of granulocyte-macrophage colony-forming units and erythroid colony-forming units in vitro.12Subsequently, we purified the factor from the conditioned media of an HCL cell line and found it to have the amino-terminal sequence identical to that of Ub.13 Furthermore, we demonstrated that the exogenous addition of purified Ub significantly inhibited colony formation of normal myeloid and erythroid hematopoietic progenitor cells.14 These results suggested that extracellular Ub could affect the growth of hematopoietic cells. However, the biologic significance and molecular mechanism of growth inhibition by extracellular Ub have yet to be determined. In this study, we examined the effects of exogenous Ub on various types of human hematopoietic cells lines, especially on a human interleukin-6 (IL-6)–dependent cell line KT-3, because IL-6 receptor (gp130) is known to mediate signaling critical for various aspects of hematopoiesis. Treatment with Ub was found to result in growth suppression of various hematopoietic cells through the induction of apoptosis. The Ub-induced apoptosis was proteasome dependent, and, in IL-6–dependent KT-3 cells, Ub-induced growth suppression and apoptosis was accompanied by proteasome-dependent STAT3 degradation. These results suggest that extracellular Ub may suppress normal hematopoiesis through the selective degradation of intracellular proteins such as STAT3.
Materials and methods
Reagents and antibodies
Highly purified recombinant human (rh) IL-6 and recombinant murine (rm) IL-3 were provided by Kirin Brewery Company (Tokyo, Japan). Bovine Ub and phytohemagglutinin (PHA) were purchased from Sigma (St. Louis, MO). Proteasome inhibitors MG132 and PSI were purchased from Peptide Institute (Osaka, Japan). A murine anti-phosphotyrosine monoclonal antibody (mAb), 4G10, was supplied by Dr B. Druker (Oregon Health Science University, Portland, OR). Rabbit anti-STAT1, anti-STAT3, and anti-STAT5b polyclonal antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Rabbit anti-phospho MAPK and anti-MAPK polyclonal antibodies were purchased from New England BioLabs (Beverly, MA). Murine anti-Bcl-2 mAb was purchased from Transduction Laboratories (Lexington, KY).
Cells and cultures
A human promyelocytic leukemia cell line HL-60, a human IL-6–dependent T-cell lymphoma cell line KT-3,15 a human Burkitt's lymphoma cell line Daudi, and human T-cell lines MT-4, YTC-3, and MOLT-4 were cultured in RPMI 1640 (Nakarai Tesque, Kyoto, Japan) supplemented with 10% fetal calf serum (FCS) (Flow, North Ryde, Australia). KT-3 was cultured in RPMI 1640 supplemented with 10% FCS in the presence of 1 ng/mL rhIL-6. A murine pro-B cell line Ba/F3 was cultured in RPMI 1640 supplemented with 10% FCS in the presence of 1 ng/mL rmIL-3. A human kidney cells line 293T was cultured in Dulbecco's modified essential medium (Nakarai Tesque) supplemented with 10% FCS. Peripheral blood sample was obtained from a normal volunteer by venipuncture after informed consent was given, and mononuclear cells were isolated with Ficoll-Hypaque (Nycomed Pharma AS, Oslo, Norway) density-gradient centrifugation.
Radioimmunoassays for measurement of Ub concentration
Ub concentration in the sera of HCL patients and the conditioned media of the cultured cells was measured by radioimmunoassays, as previously reported.16
Cell proliferation assay
To quantitate the proliferation of cultured cells, MTT (3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyl tetrazolium bromide) (Sigma) rapid colorimetric assay was used as previously reported.17 In brief, triplicate aliquots of cells (3 × 104 cells) were resuspended in 100 μL ASF103 medium (Ajinomoto, Kawasaki, Japan) and cultured in 96-well flat-bottom microtiter plates for 48 hours at 37°C in the conditions as indicated. Normal peripheral blood mononuclear cells were stimulated with 5 μg/mL PHA with or without Ub. MTT (10 μL a 5-mg/mL solution in phosphate-buffered saline [PBS]) was added for the final 4 hours of culture, and 100 μL acid isopropanol (0.04 N HCl in isopropanol) was added to all wells and mixed. The optical density was then measured on the MicroELISA plate reader (Corona Electric, Ibaragi, Japan) with a test wavelength of 595 nm and a reference wavelength of 620 nm. This assay was found to give equivalent results obtained by 3H-thymidine incorporation or cell enumeration as described previously.17
DNA content analysis
The DNA content of cultured cells was analyzed by staining with propidium iodide (PI) as previously described.18 Briefly, 1 × 106 cells were washed with ice-cold PBS twice and fixed by 70% ethanol at −20°C for 30 minutes. The fixed cells were incubated in 500 μL staining buffer (1 mg/mL RNase, 20 mg/mL PI, and 0.01% NP-40 in PBS) at 37°C for 10 minutes and then analyzed on FACSort (Becton Dickinson, Oxnard, CA) with a program Modfit LT2.0 (Becton Dickinson).
TUNEL assays
TUNEL assays were performed with the In Site Cell Death Detection Kit (Boehringer Mannheim, Indianapolis, IN). Briefly, cells were fixed in 4% paraformaldehyde in PBS for 30 minutes, transferred to permeabilization solution (0.1% Triton X-100 in 0.1% sodium citrate), and incubated on ice for 2 minutes. After washing with PBS, the cells were resuspended in TUNEL reaction mixture containing TdT enzyme and digoxigenin nucleotide. Incorporation of nucleotides into 3′-DNA fragmented ends was detected by flow cytometry.
Annexin-V staining
Cells were washed with RPMI 1640 twice and resuspended in 100 μL labeling solution containing avidin–annexin-V conjugates at room temperature for 30 minutes. The cells were rinsed and developed with fluorescein-conjugated avidin (Becton Dickinson) at 4°C for 30 minutes. The stained cells were analyzed by flow cytometry.
Assays for caspase-3 activities
Caspase-3 activities were measured with PhiPhiLux-G1D2 kit (OncoImmunin, College Park, MD). Briefly, cells were washed with PBS and resuspended in 50 μL substrate solution supplied by the manufacturer containing the caspase-3–specific substrate with amino acid sequence GDEVDGI. After 60-minute incubation in a 5% CO2 incubator at 37°C, the cells were suspended in 500 μL dilution buffer supplied by the manufacturer and subjected to flow cytometry. In this system, caspase-3 activities are measured by fluorescence derived from the cleaved substrate specific for caspase-3.
In vitro biotinylation of ubiquitin
Ub was biotinylated with EZ-Link Sulfo-NHS-LC-Biotin (Pierce, Rockford, IL) according to the manufacture's protocol, and unreacted biotin was removed by NAP-10 column (Pharmacia Biotech, Uppsala, Sweden) as previously described.19
Northern blot analysis
Northern blot analysis was performed as previously reported.20 Briefly, total cellular RNA was isolated with Trisol reagent (Gibco BRL, Gaithersburg, MD). For Northern blot analysis, equal amounts of RNA (15 μg) were size fractionated by electrophoresis through 1% formaldehyde agarose gels. After blotting to the nylon membrane (GeneScreen Plus; NEN, Boston, MA), the filters were prehybridized and then hybridized with random32P-labeled probe in rapid hybridization buffer (Amersham, Tokyo, Japan) for 2 hours at 65°C. The filters were washed and autoradiographed at −70°C with 2 intensifying screens for 1 to 2 days.
Immunoprecipitation and immunoblotting
Isolation of total cellular lysates, immunoprecipitation, gel electrophoresis, and immunoblotting were performed according to the methods described previously.21 Briefly, the cultured cells were lysed in lysis buffer (20 mmol/L Tris-HCl, pH 8.0, 137 mmol/L NaCl, 10% glycerol, 1% NP-40, 10 mmol/L EDTA, and 100 mmol/L NaF) containing protease and phosphatase inhibitors. Insoluble material was removed by centrifugation at 10 000g for 20 minutes at 4°C. For immunoprecipitation, the lysates were precleared with protein-G Sepharose beads (Pharmacia Biotech) for 2 hours at 4°C. The precleared lysates were incubated with 1 μg anti-STAT3 polyclonal antibody followed by the addition of protein-G Sepharose beads (Pharmacia Biotech). For Western blotting, immunoprecipitated proteins or total cellular lysates were subjected to sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and electrophoretically transferred to a polyvinylidene difluoride membrane (Immobilon; Millipore, Bedford, MA). After blocking residual binding sites on the filter by incubation in TBS (10 mmol/L Tris-HCl, pH 8.0, 150 mmol/L NaCl) containing 1% gelatin (Bio-Rad Laboratories, Richmond, CA), immunoblotting was performed with an appropriate antibody. Immunoreactive proteins were visualized with an enhanced chemiluminescence (ECL) detection system (DuPont NEN, Boston, MA). To detect biotin-labeled Ub, the blotted membrane was incubated with ExtrAvidin peroxidase (Sigma) at room temperature for 1 hour and washed. Then biotin-labeled Ub was visualized with the ECL detection system. In some experiments, the filters were stripped and reprobed with the anti-STAT3 or anti-MAPK antibody to examine the amounts of proteins.
Transient transfection into the cells
293T cells (1 × 106 cells) were seeded in a 60-mm dish, cultured for 24 hours, and cotransfected with 10 μg pMT123 (an expression vector of HA-tagged Ub, kindly provided from Dr D. Bohmann, Heidelberg, Germany)22 and 1 μg pMX-GFP (an expression vector of green fluorescence protein [GFP], kindly provided by Dr T. Kitamura, Tokyo University, Tokyo, Japan) by calcium phosphate coprecipitation method. Transfection into Ba/F3 cells was performed by electroporation method as previously described.21 Briefly, 1 × 107 cells were transfected with 30 μg pMT123 together with 30 μg pMX-GFP by electroporation (350V, 960 μFD) (Bio-Lad Laboratories, Richmond, CA). After 12 hours, transfected cells were washed, serum deprived, and cultured for 24 hours. Then transfection efficiencies were monitored by the GFP expression by flow cytometric analyses. Ub concentrations in the conditioned media were measured as described above.
Results
Intracellularly synthesized ubiquitin is secreted outside the cells
In previous studies, we found that intracellular Ub secreted from HCL cells would inhibit normal hematopoiesis in patients with HCL.12-14 In this study, we initially examined whether intracellularly synthesized Ub could be secreted outside the cells. For this purpose, we transfected an expression vector of Ub along with that of GFP into a murine pro-B cell line, Ba/F3, and a human kidney cell line, 293T. After 12 hours, the transfected cells were washed and cultured in serum-deprived conditions for 24 hours. After 36 hours from the transfection, approximately 60% of Ba/F3 cells and 95% of 293T cells were found to be positive for GFP expression by flow cytometric analysis (data not shown). The supernatant obtained from Ub-transfected Ba/F3 and 293T cells was found to contain more increased levels of Ub than those obtained from corresponding mock-transfected or untreated cells (Ub concentration in the supernatant, Ub-transfected Ba/F3 80 ng/mL, mock-transfected Ba/F3 12.4 ng/mL, and untreated Ba/F3 10.7 ng/mL; Ub-transfused 293T 141 ng/mL, mock-transfected 293T 19.7 ng/mL, and untreated 293T 12.4 ng/mL). These results indicated that intracellularly synthesized Ub could be secreted outside the cells.
Addition of exogenous ubiquitin leads to growth suppression of hematopoietic cells
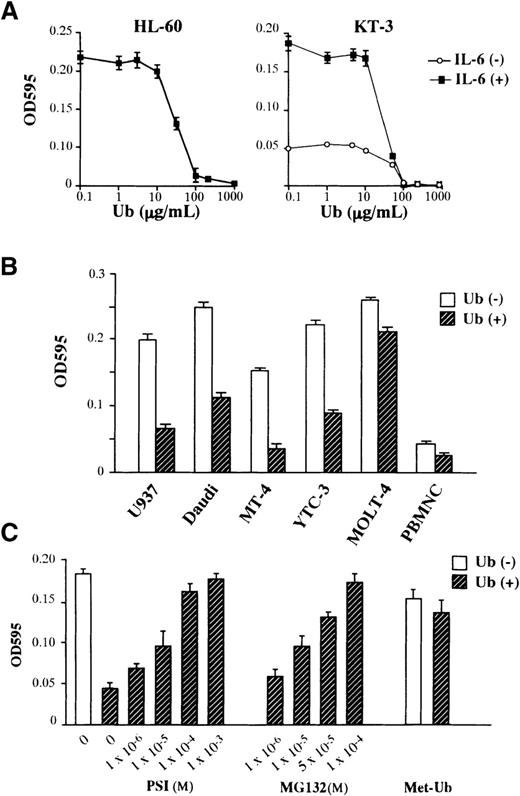
We next examined the effects of exogenously added Ub on the growth of 2 human hematopoietic cell lines, a human promyelocytic leukemia cell line HL-60 and a human IL-6–dependent T-cell line KT-3. As shown in Figure 1A, the treatment with Ub suppressed the factor-independent growth of HL-60 cells and the IL-6–dependent and –independent growth of KT-3 cells. The suppressive effect of Ub was dose dependent, with readily detectable activity at more than 10 μg/mL Ub and maximal activity at 100 μg/mL. We next examined the effects of Ub (100 μg/mL; we used this concentration in the following experiments) on the growth of various types of hematopoietic cells, including a human monocytic leukemia cell line U937, a human Burkitt's lymphoma cell line Daudi, human T-cell lines MT-4, YTC-3, and MOLT-4, and PHA-stimulated normal peripheral blood mononuclear cells. As shown in Figure 1B, the treatment with Ub induced growth suppression in all cell types tested, though a considerable difference was observed in the rate of growth suppression between target cells. To determine further whether proteolysis mediated by the Ub–proteasome pathway was involved in the growth suppression, we investigated the effects of proteasome inhibitors PSI and MG132 on the Ub-induced growth suppression of KT-3 cells (Figure 1C). The suppressive effect of Ub on IL-6–induced proliferation of KT-3 cells was rescued by the addition of either PSI or MG132 in a dose-dependent manner, suggesting the involvement of the proteasome pathway in the growth inhibition by Ub. To further define the roles of extracellular Ub and to deny the effects of toxic contaminants, we examined the effects of methylated Ub (Met-Ub) (Sigma) on Ub-induced growth suppression of KT-3 cells. Met-Ub can be ligated to the target proteins but cannot form polyubiquitin chains. Thus, it has been shown to act as an specific inhibitor of ubiquitin-dependent protein degradation in a previous article.23 KT-3 cells were preincubated with or without Met-Ub for 3 hours and subjected to culture with Ub. As shown in Figure 1C, the pretreatment with Met-Ub alone showed little effect on the growth of KT-3 cells, whereas it restored Ub-induced growth suppression nearly completely, suggesting that extracellular Ub would induce growth suppression in KT-3 cells through the polyubiquitin chain formation on target proteins as would intracellular Ub.
Effects of Ub on the growth of hematopoietic cells.
(A) Dose-response of HL-60 and KT-3 to Ub. Triplicate aliquots of the cells were cultured in serum-free ASF103 medium with various concentrations of Ub for 48 hours, followed by measurement of cell proliferation with an MTT assay. (B) Effects of Ub on various types of hematopoietic cells. The cells were cultured in ASF103 medium in the presence or the absence of 100 μg/mL Ub for 48 hours and were subjected to an MTT assay. (C) Effects of proteasome inhibitors PSI, MG132, and methylated Ub (Met-Ub) on Ub-induced growth suppression. KT-3 cells were resuspended in ASF103 medium containing 1 ng/mL rhIL-6 with or without 100 μg/mL Ub and various concentrations of PSI or MG132 as indicated, cultured for 48 hours, and subjected to an MTT assay. In addition, KT-3 cells were preincubated with 100 μg/mL Met-Ub for 3 hours, cultured with or without Ub for 48 hours, and subjected to an MTT assay. The results are shown as the mean ± SD of triplicate cultures.
Effects of Ub on the growth of hematopoietic cells.
(A) Dose-response of HL-60 and KT-3 to Ub. Triplicate aliquots of the cells were cultured in serum-free ASF103 medium with various concentrations of Ub for 48 hours, followed by measurement of cell proliferation with an MTT assay. (B) Effects of Ub on various types of hematopoietic cells. The cells were cultured in ASF103 medium in the presence or the absence of 100 μg/mL Ub for 48 hours and were subjected to an MTT assay. (C) Effects of proteasome inhibitors PSI, MG132, and methylated Ub (Met-Ub) on Ub-induced growth suppression. KT-3 cells were resuspended in ASF103 medium containing 1 ng/mL rhIL-6 with or without 100 μg/mL Ub and various concentrations of PSI or MG132 as indicated, cultured for 48 hours, and subjected to an MTT assay. In addition, KT-3 cells were preincubated with 100 μg/mL Met-Ub for 3 hours, cultured with or without Ub for 48 hours, and subjected to an MTT assay. The results are shown as the mean ± SD of triplicate cultures.
Ubiquitin induces apoptosis in HL-60 and KT-3 cells
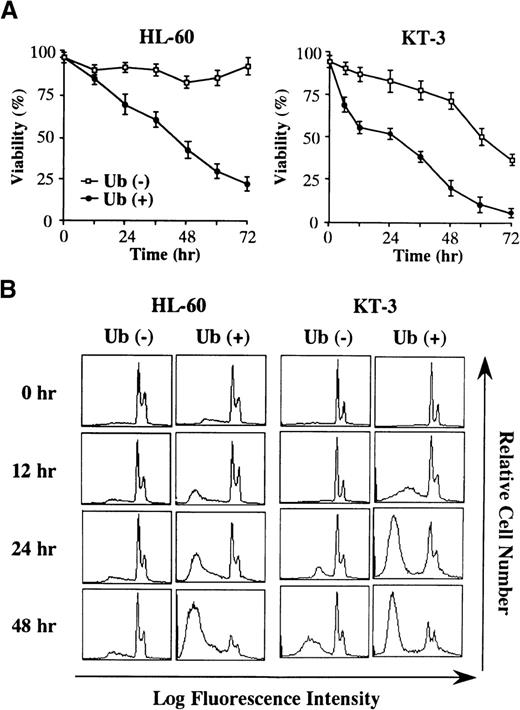
To elucidate the mechanism of Ub-induced growth suppression, HL-60 and KT-3 cells were cultured in serum-deprived conditions with or without Ub treatment, and cell viability was quantitated by trypan blue dye exclusion method at various times after the addition of exogenous Ub. Although viability of HL-60 cells was not significantly changed in the absence of Ub (Figure 2A, left panel), Ub treatment of HL-60 cells led to a decrease in the proportion of viable cells. When KT-3 cells were cultured in a serum-deprived medium containing rhIL-6, their viability gradually decreased with the lapse of time even in the absence of Ub (Figure 2A, right panel). However, the decrease in cell viability of KT-3 cells was more rapidly and remarkably induced by Ub treatment. In accord with these findings, DNA content analysis revealed that Ub treatment led to a more marked increase in the proportions of apoptotic cells in both cell types, which were detected as a subdiploid fraction by PI staining (% subdiploid fraction at 48 hours: HL-60, Ub − 8% vs. Ub + 85%; KT-3, Ub − 37% vs. Ub + 73%) (Figure 2B).
Changes in cell viability during the culture with or without Ub.
(A) HL-60 and KT-3 cells were resuspended in ASF103 and ASF103 containing 1 ng/mL rhIL-6, respectively, and cultured in the presence or absence of 100 μg/mL Ub for 72 hours. Changes in cell viability were quantitated by trypan blue dye exclusion method at the times indicated. The results are shown as the mean ± SD of triplicate cultures. (B) Flow cytometric analyses of HL-60 and KT-3 cells during the culture with or without Ub. The DNA content of the cultured cells was examined by PI staining and analyzed on FACSort.
Changes in cell viability during the culture with or without Ub.
(A) HL-60 and KT-3 cells were resuspended in ASF103 and ASF103 containing 1 ng/mL rhIL-6, respectively, and cultured in the presence or absence of 100 μg/mL Ub for 72 hours. Changes in cell viability were quantitated by trypan blue dye exclusion method at the times indicated. The results are shown as the mean ± SD of triplicate cultures. (B) Flow cytometric analyses of HL-60 and KT-3 cells during the culture with or without Ub. The DNA content of the cultured cells was examined by PI staining and analyzed on FACSort.
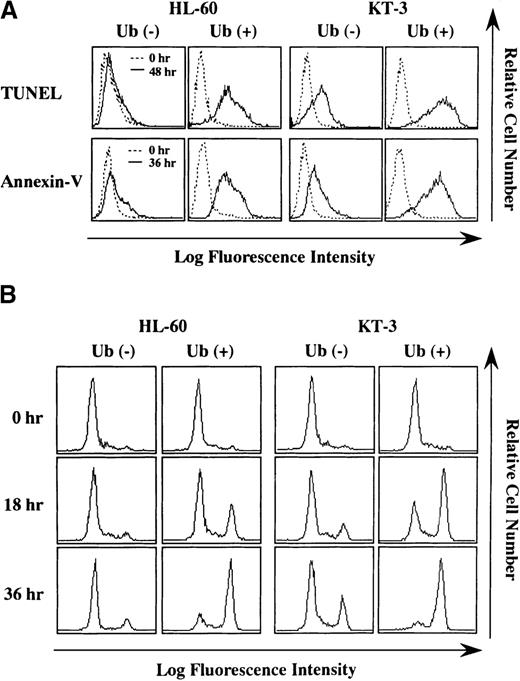
To further characterize Ub-induced cell death, we performed TUNEL assay and annexin-V staining, both of which are useful methods for detecting apoptosis. As shown in Figure 3A, more increased numbers of HL-60 and KT-3 cells became positive for TUNEL-staining after 48 hours of Ub treatment (% TUNEL-positive cells at 48 hours: HL-60, Ub − 2% vs. Ub + 86%; KT-3, Ub − 40% vs. Ub + 95%) (Figure 3A, upper panels). In addition, annexin-V–positive cells markedly increased after 48-hour Ub treatment in KT-3 and HL-60 cells (% annexin-V–positive cells at 36 hours: HL-60, Ub − 10% vs. Ub + 84%; KT-3, Ub − 38% vs. Ub + 89%) (Figure 3A, lower panels). Together, these results indicated that the exogenous addition of Ub induced apoptosis of HL-60 and KT-3 cells.
Staining for TUNEL and annexin-V before and after culture with or without Ub treatment.
(A) HL-60 and KT-3 cells were cultured under the conditions described in Figure 2. The cells were subjected to staining for TUNEL and annexin-V and analyzed on FACSort. (B) Flow cytometric analyses on caspase-3 activities. Caspase-3 activities were measured by a fluorescence intensity that derives from the caspase-3–cleaved substrate by flow cytometry. A height of right fluorescent peak indicates the degree of caspase-3 activation.
Staining for TUNEL and annexin-V before and after culture with or without Ub treatment.
(A) HL-60 and KT-3 cells were cultured under the conditions described in Figure 2. The cells were subjected to staining for TUNEL and annexin-V and analyzed on FACSort. (B) Flow cytometric analyses on caspase-3 activities. Caspase-3 activities were measured by a fluorescence intensity that derives from the caspase-3–cleaved substrate by flow cytometry. A height of right fluorescent peak indicates the degree of caspase-3 activation.
Because caspase-3 is reported to be activated in many types of apoptosis (reviewed by Thornberry and Lazebnik24), we also examined whether caspase-3 was implicated in Ub-induced apoptosis of HL-60 and KT-3 cells. Caspase-3 activity was evaluated by measuring fluorescence intensity derived from the caspase-3–cleaved substrate. Flow cytometric analysis showed that, before culture, caspase-3 activity detected as a high-fluorescence intensity (right) peak was hardly observed in HL-60 and KT-3 cells (Figure 3B). After 18- to 36-hour culture, Ub treatment distinctly up-regulated caspase-3 activities in HL-60 and KT-3 cells, whereas only a limited level of caspase-3 activation was detected in Ub-untreated HL-60 and KT-3 cells (Figure 3B).
Treatment of ubiquitin leads to degradation of STAT3 in IL-6–stimulated KT-3 cells
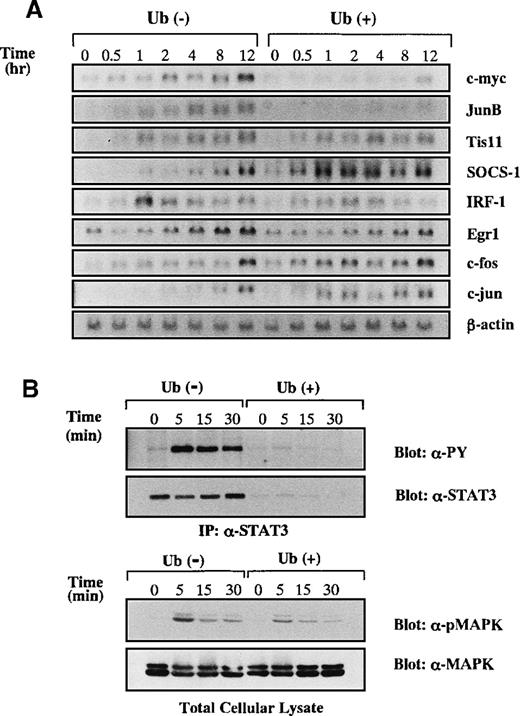
A possible explanation for the apoptosis-inducing activity of Ub was that Ub treatment resulted in the impediment of anti-apoptotic signaling in the cells. Because IL-6 was known to mediate anti-apoptotic and proliferation-inducing signals in KT-3 cells,15 we examined the effects of Ub on IL-6–mediated signal transduction. After the deprivation of rhIL-6 for 12 hours with or without Ub pretreatment, KT-3 cells were cultured with rhIL-6 in the presence or absence of Ub, and then induction levels of IL-6–responsive genes25-29 were investigated by Northern blot analysis (Figure 4A). Without Ub treatment, expressions of c-myc, JunB, TIS11, SOCS-1, IRF-1, Egr-1, c-fos, and c-jun mRNA were induced by rhIL-6, though they showed differential induction patterns. When KT-3 cells were treated with Ub, the induction of c-myc and JunB mRNA was suppressed dramatically, whereas that of TIS11, IRF-1, Egr-1, c-fos, and c-jun mRNA was not. These results indicated that IL-6–mediated signaling pathways were not totally, but only partially, abrogated by Ub.
Effects of Ub on induction of IL-6-responsive genes.
(A) KT-3 cells were serum- and rhIL-6–deprived for 12 hours and were cultured with 10 ng/mL rhIL-6 for the time indicated. To examine the effects of Ub, the cells were pretreated with 100 μg/mL Ub for the last 3 hours of the starvation period and cultured with Ub during the test period. Induction of IL-6–responsive genes was examined by Northern blot analysis. (B) Effects of Ub on IL-6–induced tyrosine phosphorylation of STAT3 and MAPK activation. KT-3 cells were cultured and treated with rhIL-6 as described above. To detect tyrosine phosphorylation of STAT3, STAT3 was immunoprecipitated from total cellular lysates with anti-STAT3 antibody and separated by SDS-PAGE, and the blot was probed with anti-phosphotyrosine mAb, 4G10. Then the filter was stripped and reprobed with anti-STAT3 antibody. MAPK activation was examined by Western blot analysis on total cellular lysates with anti-phospho MAPK antibody, which primarily recognizes activated MAPK. The filter was then reprobed with anti-MAPK antibody.
Effects of Ub on induction of IL-6-responsive genes.
(A) KT-3 cells were serum- and rhIL-6–deprived for 12 hours and were cultured with 10 ng/mL rhIL-6 for the time indicated. To examine the effects of Ub, the cells were pretreated with 100 μg/mL Ub for the last 3 hours of the starvation period and cultured with Ub during the test period. Induction of IL-6–responsive genes was examined by Northern blot analysis. (B) Effects of Ub on IL-6–induced tyrosine phosphorylation of STAT3 and MAPK activation. KT-3 cells were cultured and treated with rhIL-6 as described above. To detect tyrosine phosphorylation of STAT3, STAT3 was immunoprecipitated from total cellular lysates with anti-STAT3 antibody and separated by SDS-PAGE, and the blot was probed with anti-phosphotyrosine mAb, 4G10. Then the filter was stripped and reprobed with anti-STAT3 antibody. MAPK activation was examined by Western blot analysis on total cellular lysates with anti-phospho MAPK antibody, which primarily recognizes activated MAPK. The filter was then reprobed with anti-MAPK antibody.
Among various signaling molecules located at the downstream of IL-6 receptor, STAT3 was reported to transmit anti-apoptotic signals from IL-6.30 We therefore examined the effect of Ub on rhIL-6–induced tyrosine phosphorylation of STAT3 (Figure 4B, upper panel). Without Ub pretreatment, rhIL-6 was capable of inducing tyrosine phosphorylation of STAT3 in KT-3 cells. In contrast, STAT3–tyrosyl phosphorylation was barely observed in Ub-pretreated cells. When the same filter was reprobed with anti-STAT3 antibody, STAT3 protein bands were hardly detectable in Ub-pretreated cells, suggesting that STAT3 was degraded by Ub treatment in KT-3 cells. Next, we examined whether the Ras/MAPK pathway, which is another major signaling pathway of IL-6, was affected by Ub. MAPK activation was assessed by Western blot analysis on the whole lysates with anti-phospho MAPK antibody, which recognizes only an activated form of MAPK (ie, phosphorylated MAPK at tyrosine and serine residues). As shown in Figure 4B (lower panel), rhIL-6 was able to activate MAPK in Ub-untreated KT-3 cells and, albeit to a slightly lesser degree, in Ub-pretreated KT-3 cells. In addition, expression levels of MAPK proteins did not show an apparent difference between the Ub-untreated and Ub-pretreated cells, suggesting that Ub had little effect on IL-6–induced MAPK activation.
STAT3 is specifically degraded by extracellular ubiquitin
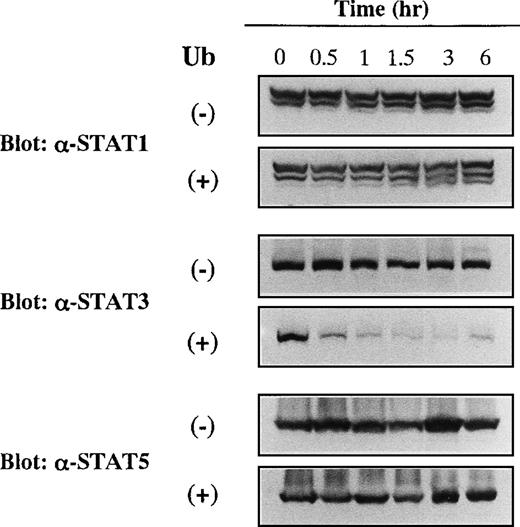
We further analyzed the effects of Ub on the expression levels of other members of STAT proteins. KT-3 cells were cultured with or without Ub for the time indicated, and expression levels of STAT1, STAT3, and STAT5 were investigated by Western blot analyses on the whole cellular lysates. As shown in Figure5, STAT1, STAT3, and STAT5 proteins were expressed constantly in Ub-untreated KT-3 cells throughout the culture period. In addition, expression levels of STAT1 and STAT5 proteins were not affected by Ub treatment. In contrast, expression of STAT3 protein began to decline as early as 0.5 hour and decreased to a barely detectable level thereafter.
Effects of Ub on expression of STAT1, STAT3, and STAT5 proteins in KT-3 cells.
KT-3 cells were cultured with or without 100 μg/mL Ub for the times indicated. Total cellular lysates were obtained at the time indicated and subjected to SDS-PAGE. The filters were probed with anti-STAT1, anti-STAT3, or anti-STAT5b antibody as indicated.
Effects of Ub on expression of STAT1, STAT3, and STAT5 proteins in KT-3 cells.
KT-3 cells were cultured with or without 100 μg/mL Ub for the times indicated. Total cellular lysates were obtained at the time indicated and subjected to SDS-PAGE. The filters were probed with anti-STAT1, anti-STAT3, or anti-STAT5b antibody as indicated.
Exogenously added ubiquitin conjugates to and destroys STAT3 in a proteasome-dependent manner
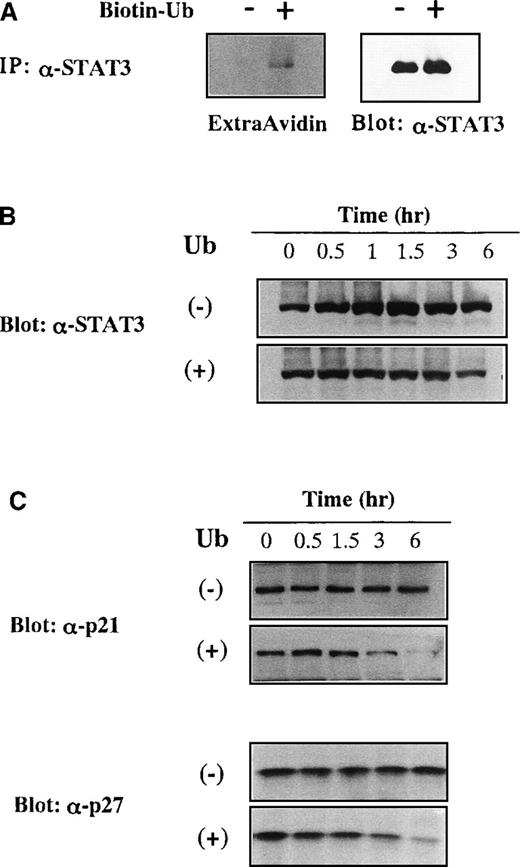
We examined whether exogenously applied Ub could actually conjugate to STAT3 by using biotinylated Ub. KT-3 cells were cultured with or without biotinylated Ub for 1 hour in the presence of proteasome inhibitor MG132, and then STAT3 was immunoprecipitated from the total cellular lysates. When biotinylated Ub was visualized with peroxidase-conjugated avidin, extracellular biotinylated Ub was found to be coimmunoprecipitated with STAT3 (Figure6A, left panel). Because Ub-induced growth suppression was canceled efficiently by proteasome inhibitor in KT-3 cells (Figure 1C), we examined whether proteasome inhibitor could prevent the decreased expression of STAT3 by Ub. KT-3 cells were pretreated with a proteasome inhibitor MG132 for 2 hours and cultured with or without Ub in the presence of MG132. When treated with MG132, STAT3 protein was expressed at an almost constant level throughout the culture with Ub (Figure 6B, lower panel), and its expression was slightly up-regulated in the absence of Ub (Figure 6B, upper panel). Together these results suggested that extracellular Ub could be transported into the cells and could participate in the destruction of STAT3 in a proteasome-dependent manner, just as intracellular Ub.
Degradation of STAT3, p21WAF1, and p27Kip1 by incorporated extracellular Ub.
(A) Binding of biotinylated extracellular Ub to STAT3 in KT-3 cells. KT-3 cells were incubated with or without biotinylated Ub for 1 hour in the presence of proteasome inhibitor MG132, and then STAT3 was immunoprecipitated from the total cellular lysates. Biotinylated Ub was visualized by peroxidase-conjugated avidin with ECL detection system (left panel). The filter was stripped and reprobed with anti-STAT3 antibody (right panel). (B) Effects of MG132 on Ub-induced STAT3 degradation in KT3 cells. KT-3 cells were pretreated with 10 μmol/L MG132 for 2 hours and then cultured with or without Ub in the presence of MG132. Total cellular lysates were obtained at the times indicated and subjected to SDS-PAGE. The filters were probed with anti-STAT3 antibody. (C) Effects of Ub on protein expression levels of p21WAF1 and p27Kip1 in KT-3 cells. KT-3 cells were cultured with or without 100 μg/mL Ub for the times indicated. Total cellular lysates were obtained at the times indicated and subjected to SDS-PAGE. The filters were probed with anti-p21WAF1 or anti-p27Kip1 Ab.
Degradation of STAT3, p21WAF1, and p27Kip1 by incorporated extracellular Ub.
(A) Binding of biotinylated extracellular Ub to STAT3 in KT-3 cells. KT-3 cells were incubated with or without biotinylated Ub for 1 hour in the presence of proteasome inhibitor MG132, and then STAT3 was immunoprecipitated from the total cellular lysates. Biotinylated Ub was visualized by peroxidase-conjugated avidin with ECL detection system (left panel). The filter was stripped and reprobed with anti-STAT3 antibody (right panel). (B) Effects of MG132 on Ub-induced STAT3 degradation in KT3 cells. KT-3 cells were pretreated with 10 μmol/L MG132 for 2 hours and then cultured with or without Ub in the presence of MG132. Total cellular lysates were obtained at the times indicated and subjected to SDS-PAGE. The filters were probed with anti-STAT3 antibody. (C) Effects of Ub on protein expression levels of p21WAF1 and p27Kip1 in KT-3 cells. KT-3 cells were cultured with or without 100 μg/mL Ub for the times indicated. Total cellular lysates were obtained at the times indicated and subjected to SDS-PAGE. The filters were probed with anti-p21WAF1 or anti-p27Kip1 Ab.
p21WAF1 and p27Kip1 are also degraded by extracellular ubiquitin
Next we examined whether extracellular Ub could affect protein expression levels of p21WAF1 and p27Kip1 that are target molecules of the Ub–proteasome pathway.31 32KT-3 cells were cultured with or without Ub for up to 6 hours, and expression levels of p21WAF1 and p27Kip1 were examined by Western blot analysis on the whole cell lysates. Although an apparent change in the protein expression levels of p21WAF1 and p27Kip1 was not detected during the culture without Ub, their expression levels were significantly reduced by Ub treatment (Figure 6C).
IL-6–induced Bcl-2 expression was inhibited by ubiquitin treatment in KT-3
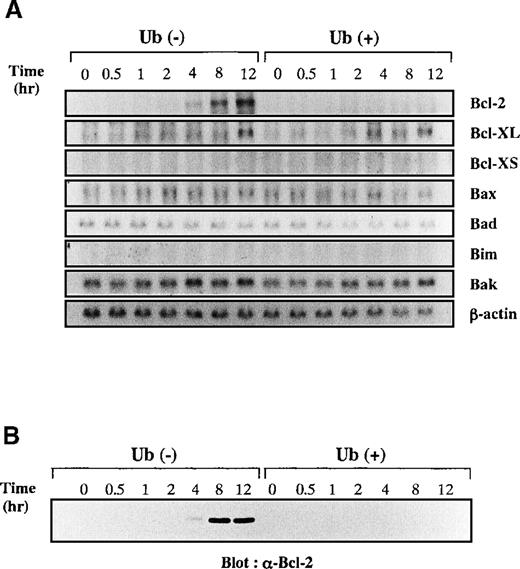
To clarify further the mechanisms underlying Ub-induced apoptosis of KT-3 cells, we examined changes in expression of apoptosis-regulating genes by Northern blot analysis (Figure7A). After starvation of rhIL-6, KT-3 cells were stimulated with rhIL-6 in the presence or absence of Ub. After the addition of rhIL-6, expression of Bcl-2 mRNA was gradually induced at 4 to 12 hours in Ub-untreated KT-3 cells, whereas little or no induction was detected in Ub-treated cells. In contrast, expression of Bcl-XL mRNA was induced similarly in both cultures. Expression of Bcl-XS was not detectable in either culture. Bax, Bad, Bim, and Bak mRNA were expressed at basal levels in both cultures, and their expression was not influenced by rhIL-6 or Ub. Furthermore, consistent with data derived from Northern blot analysis, Western blot analysis demonstrated that Bcl-2 protein was induced by rhIL-6 in Ub-untreated KT-3 cells but not in Ub-treated cells (Figure 7B).
Effects of Ub on IL-6–induced expression of apoptosis-regulating genes.
(A) KT-3 cells were rhIL-6–deprived for 12 hours and then cultured with 10 ng/mL rhIL-6. To examine the effects of Ub, the cells were pretreated with 100 μg/mL Ub for the last 3 hours of the starvation period and were cultured with Ub during the test period. Total cellular RNA was extracted at the times indicated and subjected to Northern blot analysis. (B) Expression of Bcl-2 protein during the culture with or without Ub treatment. Total cellular lysates were isolated from the cultured cells at the times indicated, and the expression levels of Bcl-2 proteins were examined by Western blot analysis.
Effects of Ub on IL-6–induced expression of apoptosis-regulating genes.
(A) KT-3 cells were rhIL-6–deprived for 12 hours and then cultured with 10 ng/mL rhIL-6. To examine the effects of Ub, the cells were pretreated with 100 μg/mL Ub for the last 3 hours of the starvation period and were cultured with Ub during the test period. Total cellular RNA was extracted at the times indicated and subjected to Northern blot analysis. (B) Expression of Bcl-2 protein during the culture with or without Ub treatment. Total cellular lysates were isolated from the cultured cells at the times indicated, and the expression levels of Bcl-2 proteins were examined by Western blot analysis.
Discussion
Ubiquitin-mediated degradation of cellular proteins plays a critical role in many cellular processes, including cell-cycle progression, signal transduction, transcriptional regulation, apoptosis, receptor down-regulation, and endocytosis.9,10 Because the ubiquitin–proteasome pathway is abundant and ubiquitous and participates in the degradation of cellular proteins located primarily in the cytosol and the nucleus, only a limited number of studies have been directed to the function of extracellular Ub. However, extracellular Ub secreted by activated T cells was shown to inhibit platelet activities.33 In addition, extracellular Ub was reported to suppress IgG production in lipopolysaccharide-stimulated splenocytes.34 Furthermore, we previously demonstrated that Ub secreted from hairy cells had an inhibitory effect on the growth of normal hematopoietic progenitor cells.12-14 In our preliminary experiments, serum Ub concentration in patients with HCL ranged from 0.08 to 1.7 μg/mL (0.44 ± 0.52 mg/mL, mean ± SD; n = 8), while those in normal controls were from 0.02 to 0.150 μg/mL (0.08 ± 0.02 μg/mL, mean ± SD; n = 64). In addition, Ub concentration in the conditioned media of an HCL-derived cell line,13 from which we purified Ub as an inhibitor of clonogenic growth of hematopoietic cells, was 0.34 μg/mL. To inhibit the growth of KT-3 and HL-60 cells in short-term cultures, Ub concentration was required to be raised more than 20 μg/mL, which is far higher than that detected in sera of patients with HCL; however, we previously found that extracellular Ub could inhibit the growth of hematopoietic cells from a concentration of 0.5 μg/mL in long-term (7 to 14 days) clonogenic assays.14Moreover, it was speculated that Ub might be present at a higher concentration in confined spaces such as the spleen and bone marrow, in which HCL cells primarily proliferate, than in the serum. Therefore, we assumed that extracellular Ub could affect hematopoiesis in vivo of patients with HCL.
In this study, we found that the exogenous addition of Ub led to apoptosis of various types of hematopoietic cells, resulting in the suppression of cell growth. The Ub-induced apoptosis and growth suppression appeared to be proteasome dependent because the Ub effects were considerably abrogated by the incubation with proteasome inhibitors. It has been reported that proteasomes do not digest cellular proteins indiscriminately but that they participate in the regulated breakdown of selective proteins. In accordance with these findings, we found that among various signaling molecules activated by IL-6, the treatment with Ub led to selective degradation of STAT3 in IL-6–dependent KT-3 cells. Moreover, the Ub-induced STAT3 degradation and apoptosis in KT-3 cells were erased by the addition of proteasome inhibitor. These findings suggest that extracellular Ub may play a role in mediating apoptosis of hematopoietic cells through proteasome-dependent degradation of selective cellular proteins such as STAT3.
In this study, the treatment with Ub enhanced the IL-6–induced expression of SOCS-1 in KT-3 cells despite severe STAT3 degradation. This finding seems to be at variance with the previous report27 showing that the expression of SOCS-1 was regulated by STAT3 directly. However, it remains unknown whether only STAT3 is responsible for SOCS-1 transcription because its mRNA has been shown to be induced by a variety of cytokines such as IL-3, IL-4, IL-13, granulocyte macrophage–colony-stimulating factor, erythropoietin, and interferon-γ.27 In our preliminary experiment, rhIL-6 was able to activate both STAT1 and STAT3 in KT-3 cells (data not shown); therefore, we speculated that rhIL-6 might induce SOCS-1 expression through the activation of STAT1 in Ub-treated KT-3 cells. In addition, because SOCS-1 can be ubiquitinylated,35 SOCS-1 and SOCS-1 associating proteins that turn off the IL-6 signaling might be degraded by Ub treatment, thereby leading to the continued IL-6 signaling and the enhanced SOCS-1 transcription in Ub-treated KT-3 cells. In addition to SOCS-1, c-myc and JunB have been proved to be direct target molecules of STAT3 in previous papers.25 26 Supporting our observation that STAT3 is severely degraded by Ub treatment, IL-6–induced expression was intensely inhibited in Ub-treated KT-3 cells (Figure 4A). Although IL-6 was found to induce IRF-1, Egr1, c-fos, and c-jun expression in Ub-treated and Ub-untreated KT-3 cells with similar efficiency (Figure 4A), the signaling molecules responsible for these inductions have not been determined. Particularly, the induction of Egr1, c-fos, and c-jun was observed in the later phases (4 to 12 hours after the addition of IL-6). Thus, it was speculated that this expression was induced in an indirect manner, possibly through the mediation of other signaling molecules such as Ras/MAPK, PI-3K/Akt, or other STAT members.
Although this study provided unique evidence that extracellular Ub could affect the fate of STAT3, it remains to be elucidated how extracellular Ub mediates the targeting of STAT3 for degradation by proteasome(s). It was previously reported that γ-irradiation–induced apoptosis of human lymphocytes was accompanied by increased Ub mRNA and ubiquitinylated nuclear proteins.36 The expression of Ub sequence-specific antisense oligonucleotides was shown to induce a significant decrease in the proportion of apoptotic cells. It is therefore possible that extracellular Ub may be incorporated in the cells and that the increased amounts of intracellular Ub may yield some cellular proteins, such as STAT3, susceptible to degradation by the proteasome pathways. This possibility may be supported, at least partially, by our preliminary results showing that transfection of Ub in HepG2 hepatoma cells led to the inhibition of IL-6–induced STAT3 transcriptional activities, dependent on the overexpression levels of Ub (data not shown).
Recent results indicate that some proteins, such as CDK inhibitor Sic1P, are targeted for degradation by phosphorylation.37In the case of STAT transcriptional factors, IFN-γ–activated STAT1 was reported to be a target of a Ub–proteasome system in a phosphorylation-dependent manner, whereas STAT5 was not exposed to this system even in a phosphorylated form.38-40 With regard to STAT3, Malek and Halvorsen41 recently showed that only a tyrosyl phosphorylated form of STAT3 was degraded by proteasomes after stimulation with ciliary neurotrophic factor. In contrast, they also revealed that treatment with phorbol ester led to the targeting of STAT3 for degradation by proteasomes, but this process was irrespective of STAT3-tyrosyl phosphorylation.41 In this study, STAT3 was suggested to be degraded by exogenous Ub regardless of its tyrosyl phosphorylation because STAT3 degradation was observed in both IL-6–stimulated and IL-6–starved KT-3 cells in which STAT3-tyrosyl phosphorylation was obviously and scarcely detectable, respectively.
The selective degradation of STAT3 in IL-6–dependent KT-3 cells is of great interest because STAT3 is the major mediator of gp130 signals. The receptor for IL-6 is composed of an IL-6–specific ligand-binding subunit, α chain, and a signal-transducing subunit, gp130, which is shared by the receptors for ciliary neurotrophic factor, leukemia inhibitory factor, oncostatin M, and cardiotropin 1 (reviewed by Hirano42 and Hirano et al43). Ligand-induced assembly of gp130 leads to activation of JAK–STAT and Ras–MAPK and PI-3K pathways. These gp130-mediated signals are involved in a variety of cellular responses, including self-renewal of embryonic stem cells,44 expansion of multipotential hematopoietic stem cells in bone marrow, aorta–gonad–mesonephros region,45and differentiation of M1 cells.28,29 Recent cumulative evidence implies that STAT3 plays an important role in hematopoiesis as a downstream effector of gp130. For example, STAT3 was shown to be linked with anti-apoptotic signals and cell-cycle regulation through the induction of Bcl-2 and c-myc in a murine IL-3–dependent Ba/F3.26,30,46 Furthermore, Takeda et al47disrupted the STAT3 gene specifically in T cells by conditional targeting and showed that IL-6 could not protect STAT3-deficient T cells from apoptosis. In the case of Ub-treated KT-3 cells, IL-6 failed to induce the up-regulation of c-myc and JunB mRNA. IL-6–induced Bcl-2 expression was not observed in Ub-treated KT-3 cells. These results suggested that STAT3 degradation may be a major cause of Ub-induced apoptosis in KT-3 cells. In contrast, the mechanisms responsible for Ub-induced growth suppression and apoptosis of factor-independent cell lines such as HL-60 and U937 remain to be determined, though expression of Bcl-XL protein was down-regulated in Ub-treated HL-60 cells (data not shown).
Our results suggest that extracellular Ub may play a role in the control of hematopoiesis. In addition to HCL cells, a human melanoma cell line HTZ-19 was also reported to secrete Ub.48 These findings raise the possibility that extracellular Ub may be involved in impaired hematopoiesis and immune response in patients with various malignancies. Furthermore, because abnormal accumulations of ubiquitylated proteins in senile plaque, lysosome, endosome, or inclusion body were observed in neurodegenerative disorders such as Alzheimer disease and Parkinson disease,49 50 the Ub system may contribute to the impaired function of hematopoietic cells. Further studies will be necessary to understand the precise mechanisms by which the Ub system regulates normal and abnormal hematopoiesis.
Acknowledgments
We thank Dr A. Yoshimura for providing us with SOCS-1 cDNA and Fujisaki Cell Center for providing KT-3.
Supported in part by grants from the Japanese Ministry of Education, Science, Sports and Culture, the Japanese Ministry of Health and Welfare, Senri Life Science Foundation, Uehara Memorial Foundation, Naito Foundation, and the Japan Medical Association.
Reprints:Yuzuru Kanakura, Department of Hematology and Oncology, Osaka University Medical School, 2-2, Yamada-oka, Suita, Osaka 565-0871, Japan; e-mail: kanakura@bldon.med.osaka-u.ac.jp.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal